Abstract
The relationship between the SLC31A1 (protein: copper transporter 1) rs10981694 A > C and ATP7B (protein: P-type adenosine triphosphatase 7B) rs9535828 A > G polymorphisms on the overall survival and disease-free survival of 104 Japanese patients with esophageal squamous cell carcinoma (ESCC) receiving neoadjuvant chemoradiotherapy (CRT) was investigated. Chemotherapy consisted of protracted infusion of 5-fluoracil (800 mg/m2/day) on days 1–5 and cisplatin or nedaplatin (80 mg/m2/day) on day 1. The median (range) follow-up was 47 (6–127) months. The 5-year overall and disease-free survival rates were 71.2% and 60.6%, respectively. The 5-year overall survival rate was significantly higher in patients with the SLC31A1 rs10981694 C allele compared with the rs10981694 A/A genotype (91.7% vs. 65.0%, P = 0.018). The 5-year disease-free survival rate was significantly higher in patients with the SLC31A1 rs10981694 C allele compared with the rs10981694 A/A genotype (79.2% vs. 55.0%, P = 0.043). In addition, univariate and multivariate analyses showed the SLC31A1 rs10981694 A > C polymorphism to be a significant prognostic factor affecting 5-year overall survival after neoadjuvant CRT. However, the overall and disease-free survival rates after surgery did not differ significantly among the ATP7B rs9535828 genotypes. In conclusion, only the SLC31A1 rs10981694 A/A genotype was an independent predictor of a poorer 5-year overall survival. Therefore, in neoadjuvant CRT for ESCC patients, the effect of platinum was affected by the SLC31A1 rs10981694 A > C polymorphism. The presence of this polymorphism should be considered when devising neoadjuvant CRT regimens or treatment strategies for ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Although esophageal squamous cell carcinoma (ESCC) has a high malignant potential and poor prognosis [1, 2], it often responds well to chemotherapy and/or radiotherapy [3, 4]. Chemoradiotherapy (CRT) is now a standard treatment for ESCC [3, 4]; however, the clinical outcomes after neoadjuvant CRT differ among patients with ESCC. Therefore, biomarkers predicting the responsiveness of personalized CRT regimens for ESCC are necessary.
Platinum-based chemotherapy is widely used for many cancer types such as lung, bladder, head, and neck cancers [5,6,7]. In ESCC, combination therapy consisting of cisplatin, as a platinum-based anticancer drug, and 5-fluoracil (5-FU) is used as the key regimen to treat various stages of this disease in Japan [8,9,10]. Platinum-based anticancer drugs act by binding to DNA with intra- or inter-strand crosslinks, and the platinum–DNA adducts lead to DNA lesions, activation of multiple pathways, and ultimately cell apoptosis [11, 12]. However, the clinical outcomes of patients receiving platinum-based anticancer drugs are affected by the development of resistance to these drugs. The mechanisms of platinum resistance are thought to include decreased membrane transport of the drugs, detoxification in the cytoplasm, inactivation of DNA repair pathways, increased DNA repair, and induction of apoptosis due to DNA damage [11, 12].
Platinum-based anticancer drugs are taken up by cancer cells via copper transporter (CTR) 1 (gene: SLC31A1) and are excreted from cancer cells via copper-transporting P-type adenosine triphosphatase 7B (ATP7B) [13,14,15]. The expression levels of CTR1 and ATP7B have been reported to affect platinum resistance in patients with non-small cell lung cancer or ovarian carcinoma treated with platinum-based anticancer drugs [16, 17]. However, it is not clear how the drug transporters CTR1 and ATP7B contribute to platinum tolerance, among the various mechanisms of platinum resistance [11, 12], or their clinical effects on platinum resistance in patients with ESCC.
Several single nucleotide polymorphisms (SNPs) have been identified in SLC31A1 and ATP7B [18,19,20]. These SNPs may explain the inter-individual differences in the response to, and toxicity of, CRT. The SLC31A1 rs10981694 A > C and ATP7B rs9535828 A > G polymorphisms have been reported to affect platinum resistance or the occurrence of adverse events in lung and ovarian cancer patients treated with platinum-based anticancer drugs [18,19,20]; however, no such evidence is available in ESCC patients.
The aim of the present study was to determine whether the survival of 104 patients with ESCC receiving neoadjuvant CRT was affected by the SLC31A1 rs10981694 A > C or ATP7B rs9535828 A > G polymorphism.
Methods
Protocol
This study was approved by the Ethics Committee of Akita University Graduate School of Medicine (nos. 419 and 600). The study participants were 104 Japanese patients treated with neoadjuvant CRT for ESCC at Akita University Hospital between 2008 and 2018. For all patients, the clinical cancer stage and treatment strategy were defined at an esophageal cancer conference attended by physicians, radiologists, surgeons, and pharmacists. In addition, the disease was classified according to the UICC International Union Against Cancer tumor/node/metastasis Classification of Malignant Tumors (7th edition). Chemotherapy consisted of protracted infusion of 5-FU (800 mg/m2/day) on days 1–5 and cisplatin or nedaplatin (80 mg/m2/day) on day 1. This protocol was repeated once or twice every 3–5 weeks. Radiotherapy involved high-energy 10 MV X-rays delivered in 1.8–2 Gy daily fractions for a total of 40–41.2 Gy. All patients received a three-dimensional radiation treatment plan.
