Abstract
Sugarcane plant is a glycophyte, hence its growth and sucrose contents are severely affected by drought and salinity stresses. Bioengineering approaches offer a plausible and rapid solution to mitigate these losses. Therefore for genetic improvement of sugarcane against these stresses, the present study was conceived to transform Arabidopsis Vacuolar Pyrophosphatase (AVP1) gene—confers tolerance against drought and salinity—into sugarcane through Agrobacterium. For this purpose, highly regenerable apical buds of sugarcane variety CP77-400 were used as explants. EHA105 strain of Agrobacterium harboring pGreen0029 vector containing AVP1 gene driven under 35SCaMV promoter was employed for transformation. The key factors studied include application of acetosyringone, cefotaxime, kanamycin, and co-cultivation period for successful transformation. Maximum regeneration frequency of 77.5 % was achieved on MS media containing 1 mg/l BAP, 1 mg/l Kn, 1 mg/l GA3, 0.25 mg/l NAA, 50 μM acetosyringone, 500 mg/l cefotaxime, and 150 mg/l kanamycin on 3 days of co-cultivation. The results revealed that apical buds are distinctive viable tissues for sugarcane transformation and regeneration to produce a large number of CP77-400 transgenic plants in shorter period of time without intervening mosaics and chimeras. The AVP1 transcripts expression in transgenic lines at various levels was detected by RT-PCR. Longer and profuse root system was observed in transgenic plants in comparison with control plants. Concomitantly, only transgenic plants were able to withstand higher NaCl salt stress as well as scarcity of water thus, showing tolerance against salinity and drought stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stresses gradually lead to plant yield losses on a large scale under diverse agricultural production systems. Abiotic stresses may decline the potential yields of crop plants by 70 % [1]. Among various abiotic stresses, plants mostly encounter with temperature extremes, drought, and salinity. Globally, about 22 % of the agricultural land is saline [2] and the area under drought has been reported to increase in future [3]. Drought and salinity are the two major stress constraints that threaten crop plants very frequently [4].
Sugarcane (Saccharum officinarum L.) is cultivated in almost all the tropical and subtropical areas of the world, also including regions where water availability is limited or highly inconsistent [5–7]. Sugarcane is considered an unusual plant with some peculiarities such as the multiple genome copies that introduce hurdles for physiological, genetical, and biochemical approaches for its improvement. The enormous impact of biofuel on the economy, the successful exploitation of sugarcane in bio-factory, and the climatic changes have changed the global focus and identified the inevitable need for extensive research on exotic plant species particularly, sugarcane [7].
Plants have developed complex mechanisms for adaptation to ionic and osmotic stresses caused by salinity concentration, and adjusting stress through an amassing of compatible solutes, but still the severe sensitivity of sugarcane to high salt concentration at various growth phases is clearly manifested by a considerable growth rate reduction [8]. Salt stress severely affects the crop productivity and quality of the product, mostly resulting in reduction of photosynthetic activity [9] that eventually declines the sucrose concentration in the cane stalk [10]. This is mainly manifested by a disturbance in the homeostasis of plant water potential and ions distribution at the cellular and the whole plant level as well [11]. The first response of plant in water deficit condition is reduction in plant growth [12, 13]. There exists a direct correlation, as the water potential of plants reduces; the photosynthetic rate is also decreased. Moreover, sugarcane is a typical glycophyte and therefore exhibits stunted growth or even no growth under salinity. Its yield declines to 50 % or even lesser than its actual potential [14]. For sustainable sugarcane production, an improved productivity, tolerance to biotic and abiotic stresses, and improved sugar recovery are the major challenges.
The advancements in genomics and genetic engineering have enabled the scientists to identify, isolate, and insure transfer and integration of target genes controlling desirable traits into sugarcane plant [15]. The Arabidopsis AVP1 gene codes for Vacuolar Pyrophosphatase protein and executes its function in conferring resistance against higher concentrations of NaCl and water deprivation. It is recruited in enhancement of cation uptake in the vesicle. An increase in vacuolar proton gradient results in increased solute accumulation and water retention [16, 17]. Over expression of AVP1 gene improves vegetative development of roots that provide higher water absorption and retention capacity from the soil. Transgenic tomato transformed with AVP1 gene take up greater amounts of water during the imposed water deficit stress, resulting in a more favorable plant water status and less injury [18].
