Abstract
The NRAMP gene family encodes integral membrane protein and mediates the transport of Fe, however, its function in transport of toxic metal ions is not very clear in plants. TcNRAMP3 was isolated from Thlaspi caerulescens, and encoded a metal transporter member of the NRAMP family. TcNRAMP3 was predominantly expressed in roots of T. caerulescens by semi-quantitative RT-PCR. The expression of TcNRAMP3 was induced by iron starvation and by the heavy metals Cd and Ni in roots. TcNRAMP3 was able to rescue growth of an iron uptake fet3fet4 mutant yeast strain, suggesting a possible role in iron transport. Expression of TcNRAMP3 in yeast increased Cd sensitivity and Cd content, while it enhanced the Ni resistance and reduced Ni accumulation, indicating that TcNRAMP3 could accumulate Cd and exclude Ni in yeast. Furthermore, overexpression of TcNRAMP3 in tobacco resulted in slight Cd sensitivity of root growth and did not influence Ni resistance. These results suggested that TcNRAMP3 played a role in metal cation homeostasis in plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metal cations are essential for plant nutrition, for example, iron, manganese, and copper are cofactors for many enzymes, such as proteases and superoxide dismutases. However, plants need the ability to prevent excessive accumulation of essential cations and toxic heavy metals, such as zinc, copper, cadmium, lead, mercury, and arsenic. The metal cation transporters played a role in maintaining metal homeostasis [1]. With the progress of molecular genetic analysis, it has been revealed that many kinds of membrane proteins, including channel, pump, and transporter proteins, participate in metal transport and homeostasis, and among them the NRAMP family genes have important roles in metal transport [2].
The first member of the NRAMP family was named natural resistance associated macrophage protein 1 (NRAMP1), because mutations in this macrophage-specific protein confer increased sensitivity to intracellular bacterial pathogens [3]. The metal transport function of NRAMP was assigned later, when Supek et al [4] characterized SMF1, a yeast NRAMP homologue, as an Mn uptake system. NRAMP proteins are now recognized as a ubiquitous family of metal transporters with homologues in fungi, yeast, animals, plants, and bacteria [5].
NRAMP genes have been identified in many plant species. To date, six members of the NRAMP gene family in Arabidopsis thaliana have been assigned and partially characterized [6]. Functional studies have shown that overexpression of AtNRAMP1 decreases sensitivity to toxic Fe levels, and overexpression of AtNRAMP3 leads to Cd hypersensitivity and Fe overaccumulation in A. thaliana [7, 8]. However, the expression and function of NRAMP under heavy metal stress remains limited in plants.
Thlaspi caerulescens is a heavy metal hyperaccumulator that grows in soils contaminated with Zn, Pb, and Cd [9]. In this study, TcNRAMP3, a T. caerulescens homologue of the NRAMP family transporters identified in our group previously [10], was used for heterologous expression in yeast and plants, in order to reveal the NRAMP function in yeast and plants.
Materials and Methods
Plant Materials and Treatments
Thlaspi caerulescens was cultured in MS medium (pH 5.7) in a growth chamber at 22°C on a 16 h light/8 h dark cycle for 8 weeks, then transferred onto a medium containing 1,000 μM Ferrozine®, or 500 μM CdCl2, or 600 μM NiCl2. Roots were harvested at time zero, 2 h, 4 h, 8 h, 12 h, 24 h, 48 h, 72 h, and 96 h. All treatments were done in triplicate.
Southern Blot Analysis
Genomic DNA from T. caerulescens leaves was isolated via a modified CTAB extraction method. Genomic DNA hybridization was performed as described [11], with a full-length TcNRAMP3 cDNA probe labeled with digoxigenin–dUTP.