The Agrobacterium-mediated transformation has the potential advantages over biolistic method particularly in simple methodology and a high efficiency of transgene events integration. Grasses are not the natural hosts of Agrobacterium therefore conditions for reliable transformation have been established for several plant species including some of the major cereal crops [19, 20] and switchgrass [21]. It is an uphill task to determine optimum parameters in plants with low transformation rate and considerable variability among cultures, as large-scale and repeated experiments are a requisite to get consistent results.
This study provides an efficient protocol for Agrobacterium-mediated transformation of a sugarcane cultivar CP-77-400 with AVP1 gene using apical meristem as the target tissue. Moreover, we also posit that transgenic plants expressing higher levels of AVP1 transcripts in sugarcane are able to withstand salt and drought stress regimes probably due to profuse root system development in these plants.
Materials and Methods
Plant Material and Culture Conditions
Young tops of sugarcane cultivar CP-77-400 were collected from the Sugar Crops Research Program, Crop Sciences Institute, National Agricultural Research Center, Islamabad, Pakistan. The top outermost leaves were cleaned with 70 % ethanol and outer leaves from the top were removed. From the immature leaf whorl apical meristem buds of 0.5–1.0 cm were excised under aseptic condition. The excised apical buds were surface sterilized with 100 % Clorox (commercial bleach, containing 5.25 % v/v sodium hypochlorite as an active ingredient) for 10 min. Finally, the explants were 4–5 times rinsed with autoclaved distilled water.
Without a special note Murashige and Skoog [22] basal medium was used for all the tissue culture experiments, i.e., shoot induction, shoot multiplication, and root induction. The cultures were incubated at 25 ± 2 ºC with light intensity of 2,100 lx and 16:8 h (light:dark) photoperiods in the growth room.
Transgene Construct
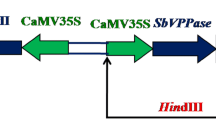
Transformation was performed using Agrobacterium tumefaciens strain EHA105. The strain harbors a binary vector pGreen0029 containing AVP1 gene (with exons and introns) under the transcriptional control of 35S constitutive promoter from cauliflower mosaic virus (Fig. 1). The construct was kindly provided by Roberto Gaxiola, Arizona State University, USA.
Bacterial Infection and Co-Cultivation
A single colony of Agrobacterium strain EHA105 harboring AVP1 target gene was suspended in 10 ml of liquid yeast extract broth (YEB) media containing of 50 mg/l each of kanamycin and rifampicin. The bacterial culture was incubated at 28 ºC at 200 rpm in a rotary shaker under dark until culture reached to density A600 = 1. The culture was pelleted through centrifugation at 5,000 rpm for 10 min at 4 ºC. Pellet was resuspended in MS basal liquid medium (pH 5.8) containing 25, 50, 75, and 100 μM of acetosyringone with final optical density of 4 × 108 cells. Apical meristematic buds were used as target tissue for Agrobacterium-mediated transformation of AVP1 gene. After sterilization, explants were slightly injured with sterilized hypodermic syringe needle 3–4 times. Finally, the injured explants were immersed in the bacterial suspension and subjected to vacuum infiltration for 10, 20, or 30 min. After that explants were blotted dried on sterile Whatman No. 1 filter paper and inoculated on the plates containing MS basal solid media. The co-cultivation period ranged from 2–4 days under dark at 28 ºC. Finally, the explants were thoroughly washed with sterilized distilled water containing cefotaxime (500 mg/l), blotted dried, and inoculated on pre-selection media.
Regeneration and Selection of Transformation Events
The regeneration protocol for sugarcane plant was employed without any modification as previously reported [23]. Co-cultivated apical buds were inoculated (one explant per flask) in the solid pre-selection/regeneration media (SIM: 1 mg/l BAP + 1 mg/l Kn + 1 mg/l GA3 + 0.25 mg/l NAA) containing kanamycin (50, 100, 150, and 200 mg/l) for selection of transformation events. The cultures were maintained under 16/8 h (light/dark) photoperiods at 25 ± 1 ºC with 2,100 lx of light intensity for 3 weeks. Similar selection pressure was maintained by frequent subculturing of the apical buds on the same media. The emerged primary shoots were trimmed and dead tissues were eliminated from the base. These plantlets were subcultured into the same solid shoot induction media containing kanamycin for shoot multiplication. After a period of 4 weeks the secondary shoots without chimerics and escapes were developed through three successive cycles of shoot multiplication. During shoot multiplication, the chimerics and escapes were eliminated and discarded weekly as these were unable to survive.
The well-developed shoots were transferred onto RIM media (half strength MS solid with kanamycin and 6 % sucrose) for root induction. The profusely rooted plantlets were shifted to glasshouse for hardening in the pots containing sandy clay loam soil (1:1:1) for 2 weeks. This was called as first generation of putative transgenic sugarcane plants and named as the first regenerated (R0) clones.
The biomass and length of roots of transgenic and control plants were measured. For this purpose, the potting mix was removed from the intact root system of plants before the stress application by rinsing with water. The washed root mass was dried at 80 °C for 1 week before measurement of dry weight (DW). Simple scale was used to measure the length of the roots.
Molecular Analysis of Putative Transgenic Plants
To confirm the insertion of the target gene into the genomic DNA of putative transgenic plants, standard PCR was performed. For this purpose, genomic DNA from randomly selected transgenic plants and control/wild type plants was extracted by CTAB method [24]. The gene-specific primers for PCR and RT-PCR analyses were manually designed by retrieving the AVP1 sequence (NM_101437) from NCBI database. The forward and reverse primers sequences used for RT-PCR were 5′-ATGGTGGCGCCTGCTTTGTTACCG-3′ and 5′-GAACAGAGGTAACAGCACCA-3′, respectively. PCR was carried out in 25 μl reaction volume. The PCR mixture was initially denatured at 95 ºC for 5 min followed by 34 cycles of denaturing at 95 ºC for 1 min, annealing at 58 ºC for 30 s, extension at 72 ºC for 1 min, and a final extension at 72 ºC for 12 min.
For reverse transcription-polymerase chain reaction (RT-PCR) analysis, total RNA was isolated from 1 gram of fresh leaves of randomly selected transgenic as well as wild-type sugarcane plants using PureLink™ RNA Mini kit (Invitrogen). High-quality total RNA was used as template for synthesizing first strand cDNA utilizing RevertAid™ Reverse Transcriptase kit (Fermentas) according to the manufacturer’s instructions. RT-PCR was carried out in 20 μl reaction volumes using Taq polymerase (Fermentas) for AVP1 primer pairs for 14 cycles and continued to another 16 cycles after the addition of 18S rRNA primers. Normalization was done using constitutively expressed 18S rRNA gene [25]. The PCR products were resolved on 1.5 % agarose gel and visualized by gel documentation system. The experiment was repeated three times for each of the technical as well as biological replicates.
Salt and Drought Trials
PCR confirmed transgenic plants (n = 10) of line L from R0 generation plants and wild-type plant (nontransformed) of the same age and height were selected for drought and salt stress treatments. The salinity trial was imposed for 1 week after transplantation. For salt treatment, different concentrations of NaCl (50, 100, 150, and 200 mM) were applied to plants at regular intervals with photoperiod adjusted to 16/8 (light/dark) at 25 ± 2 ºC in the glasshouse. Starting with 50 mM, the concentration of NaCl was increased to next higher concentrations, i.e., 100, 150, and 200 mM, after every 3 days time interval. After 200 mM of NaCl treatment, the plants were shifted to normal growth conditions to record the survival frequency of the treated plants.
To evaluate the performance of the transgenic plants under drought conditions, selected plants from R0 generation were grown under water condition in the pots for 1 week with same photoperiod and temperature as adjusted for salt-treated plants. These plants were subjected to dry spell conditions for 2 weeks and then shifted to normal growth conditions to observe their performance in both conditions. The data were recorded in the form of plant survival efficiency under salt and drought stress treatments.
Results
Infection Time and Co-Cultivation Greatly Affect the Plant Transformation
The bacterial infection and co-cultivation are the critical steps during Agrobacterium-mediated transformation. In this experiment, three different co-cultivation periods (2, 3, and 4 days) were applied for explants co-cultivation with three different infections regimes (10, 20, and 30 min) for the production of transgenic plants. Table 1 demonstrates that a transformation frequency of 77.5 % is achievable with 20 min of infection time on co-cultivation media for 3 days. Out of 40 explants, 31 could regenerate shoots. It was observed that any further increase in infection time and co-cultivation period adversely affected the explants survival and lead to death of explants on selection media after 3–4 days. From these results, it can be inferred that explants regeneration on selection media exhibited a correlation with infection time and co-cultivation period. Hence their optimization is essential for efficient transformation procedure.