RNA Isolation and Semi-Quantitative RT-PCR Analysis
Total RNA was isolated from shoots, roots, and leaves of control plant, and from roots treated with 1,000 μM Ferrozine®, 600 μM NiCl2, or 500 μM CdCl2 for 96 h. Reverse transcription (RT) was performed and the PCR conditions were as follows: 94°C for 2 min; 28 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min using primers FTcN3 (5′-CGACGAAACAGAGAAAGTTCACATC-3′) and RTcN3 (5′-ACTCTGCTTTAGGACGCCACGGA-3′). Actin was used as an internal control. All RT-PCR analyses were done in triplicate.
Construction of a TcNRAMP3 Expression Vector and Transformation into Yeast
Saccharomyces cerevisiae DEY1453 (MATα/MAT2 ade2/+ can1/can1 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 fet3-2::HIS3/fet3-2::HIS3 fet4-1::LUE/fet4-1::LUE) and YK44 (ura3-52 his3-200, △ZRC, △Cot1, mating type α) were used as the host cells, and plasmid pYES2 was used as an expression vector. The fet3fet4 double mutant strain DEY1453 is defective in both low-affinity and high-affinity iron uptake systems [12], and can grow only in medium containing 100 μM FeCl3. Yeast strain YK44, which was kindly donated by Prof Dietrich H. Nies of Martin-Luther University, is sensitive to Zn, Cd, Ni, and Co. The coding region of the TcNRAMP3 gene was amplified from the originally isolated clone with sense primer 5′-CGCGGATCCTCAACTAGACTCTGC-3′ and antisense primer 5′-CGCGGATCCACTAGACTCTGCTTTA-3′. The amplified product was digested with HindIII and BamHI, and ligated to pYES2 to construct the expression vector pYES2–TcNRAMP3. The plasmid was transformed into S. cerevisiae using lithium acetate method. Yeast transformed with pYES2 was used as a control. Transformed cells were selected in SD-ura medium (Difco BRL, Gaithersburg, MD, USA) and transferred to YPGAL medium (1% yeast extract, 2% peptone, 2% galactose, Difco) for further test.
Yeast Complementation Assay and Heavy Metal Tolerance Assay
The DEY1453 yeast cells harboring pYES2–TcNRAMP3 or pYES2 were grown overnight in liquid SD-ura containing 100 μM FeCl3. The cultures were diluted to OD values of 10−1–10−3 (as indicated) and spotted on YPGAL plates with 100 μM FeCl3 or without Fe.
YK44 yeast cells transformed with pYES2–TcNRAMP3 or pYES2 were grown on SD-ura medium. The cultures were diluted to ODs of 10−1–10−2 and spotted on YPGAL plates with 10 μM CdCl2 or 800 μM NiCl2. All experiments were done in triplicate.
Metal Ion Content Measurements
YK44 yeast recombined with empty or pYES2–TcNRAMP3 was pre-cultured at 30°C with shaking at 140 rpm in SD-ura medium. When cell growth reached late log phase, 1/1,000 volume of culture broth was added to YPGAL medium with 5 μM CdCl2 or 400 μM NiCl2. After incubation for 36 h, cells were harvested by centrifugation, washed twice with 50 mM EDTA in cold distilled Ultrapure water, and then dried at 80°C for 12 h. The yeast cells were used for analysis by inductively coupled plasma mass spectroscopy (ICP–MS).
Plant Transformation
The expression cassette of TcNRAMP3 cDNA was inserted into the XbaI and BamHI site of the plant expression vector pBI121 to construct the expression vector pBI121–TcNRAMP3. The recombinant plasmid pBI121–TcNRAMP3 and the empty vector pBI121 were introduced into the A. tumefaciens EHA105 strain, which was then used to transform tobacco (Nicotiana tabacum) NC89 by the Agrobacterium-mediated transformation method. Total RNAs were isolated from both the empty vector pBI121 and TcNRAMP3 transgenic plants. Northern blots were used to verify the expression of the introduced genes.