Effects of Cefotaxime and Acetosyringone on Transformation
In order to investigate the effect of acetosyringone on transformation efficiency, different concentrations (50, 75, and 100 μM) of this phenolic compound were used during co-cultivation of explants (n = 40). Remarkably, acetosyringone was found to influence the transformation efficiency. Higher concentrations of acetosyringone resulted in bacterial overgrowth and browning of explants, eventually leading to death of the explants. A lower concentration of acetosyringone, i.e., 50 μM, was found to be the paramount dose for the best transformation event. Hence, the transformation frequency is indirectly correlated with the concentration of acetosyringone (Table 2).
In order to get rid of excessive bacteria after co-cultivation from explants, three different concentrations of cefotaxime (250, 500, and 750 mg/l) were used. 500 mg/l cefotaxime not only significantly eliminated the Agrobacterium but also resulted in maximum regeneration frequency (72 %) (Table 3). On the other hand, the use of 250 and 750 mg/l of cefotaxime reduced the transformation frequency and caused browning of explants. Thus, 500 mg/l cefotaxime has a tremendous effect on controlling the bacterial over growth after infection.
Regeneration and Screening of Kanamycin-Resistant Shoots
Kanamycin sulfate, an aminoglycoside antibiotic, is used as a plant selectable marker in the genetic transformation experimentations. To optimize the lethal dose of kanamycin for effective selection of putative transgenic plants, the control/untransformed explants were subjected to MS solid media with kanamycin. At 150 mg/l and above, the explants turned brown and did not grow further; therefore, 150 mg/l was used as selection pressure for the transgene selection and co-cultivated explants (Table 4). The co-cultivated explants produced primary shoots in the solid pre-selection regeneration media containing kanamycin (150 mg/l) within 15 days. The primary shoots emerged were shifted to the same shoot induction media for shoot multiplication for 20–25 days. The secondary shoots continued to survive at higher dose of kanamycin (150 mg/l) during selection of transgenic shoots. The chimeric plants and escapes were easily eliminated from the cultures when secondary shoots were subjected to three successive subculturing cycles with 150 mg/l selection pressure of kanamycin. Nevertheless, transgenic plants showed normal growth and remained healthy. The healthy shoots were finally shifted to RIM (half strength MS with 6 % sucrose and 150 mg/l kanamycin) for 20–25 days. The profusely rooted plants were successfully acclimatized in the glasshouse with a survival frequency of 90 % in pots containing sandy clay loam soil in 3–4 weeks.
Expression Analysis
For confirmation of stable integration of AVP1 gene into sugarcane genome, PCR analysis was carried out using gene-specific primers. The expected fragment of 630 bp was amplified from transgenic plants while no such amplification was detected in wild-type plants (Fig. 2a).
Molecular analyses of transgenic sugarcane plants. a PCR amplification of AVP1 gene from transgenic sugarcane plants. 1 DNA ladder 1 kb; 2–11 transgenic plant lines showing target gene amplification; 12 wild-type nontransgenic plant used as a negative control; 13 positive control. b Expression of AVP1 transcripts in transgenic sugarcane plants through RT-PCR. NT nontransgenic plant lines; NC negative control; M 1 kb ladder; C positive control; T1, T2, T3 transgenic plant lines showing amplification of target gene after 2 weeks of dehydration; R1, R2 transgenic plant lines after 2 weeks of 200 mM NaCl salt stress application. The bands in the lower panel indicate expression of 18S rRNA gene
In order to observe the expression of the target gene in transgenic plants, RT-PCR was performed. For this purpose, the total RNA isolated from salt and drought stress treated transformed plants and nontransformed control plants was used as template for the synthesis of complementary DNA. Relative expression of AVP1 transcripts was observed by taking 18S rRNA as an internal control. A 630 bp transcript of AVP1 genes could easily be identified in transgenic plants only, indicating that AVP1 gene is not only stably integrated in the genomes but also gene is expressed to varying levels in different transgenic lines (Fig. 2b).
Next we focused on testing the salinity and drought-resistant potential of AVP1 transgenic plants.
Drought and Salinity Trials
To assess the salinity tolerant potential of transformed AVP1 gene, wild-type control plant as well as transgenic plants were subjected to increasing levels of NaCl stress (50, 100, 150, and 200 mM). Both control and transgenic plants of the same age and height were given salinity stress. Phenotypic differences could be detected between control plant and transgenic plants before stress application (Fig. 3).