Seedling Development Assay
Transgenic tobacco plants were raised in growth chambers at 25°C with a 16 h light/8 h dark cycle and selected on MS medium containing 100 μg/ml of kanamycin. Primary transformants were self-fertilized and seeds were collected. Surface-sterilized seeds were then kept in 0.1% agar for 4 days, sowed onto 1/2 MS medium plates containing 50 μM CdCl2 or 800 μM NiCl2, and incubated at 25°C with a 16 h light/8 h dark cycle. Thirty seeds per replicate were examined, and all treatments were done in triplicate.
Results
The TcNRAMP3 Proteins Belongs to the NRAMP Family of Membrane Transporters
TcNRAMP3 was 1,819 bp long and contained a 1,539 nt open reading frame (ORF) encoding 512 amino acid residues. TcNRAMP3 displayed 90% amino acid identity with AtNRAMP3, 76% with TjNRAMP4, 74% with AtNRAMP4, and 41% with AtNRAMP1. Hydropathy analysis indicated that TcNRAMP3 consisted of 11 transmembrane region (TM) domains. There was a conserved transport motif (5′-GQSSTITGTYAGQFIMGGFLN-3′), common among NRAMP homologues, between transmembrane segments 7 and 8. There are two classes of NRAMP proteins in plants: AtNRAMP1 and OsNRAMP1, 3 represent one class, and AtNRAMP2-5 and OsNRAMP2 the other [6, 7], suggesting that TcNRAMP3 belonged to the second class of NRAMP proteins. A phylogenetic tree was constructed on the basis of the deduced amino acid sequences of TcNRAMP3 and other NRAMP proteins from Arabidopsis, Oryza sativa, Thlaspi japonicum, and soybean. As shown in Fig. 1, TcNRAMP3 and AtNRAMP3 were grouped together.
A phylogenetic tree of selected members of the NRAMP family found in plants: AtNRAMP1 (AF165125), AtNRAMP2 (AF141204), AtNRAMP3 (AF202539), AtNRAMP4 (AF202540), AtNRAMP5 (CAC27822), AtNRAMP6 (CAC28123), and AtEIN2 (AAD41076) from Arabidopsis; OsNRAMP1 (L41217), OsNRAMP2 (L81152), and OsNRAMP3 (U60767) from rice; TjNRAMP4 (AB115423) from Thlaspi japonicum; GmDMT1 (AY169405) from soybean (Glycine max). Amino acid sequences from NRAMP were aligned using ClustalW. The diagram shows an unrooted tree constructed with the program TREEVIEW
TcNRAMP3 is Present in T. caerulescens Genome as Low Copy Number
To determine the copy number of TcNRAMP3 in the T. caerulescens genome, Southern blot analysis was performed. Two major band patterns were observed due to an endogenous KpnI restriction enzyme site within the TcNRAMP3 gene (Fig. 2a). Furthermore, an additional weak band was observed in each digestion, which might be the homologues of TcNRAMP3. It is likely that single copy number of TcNRAMP3 is present in the T. caerulescens genome.
The organization and expression of the TcNRAMP3 gene in T. caerulescens. (a) Southern blot analysis of TcNRAMP3. Twenty micrograms of genomic DNA digested with EcoRI (external cutter) or KpnI (internal cutter). Major DNA fragments are marked with asterisks. (b) Tissue-specific expression of TcNRAMP3 in T. caerulescens seedling under normal condition. (c)–(e) Expression of TcNRAMP3 in roots exposed to various heavy metals for 96 h, such as 1,000 μM Ferrozine® (c), 600 μM NiCl2 (d) or 500 μM CdCl2 (e)
TcNRAMP3 is Upregulated by Fe Deficiency and Heavy Metals
The expression pattern of TcNRAMP3 in T. caerulescens seedling was performed by semi-quantitative RT-PCR. TcNRAMP3 was mainly expressed in root, and weakly expressed in leaf and stem tissues under normal condition (Fig. 2b). To study whether the expression of TcNRAMP3 in root was responsive to metal stress, T. caerulescens seedlings were treated with 1,000 μM Ferrozine®, 600 μM NiCl2 or 500 μM CdCl2, respectively. TcNRAMP3 mRNAs increased when exposed to 1,000 μM Ferrozine® for 2 h, peaking at 12 h, and then decreased, however, levels of mRNA were maintained for 96 h at a higher level compared to that of the control plant (Fig. 2c). The expression of TcNRAMP3 increased slowly when T. caerulescens plants were treated with 600 μM NiCl2, and reached the highest level between 24 h and 48 h (Fig. 2d). The transcripts of TcNRAMP3 were enhanced continuously until 96 h when treated with 500 μM CdCl2 (Fig. 2e). These results implied a possible role in Fe, Ni, and Cd transport.
Metal Transporting Ability of TcNRAMP3 in Yeast
To determine whether TcNRAMP3 encode functional iron NRAMP transporters, the coding sequence was cloned into the yeast expression vector pYES2. The fet3fet4 yeast (DEY1453) strain is defective in both low-affinity and high-affinity Fe uptake systems and requires high concentrations of Fe in its medium for growth. The fet3fet4 mutants transformed with pYES2 containing TcNRAMP3, or empty control vector, were spotted in different dilutions on YPGAL medium. As shown in Fig. 3, TcNRAMP3 restored the ability of fet3fet4 to grow on a medium without Fe, indicating that TcNRAMP3 was a functional Fe transporter.
The yeast YK44 strain was used to test whether TcNRAMP3 transports other heavy metals. Transformants of pYES2–TcNRAMP3 and pYES2 grew normally and without obvious difference under control conditions (Fig. 4a). However, the growth of both transformants was impaired when the medium contained 10 μM CdCl2 or 800 μM NiCl2. As shown in Fig. 4b, the yeast expressing TcNRAMP3 showed strong growth repression on the medium supplemented with 10 μM CdCl2 compared to the control. On the contrary, the growth of pYES2–TcNRAMP3-expressing yeast cells was better than that of control under 800 μM NiCl2 stress (Fig. 4c). These results indicated that TcNRAMP3 confers Cd sensitivity and Ni resistance in yeast.
Functional analysis of TcNRAMP3 in YK44 yeast cells. Growth of recombinant yeasts on (a) YPGAL medium, (b) YPGAL medium with 10 μM CdCl2 or (c) YPGAL medium with 800 μM NiCl2. (d) The Cd content of YK44 yeast cells containing TcNRAMP3 or the empty pYES2 grown in liquid medium with 5 μM CdCl2 and (e) the Ni content in liquid medium with 400 μM NiCl2. Data are expressed as mean ± S.E. * P< 0.05 vs. control group
To elucidate the Cd2+ and Ni2+ accumulation activity of TcNRAMP3 in yeast, the contents of Cd or Ni in yeasts transformed with TcNRAMP3 or pYES2 were determined. The Cd2+ content of yeast expression in TcNRAMP3 was about twice of the control after 36 h of cultivation with 5 μM CdCl2 (Fig. 4d), whereas the control accumulated 8.5-fold amount of Ni2+ than that of TcNRAMP3-expressing yeast after cultivation with 400 μM NiCl2 (Fig. 4e). All these results suggested that TcNRAMP3 transported Cd2+ into the yeast cells and excluded Ni2+ from yeast cells.