However, with the increase in salt concentration, the phenotypic differences between control plant and transgenic plants became clearly obvious. Control plant showed retarded growth at 200 mM of NaCl after 1 week of treatment. Nonetheless, 200 mM salt concentration completely abolished the control plants growth. On the contrary the transgenic plants remained unaffected by higher salt stress and exhibited normal growth and phenotype.
For drought stress treatment, the control and transgenic plants were grown under normal growth condition for 1 week (Fig. 4). Plants were irrigated with hand shower normally during this period. The drought stress was imposed by withholding the water for 2 weeks. Control plants showed signs of wilting after 4–5 days of water scarcity. After 14–15 days most of leaves turned brownish, dried up, and eventually died. The situation was totally different in case of transgenic plants having AVP1 gene. These plants remained healthy and survived under extreme drought period. After 2 weeks of dry spell when plants were shifted to normal growth conditions, remarkably, the transgenic plants reverted back to normal phenotype, produced new leaves, and showed enhanced growth but control plants failed to show normal growth (Fig. 4).
Transgenic Plants Exhibit Longer and Profuse Root System
As root is the primary organ affected by the stress, therefore, roots system of transgenic and controlled plants was analyzed. Visual observations showed a more profuse and extensive root system in transgenic plants in comparison with controlled plants (Fig. 5a, b). Figure 5c exhibits the graph showing increase in length in the transgenic plants. The maximum length attained by the transgenic plants is 14 cm against 8 cm of the nontransgenic plants. Hence there was an obvious enhancement in the root system in the transgenic plant before stress treatment. Moreover, the shoots also look profuse and healthier in comparison with control. By analogy the root biomass of the transgenic plants is also enhanced accordingly. The DW of transgenic root was 6 g. On the other hand, it remained only 3 g in control plants. The longer roots might enable the plants to improve the water holding capacity, which in turn help in coping with stress inflictions.
Alterations in root phenotypes of AVP1 transgenic sugarcane plants. a, b The intact root system is exhibited for control and transgenic plants. c, d Graphs showing comparison of root length and biomass (dry weight) in control and transgenic sugarcane plants. Error bars represent standard deviation of the mean
Cumulative data from transformation, gene confirmation, and screening of stress resistant lines experiments revealed that apical buds are suitable explants for transformation and AVP1 gene is not only expressed in transgenic plants but also effectively cope with salt and drought stress.
Discussion
Continuous supply of food to ever increasing population through sustainable agriculture is a major goal worldwide. To come up against this challenge, it will be inevitable to increase the productivity of land already under cultivation and to reclaim of farmlands lost to salinization and lack of rainfall [26, 27]. Transgenic crops engineered to tolerate some measure of salinity and drought constraints hold a grate promise to significantly increase the yield under all soil conditions, particularly for developing countries to alleviate threat of hunger [28].
A voluminous body of research relevant to salt and drought stress adaptation processes has enabled scientists to engineering plants with improved tissue tolerance to dehydration [29–32]. In this study we have shown that overexpression of the H-pyrophosphatase (H-PPase) AVP1 results in generation of salt and water stress tolerant sugarcane plants. Sugarcane is the most efficient biomass plant and considered as a renewable energy source therefore, efforts are now targeted to develop it as an energy plant [33]. Sugarcane genetics has been given little consideration in comparison with other economically important crops due to its high polyploidy level, heterozygosity, repeatedly aneuploid nature, poor reproduction, complex genome, and lengthy propagation period and selection cycles [34].
The present study was envisaged to improve drought and salt tolerance in sugarcane through ectopic expression of AVP1 gene of Arabidopsis using Agrobacterium-mediated transformation system. Direct regeneration using apical bud as explants resulted in minimal genetic changes, therefore production of true to type progeny thereby reducing the chances of somaclonal variations. For the successful transformation of AVP1 gene in sugarcane cultivar CP-77-400 through Agrobacterium system, various factors were optimized. These factors include acetosyringone concentration, co-cultivation period and infection time, cefotaxime and kanamycin concentrations. Remarkably, the highest transformation frequency of 77.5 % was achieved using apical bud as explants with co-cultivation period of 3 days and 20 min infection time. In our experiment the optimum length of the co-cultivation period of sugarcane explants with Agrobacterium was 48 h. Co-cultivation beyond this period affected the transformation frequency due to leaching of bacterial overgrowth. During co-cultivation period, the explants suffer wounding, therefore pathogen infection and environmental stress occurred. Concomitantly, a series of wound and pathogen defense response pathways become active. This defense mechanism function by ejecting phytoalexins and some secondary metabolites those act as repellants or antibacterial agents. Thus induced cell death appeared as a barrier to protect the adjoining healthy tissue [35]. Similar results were also obtained in melon [36] and rice [37].