The Response of TcNRAMP3 Transgenic Tobacco Plant Under Heavy Metal Stress
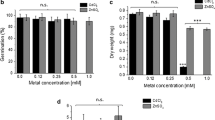
To investigate the physiological function of TcNRAMP3 in plants, TcNRAMP3 under the control of the strong CaMV 35S promoter were expressed in transgenic tobacco plants. Northern blot analysis indicated that TcNRAMP3 was expressed constitutively in transgenic tobacco plants (Fig. 5a). The root lengths of both TcNRAMP3-over-expressing and empty vector control plants were compared in the presence of 50 μM CdCl2 or 800 μM NiCl2. As shown in Fig. 5, root development was not noticeably different between TcNRAMP3-overexpressing and control seedlings under normal conditions (Fig. 5b), while root development of both TcNRAMP3-overexpressing and control plantlets was inhibited significantly under heavy metal stress (Cd or Ni) (Fig. 5c–e). However, Fig. 5c shows that the root length of TcNRAMP3-overexpressing lines was slightly shorter than that of control plants under CdCl2 stress. Nevertheless there was no difference in root growth of both TcNRAMP3 transgenic tobacco and control plant under Ni stress (Fig. 5d). The Ni transport capability of TcNRAMP3 revealed by yeast does not lead to increased Ni tolerance in transgenic tobacco.
The root elongation of the TcNRAMP3 overexpression transgenic tobacco plants in response to Cd or Ni stress. (a) Northern blot analysis of an over-expressing line. (b)–(d) Wild type and transgenic plant were grown in 1/2 MS medium (b) or with 50 μM CdCl2 (c) or 800 μM NiCl2, (d) for 2-week. (e) Diagram for root length of wild type and transgenic tobacco seedlings
Discussion
NRAMP proteins are broad-range, membrane-bound metal transporters in both eukaryotes and prokaryotes [13]. To date, NRAMP family genes have been identified in many plant species, including Arabidopsis, rice, tomato, soybean, and T. japonicum [7, 14–17]. Here, we report the functional characterization of TcNRAMP3 from the heavy metal accumulator T. caerulescens.
TcNRAMP3 was expressed preferentially in roots, whereas the Arabidopsis homologue AtNRAMP3 was expressed in roots and aerial parts at similar levels. NRAMP family genes were upregulated under Fe deficiency or sufficiency conditions. In this study, TcNRAMP3 was induced by Fe starvation in the roots of T. caerulescens. Its ability to rescue growth of the yeast fet3fet4 mutant strain under Fe-limiting conditions makes TcNRAMP3 a strong candidate for iron transporter (Fig. 3). This result was consistent with other NRAMP proteins in plants. AtNramp3 and AtNramp4 complement the phenotype of fet3fet4 mutant yeast as effectively as the well-characterized Arabidopsis Fe uptake transporter IRT1 [18]. All these results underscored the conservation in the function of this family of proteins of transporting Fe in plants. The expression of TcNRAMP3 in the roots of T. caerulescens was upregulated by CdCl2 and NiCl2 stress (Fig. 2). Tomato LeNRAMP1, 2, and 3 was induced by heavy metal stress [19], suggesting that NRAMP proteins are also involved in heavy metal transport in plant.
Overexpression of TcNRAMP3 led to an increase of Cd content in yeast cells and inhibited the growth of yeast when treated with CdCl2 (Fig. 4). However, overexpression of TcNRAMP3 in transgenic tobacco plants, slightly reduced the lengths of tobacco seedling roots (Fig. 5). This was different from that of overexpression of AtNRAMP3 in Arabidopsis, which led to Cd hypersensitivity. The NRAMP family is localized mainly at the vacuolar membrane and acts to control and maintain homeostasis of metals, such as Fe and Cd [20, 21]. The Cd hypersensitivity induced by overexpression of AtNRAMP3 or AtNRAMP4 was proposed to be due to increased remobilization of Cd from the vacuole [20, 22]. As a homologue of AtNRAMP3, TcNRAMP3 might participate in the control of metal ion homoeostasis in plant cell. Furthermore, TcNRAMP3 was isolated from the heavy metal hyperaccumulating plant, and might not remobilize large amount of Cd2+ from the vacuole to cytoplast. Therefore, TcNRAMP3 could contribute to a slightly decreased cadmium resistance of root growth in transgenic lines.
TcNRAMP3-overexpression yeasts could exclude Ni2+ from the yeast cells (Fig. 4). The function of this metal ion efflux pump is not very common in the NRAMP family. An animal NRAMP1 protein is known to act as an iron efflux pump [23]. Zhou and Yang (2005) [24] suggested that OsNRAMP3 is involved in the iron efflux process. Although TcNRAMP3 exclude Ni from yeast cell, TcNRAMP3-overexpression transgenic tobacco seedling showed no significant difference on Ni tolerance when compared to the control plants (Fig. 5). These data supported our hypothesis that TcNRAMP3 mainly functioned in maintaining metal ion homeostasis in plant cells when plants were exposed to high levels of Cd or Ni. In addition, the competition with other divalent cations may prevent Ni from transporting by TcNRAMP3 in plants, and this possibility was supported by Fleming [25], who reported that, despite lack of specificity of metals transported NRAMP when expressed in a heterologous system, it is predominantly an iron transporter in vivo.
References
Nelson, N. (1999). Metal ion transporters and homeostasis. The EMBO Journal, 18, 4361–4371. doi:10.1093/emboj/18.16.4361.
Williams, L. E., Pittman, J. K., & Hall, J. L. (2000). Emerging mechanisms for heavy metal transport in plants. Biochimica et Biophysica Acta, 1465, 104–126. doi:10.1016/S0005-2736(00)00133-4.
Vidal, S. M., Malo, D., Vogan, K., Skamene, E., & Gros, P. (1993). Natural resistance to infection with intracellular parasites-isolation of a candidate for Bcg. Cell, 73, 469–485. doi:10.1016/0092-8674(93)90135-D.
Supek, F., Supekova, L., Nelson, H., & Nelson, N. (1996). A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proceedings of the National Academy of Sciences of the United States of America, 93, 5105–5110. doi:10.1073/pnas.93.10.5105.
Cellier, M., & Gros, P. (Eds.). (2004). The Nramp Family. New York: Eurekah.com and Kluwer Academic/Plenum.
Mäser, P., Thomine, S., Schroeder, J. L., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenitic relationships within cation transporter families of Adabidopsis. Plant Physiology, 126, 1646–1667. doi:10.1104/pp. 126.4.1646.
Curie, C., Alonso, J. M., Jean, M. L. E., Ecker, J. P., & Briat, J. F. (2000). Involvement of NRAMP1 from Arabidopsis thariana in iron transport. The Biochemical Journal, 347, 749–755. doi:10.1042/0264-6021:3470749.
Thomine, S., Wang, R., Ward, J. M., Crawford, N. M., & Schroeder, J. I. (2000). Cadmium and iron transport by members of a plant transporter gene family in Arabidopsis with homology to NRAMP genes. Proceedings of the National Academy of Sciences of the United States of America, 97, 4991–4996. doi:10.1073/pnas.97.9.4991.
Baker, A. J. M., & Brooks, R. R. (1989). Terrestrial higher plants, which hyperaccumulate metallic elements—a review of their distribution, ecology and phytochemistry. Biorecovery, 1, 81–126.
Lang, M. L., Zhang, Y. X., Guan, Z. Q., & Chai, T. Y. (2005). PCR-enriched cDNA pool method for cloning of gene homologues. Plant Molecular Biology Reporter, 23, 219–226. doi:10.1007/BF02772752.
Church, G. M., & Gilbert, W. (1984). Genomic sequencing. Proceedings of the National Academy of Sciences of the United States of America, 81, 1991–1995. doi:10.1073/pnas.81.7.1991.
Dix, D. R., Bridgham, J. T., Broderius, M. A., Byersdorfer, C. A., & Eide, D. J. (1994). The fet4 gene encodes the low-affinity Fe(II) transport protein of Saccharomyces cerevisiae. The Journal of Biological Chemistry, 269, 26092–26099.