The AVP1 gene codes for Arabidopsis Vacuolar Pyrophosphatase protein that confers tolerance against salinity and drought in Arabidopsis and various other plants [16, 17]. This is the first time that this gene has been transformed into sugarcane, a monocot plant with complex genome. Remarkably, the salinity and drought trials revealed that AVP1 gene is effective against NaCl stress and scarcity of water. The transgenic plants not only tolerated the stresses but also produced new leaves and enhanced growth after restoration of normal growth conditions.
Previous studies have demonstrated that in some plant species, deeper and/or more extensive root systems are associated with increased drought resistance [18, 38–45]. A more robust root system accords for water uptake to occur from a greater volume of the soil during periods of growth on scarce soil water, thus reducing the extent of plant dehydration. The development of a more vigorous root system in the AVP1 expressing plants may provide the morphological and/or physiological basis for enhanced performance of plants exposed to low soil water conditions, resulting in less severe symptoms than those observed in controls. AVP1 expression certainly increases root growth in sugarcane to help facilitate improved water deficit recovery. Consistent with these findings, AVP1 overexpressing Arabidopsis lines also developed more robust root systems than wild type [46]. In contrast, Arabidopsis avp1-1-null mutants exhibited severely disrupted root development. The authenticity of recruitment of AVP1 gene in root development in sugarcane can be best validated by the complementation of Saccharum officinarum avp1 mutant lines.
Best known for their role in gene regulation it is also well established that introns contribute in increased stability and expression of genes [47–49]. By ectopically expressing AVP1 gene amplified from DNA containing exons and introns in tobacco it was demonstrated that salinity and drought tolerance could be achieved in experiment in sand under saline and water regime, thus confirming that introns play a key role in gene expression and regulations and improve the growth of plants [50]. In our experiment the salinity and drought tolerance exhibited by transgenic plants might be the outcome of enhanced expression of AVP1 gene due to the inclusion of intronic elements in the gene construct.
Abbreviations
- BAP:
-
Benzyl amino purine
- MS:
-
Murashige and Skoog
- SIM:
-
Shoot induction medium
- RIM:
-
Root induction medium
- LB:
-
Luria broth
- NAA:
-
Naphthalene acetic acid
- Kn:
-
Kinetin
- GA3:
-
Gibberellic acid
References
Agrwal, P. K., Agarwal, P., Reddy, M. K., & Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports, 25, 1263–1274.
FAO (Food, Agriculture Organization of the United Nations). (2004). FAO production year book. Rome: FAO.
Burke, E. J., Brown, S. J., & Christidis, N. (2006). Modeling the recent evolution of global drought and projections for the twenty first century with the Hadley centre climate model. Journal of Hydrometeorology, 7, 1113–1125.
Asif, M. A., Zafar, Y., Iqbal, J., Iqbal, M. M., Rashid, U., Ali, G. M., et al. (2011). Enhanced Expression of AtNHX1, in Transgenic Groundnut (Arachis hypogaea L.) Improves Salt and Drought Tolerance. Molecular Biotechnology, 49, 250–256.
Inman-Bamber, N. G., & Smith, D. M. (2005). Water relations in sugarcane and response to water deficits. Field Crops Research, 89, 185–202.
Zhang, S. Z., Yang, B. P., Feng, C. L., Chen, R. K., Luo, J. P., Cai, W. W., et al. (2006). Expression of the Grifola frondosa trehalose synthase gene and improvement of drought-tolerance in sugarcane (Saccharum officinarum L.). Journal of Integrative Plant Biology, 48, 453–459.
Azevedo, R. A., Carvalho, R. F., Cia, M. C., & Gratao, P. L. (2011). Sugarcane under pressure: An overview of biochemical and physiological studies of abiotic stress. Tropical Plant Biology, 4, 42–51.
Plaut, Z., Meinzer, F., & Federman, E. (2000). Leaf development, transpiration and ion uptake and distribution in sugarcane cultivars grown under salinity. Plant and Soil, 218, 59–69.
Akhtar, S., Wahid, A., & Rasul, E. (2003). Emergence, growth and nutrient composition of sugarcane sprouts under NaCl salinity. Biologia Plantarum, 46, 113–116.
Rozeff, N. (1995). Sugarcane and salinity—A review paper. Sugarcane, 5, 8–19.
Munns, R., & Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review of Plant Biology, 59, 651–681.
Inman-Bamber, N. G., Bonnett, G. D., Spillman, M. F., Hewitt, M. L., & Jackson, J. (2008). Increasing sucrose accumulation in sugarcane by manipulating leaf extension and photosynthesis with irrigation. Australian Journal of Agricultural Research, 59, 13–26.
Wilkinson, S., & Davies, W. J. (2010). Drought, ozone, ABA and ethylene: new insights from cell to plant community. Plant Cell and Environment, 33, 510–525.
Suprasanna, P., Patade, V. Y., Desai, N. S., Devarumath, R. M., Kawar, P. G., Pagariya, M. C., et al. (2011). Biotechnological Developments in Sugarcane Improvement: An Overview. Sugar Technology, 13, 322–335.
Butterfield, K., Irvine, E., Valdez, G. M., & Mirkov, E. (2002). Inheritance and segregation of virus and herbicide resistance transgenes in sugarcane. Theoretical and Applied Genetics., 104, 797–803.
Gaxiola, R. A., Li, J., Undurraga, S., Dang, L. M., Allen, G. J., Alper, S. L., et al. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+pump. Proceedings of the National Academy of Sciences of the United States of America, 98, 11444–11449.
Gaxiola, R. A., Fink, G. R., & Hirschi, K. D. (2002). Genetic manipulation of vacuolar proton pumps and transporters. Plant Physiology, 129, 967–973.
Park, S., Li, J., Pittman, J. K., Berkowitz, G. A., Yang, H., Undurraga, S., et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proceedings of the National Academy of Sciences of the United States of America, 102, 18830–18835.
Cheng, M., Lowe, B. A., Spencer, T. M., Ye, X. D., & Armstrong, C. L. (2004). Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cellular and Development Biology—Plant, 40, 31–45.
Shrawat, A. K., & Lorz, H. (2006). Agrobacterium-mediated transformation of cereals: A promising approach crossing barriers. Plant Biotechnology Journal, 4, 575–603.
Li, R. Y., & Qu, R. D. (2011). High throughput Agrobacterium-mediated switchgrass transformation. Biomass and Bioenergy, 35, 1046–1054.
Murashige, T., & Skoog, K. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497.
Uzma Khan, M. R., Muhammad, A., Hussain, I., Shah, S. H., Kumar, T., Inam, S., et al. (2012). Rapid in vitro Multiplication of Sugarcane Elite Genotypes and Detection of Sugarcane Mosaic Virus through Two Steps RT-PCR. International Journal of Agriculture and Biology, 14, 870–878.
Rogers, S. O., & Bendich, A. J. (1988). Extraction of DNA from plant tissues. In S. B. Gelvin & R. A. Schilperoort (Eds.), Plant Molecular Biology Manual (pp. A1–A10). Boston: Kluwer Academic Publishers.
Khan, M. R., Khan, I., & Ali, G. M. (2012). MPF2-like MADS-box genes affecting SOC1 and MAF1 expression are implicated in flowering time control. Molecular Biotechnology, 12, 9540–9549.
Herrera-Estrella, L. (1999). Transgenic plants for tropical regions: Some considerations about their development and their transfer to the small farmer. Proceedings of the National Academy of Sciences of the United States of America, 96, 5978–5981.
Serrano, R., & Gaxiola, R. (1994). Microbial models and salt stress tolerance in plants. Critical Reviews in Plant Sciences, 13, 121–138.
Thomson, J. A. (2002). Research needs to improve agricultural productivity and food quality, with emphasis on biotechnology. Journal of Nutrition, 132, 3441S–3442S.
Chaves, M. M., & Oliveira, M. M. (2004). Mechanisms underlying plant resilience to water deficits: prospects for water-saving agriculture. Journal of Experimental Botany, 55, 2365–2384.
Penna, S. (2003). Building stress tolerance through over-producing trehalose in transgenic plants. Trends in Plant Science, 8, 355–357.
Vinocur, B., & Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Current Opinion in Biotechnology, 16, 123–132.
Zhang, J. Z., Creelman, R. A., & Zhu, J.-K. (2004). From Laboratory to Field. Using Information from Arabidopsis to Engineer Salt, Cold, and Drought Tolerance in Crops. Plant Physiology, 135, 615–621.