Forbes, J. R., & Gros, P. (2001). Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends in Microbiology, 9, 397–403. doi:10.1016/S0966-842X(01)02098-4.
Belouchi, A., Kwan, T., & Gros, P. (1997). Cloning and characterization of the OsNRAMP family from Osyza sativa, a new family of membrance proteins possibly implicated in the transport of metal ions. Plant Molecular Biology, 33, 1085–1092. doi:10.1023/A:1005723304911.
Bereczky, Z., Wang, H. Y., Schubert, V., Ganal, M., & Bauer, P. (2003). Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. The Journal of Biological Chemistry, 278, 24697–24704. doi:10.1074/jbc.M301365200.
Kaiser, B. N., Moreau, S., Castelli, J., Thomson, R., Lambert, A., Bogliolo, S., et al. (2003). The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. The Plant Journal, 35, 295–304. doi:10.1046/j.1365-313X.2003.01802.x.
Mizuno, T., Usui, K., Horie, K., Nosaka, S., Mizuno, N., & Obata, H. (2005). Cloning of three ZIP/Nramp transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+-transport abilities. Plant Physiology and Biochemistry, 43, 793–801. doi:10.1016/j.plaphy.2005.07.006.
Eide, D., Broderius, M., Fett, J., & Guerinot, M. L. (1996). A novel iron regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences of the United States of America, 93, 5624–5628. doi:10.1073/pnas.93.11.5624.
Ouziad, F., Hildebrandt, U., Schmelzer, E., & Bothe, H. (2005). Differential gene expression in arbuscular mycorrhizal-colonized tomato grown under heavy metal stress. Journal of Plant Physiology, 162, 634–649. doi:10.1016/j.jplph.2004.09.014.
Thomine, S., Lelie`vre, F., Debarbieux, E., Schroeder, J. I., & Barbier-Brygoo, H. (2003). AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. The Plant Journal, 34, 685–695. doi:10.1046/j.1365-313X.2003.01760.x.
Lanquar, V., Lelievre, F., Bolte, S., Hames, C., Alcon, C., Neumann, D., et al. (2005). Mobilization of vacuolar iron by AtNramp3 and AtNramp4 is essential for seed germination on low iron. The EMBO Journal, 24, 4041–4051. doi:10.1038/sj.emboj.7600864.
Lanquar, V., Lelie`vre, F., Barbier-Brygoo, H., & Thomine, S. (2004). Regulation and function of AtNRAMP4 metal transporter protein. Soil Science and Plant Nutrition, 50, 477–484.
Govoni, G., & Gros, P. (1998). Macrophage NRAMP1 and its role in resistance to microbial infections. Inflammation Research, 47, 277–284. doi:10.1007/s000110050330.
Zhou, X. J., & Yang, Y. N. (2005). Differential expression of rice Nramp genes in response to pathogen infection, defense signal molecules and metal ions. Physiological and Molecular Plant Pathology, 65, 235–243. doi:10.1016/j.pmpp. 2005.02.007.
Fleming, M. D., Trenor, C. C., Su, M. A., Foernzler, D., Beier, D. R., Dietrich, W. F., et al. (1997). Microcytic anaemia mice have a mutation in Mramp2, a candidate iron transporter gene. Nature Genetics, 16, 383–386.
Acknowledgments
This research was supported by the National High Technology Planning Program of China (Grant nos. 2007AA021404 and 2006AA10Z407), the Key Laboratory of Nuclear Analytical Techniques of CAS (K131) and the China National Natural Sciences Foundation (Grant nos. 30570146).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wei, W., Chai, T., Zhang, Y. et al. The Thlaspi caerulescens NRAMP Homologue TcNRAMP3 is Capable of Divalent Cation Transport. Mol Biotechnol 41, 15–21 (2009). https://doi.org/10.1007/s12033-008-9088-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-008-9088-x