Joyce, P., Kuwahata, M., Turner, N., & Lakshmanan, P. (2010). Selection system and cocultivation medium are important determinants of Agrobacterium-mediated transformation of sugarcane. Plant Cell Reports, 29, 173–183.
Gupta, V., Raghuvanshi, S., Gupta, A., Saini, N., Gaur, A., Khan, M. S., et al. (2010). The water-deficit stressand red-rot-related genes in sugarcane. Functional & Integrative Genomics, 10, 207–214.
Trifonova, A., Madsen, S., & Olesen, A. (2001). Agrobacterium-mediated transgene delivery and integration into barley under arrange of In vitro culture conditions. Plant Sciences, 161, 871–880.
Dong, J. Z., Yang, M. Z., Jia, S. R., & Chua, N. H. (1991). Transformation of melon (Cucumismelo L.) and expression from the cauliflower mosaic virus 35S promoter in transgenic melon plants. Nature Biotechnology, 9, 858–863.
Li, X. Q., Liu, C. N., Ritchie, S. W., Peng, J., Gelvin, S. B., & Hodges, T. K. (1992). Factors influencing Agrobacterium-mediated transient expression of Gus Ainrice. Plant Molecular Biology, 20, 1037–1048.
Chaves, M. M., Pereira, J. S., Maroco, J., Rodrigues, M. L., Ricardo, C. P., Osório, M. L., et al. (2002). How plants cope with water stress in the field. Photosynthesis and growth. Annals of Botany, 89, 907–916.
Costa, E., Silva, F., Shvaleva, A., Maroco, J. P., Almeida, M. H., Chaves, M. M., et al. (2004). Responses to water stress in two Eucalyptus globulus clones differing in drought tolerance. Tree Physiology, 24, 1165–1172.
Ober, E. S., & Sharp, R. E. (2003). Electrophysiological responses of maize roots to low water potentials: Relationship to growth and ABA accumulation. Journal of Experimental Botany, 54, 813–824.
Pinheiro, H. A., Damatta, F. M., Chaves, A. R., Loureiro, M. E., & Ducatti, C. (2005). Drought tolerance is associated with rooting depth and stomatal control of water use in clones of Coffea canephora. Annals of Botany, 96, 101–108.
Sharp, R. E., & LeNoble, M. E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. Journal of Experimental Botany, 53, 33–37.
Sharp, R. E., Poroyko, V., Hejlek, L. G., Spollen, W. G., Springer, G. K., Bohnert, H. J., et al. (2004). Root growth maintenance during water deficits: physiology to functional genomics. Journal of Experimental Botany, 55, 2343–2351.
Tschaplinski, T. J., Tuskan, G. A., Gebre, G. M., & Todd, D. E. (1998). Drought resistance of two hybrid Populus clones grown in a large-scale plantation. Tree Physiology, 18, 653–658.
Park, S., Jisheng, L., Jon, K. P., Gerald, A. B., Haibing, Y., Soledad, U., et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proceedings of the National Academy of Sciences of the United States of America, 102, 18830–18835.
Hirschi, K. D. (1999). Expression of Arabidopsis CAX1 in tobacco: Altered calcium homeostasis and increased stress sensitivity. Plant Cell, 11, 2113–2122.
Buchman, A. R., & Berg, P. (1988). Comparison of intron-dependent and intron-independent gene expression. Molecular and Cellular Biology, 8, 4395–4405.
Chung, S., & Perry, R. (1989). Importance of introns for expression of mouse ribosomal protein gene rpL32. Molecular and Cellular Biology, 9, 2075–2082.
Okkema, P. G., Harrison, S. W., Plunger, V., Aryana, A., & Fire, A. (1993). Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics, 135, 385–404.
Ibrahim, M., Khan, S. A., Zafar, Y., Mansoor, S., Yusuf, A., & Mukhtar, Z. (2009). Expression of a full length Arabidopsis vacuolar H+pyrophosphatase (AVP1) gene in tobacco (Nicotiana tabaccum) to increase tolerance to drought and salt stress. Journal of Phytology, 1, 433–444.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Tanweer Kumar and Uzma have contributed equally to this study.
Rights and permissions
About this article
Cite this article
Kumar, T., Uzma, Khan, M.R. et al. Genetic Improvement of Sugarcane for Drought and Salinity Stress Tolerance Using Arabidopsis Vacuolar Pyrophosphatase (AVP1) Gene. Mol Biotechnol 56, 199–209 (2014). https://doi.org/10.1007/s12033-013-9695-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-013-9695-z