The gross tumor volume of the primary tumor and metastatic lymph nodes was determined. The primary tumor was contained within the craniocaudal length of the primary tumors ranged from 3 to 5 cm. The clinical target volume was determined by setting a margin of approximately 1 cm around the gross tumor volume, and it included mediastinal lymph nodes. The following histopathological criteria were used to determine the efficacy of neoadjuvant CRT [21, 22]: grade 0 (ineffective), no recognizable cytological or histological therapeutic effect; grade 1 (slightly effective), apparently viable cancer cells (including cells with eosinophilic cytoplasm with vacuolation and swollen nuclei) making up one-third or more of the tumor tissue but with some evidence of degeneration of cancer tissue or cells; grade 2 (moderately effective), viable cancer cells making up less than one-third of the tumor tissue with severe degeneration or necrosis of other cancer cells; and grade 3 (markedly effective), no viable cancer cells evident.
Disease-free survival was defined as the period from the start of neoadjuvant CRT to the date of the last visit without recurrence. Overall survival was defined as the period from the start of neoadjuvant CRT to death from any cause or the last visit.
Genotyping
For SNP analysis, 1 mL peripheral blood samples were obtained and stored at −80 °C. DNA was extracted from the peripheral blood samples using the QIAamp Blood Kit (Qiagen, Hilden, Germany) and stored at −80 °C until analysis. The SLC31A1 rs10981694 A > C and ATP7B rs9535828 A > G genotypes were identified using PCR–restriction fragment length polymorphism [18,19,20]. The PCR mixture contained 5 μL 10 ng/μL genomic DNA, 25 μL 2× HotStarTaq Master Mix, 2.5 μL each primer (10 μM), and 15 μL RNase-free water in a final volume of 50 μL. The PCR conditions were 95 °C for 15 min, 35 cycles of 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 5 min. The genotype frequency of SLC31A1 rs10981694 A > C was 76.92% for A/A (n = 80), 22.12% for A/C (n = 23), and 0.96% for C/C (n = 1). Therefore, patients with the SLC31A1 rs10981694 A/C and C/C genotypes were analyzed as a single group.
Statistical analyses
The distribution of the continuous variables was evaluated for normality using the Shapiro–Wilk test. The values of the patient characteristics are presented as medians (range). The Kruskal–Wallis test or Mann–Whitney U test was used to compare the continuous variables between groups. Differences in patient characteristics and genotype frequency were determined using the Pearson χ2 test or Fisher’s exact test. Survival curves for overall survival and disease-free survival were analyzed using the Kaplan–Meier method. The Cox proportional hazard regression model was used to perform univariate and multivariate analyses. In the multivariate model, age, sex, lymph node metastasis, histopathological criteria, and the SLC31A1 rs10981694 A > C polymorphism were included as explanatory variables. For each patient, the genotypes of the transporters were replaced with dummy variables (in two groups, 1 and 0; in three groups, 1 and 0, 0 and 0, and 0 and 1). A two-sided P value less than 0.05 was considered statistically significant. Statistical analyses were performed using SPSS 26.0 for Windows (SPSS IBM Japan Inc., Tokyo, Japan).
Results
The patient population included 87 males and 17 females. The median (range) age of the patients was 63.5 (43–77) years. Cisplatin plus 5-FU therapy and nedaplatin plus 5-FU therapy were received by 31 and 73 patients, respectively. According to the histopathological criteria, the effect of neoadjuvant CRT was judged to be grade 1 in 34 (32%) patients, grade 2 in 38 (37%), and grade 3 in 32 (31%). The characteristics of the patients did not differ between the SLC31A1 rs10981694 A/A genotype and C allele or among the ATP7B rs9535828 A/A, A/G, and G/G genotypes (Table 1). In addition, there was no significant difference in the histopathological grade, defining the effectiveness of CRT, according to the genotype of each polymorphism (Table 1).
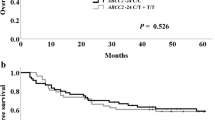
The median (range) follow-up was 47 (6–127) months. The 5-year overall survival and 5-year disease-free survival rates were 71.2% and 60.6%, respectively. Comparison of the overall survival and disease-free survival rates according to the SLC31A1 rs10981694 A > C and ATP7B rs9535828 A > G polymorphisms are shown in Fig. 1. The 5-year overall survival rates of the patients with the SLC31A1 rs10981694 A/A genotype and C allele were 65.0% (95% Cl 44.7–52.1) and 91.7% (95% Cl 52.8–61.6), respectively, and this difference was significant (P = 0.018) (Fig. 1a). On the other hand, the 5-year overall survival rates of the patients with the ATP7B rs9535828 A/A, A/G, and G/G genotypes were 77.8% (95% Cl 46.9–58.2), 68.6% (95% Cl 45.1–54.1), and 69.2% (95% Cl 43.4–56.2), respectively, but the differences were not significant (P = 0.681) (Fig. 1b).
Overall and disease-free survival curves of patients with esophageal squamous cell carcinoma according to genotype. Overall survival curves for patients with the SLC31A1 rs10981694 A > C (a) and ATP7B rs9535828 A > G (b) polymorphisms. Disease-free survival curves for patients with the genotype SLC31A1 rs10981694 A > C (c) and ATP7B rs9535828 A > G (d) polymorphisms
The 5-year disease-free survival rates of the patients with the SLC31A1 rs10981694 A/A genotype and C allele were 55.0% (95% Cl 35.3–45.5) and 79.2% (95% Cl 44.0–58.2), respectively, and this difference was significant (P = 0.043) (Fig. 1c). The 5-year disease-free survival rates of the patients with the ATP7B rs9535828 A/A, A/G, and G/G genotypes were 55.6% (95% Cl 36.6–51.7), 64.7% (95% Cl 37.0–49.7), and 57.7% (95% Cl 31.5–49.4), respectively, but the differences were not significant (P = 0.858) (Fig. 1d).
The results of univariate analyses of 5-year overall survival after neoadjuvant CRT, which included age (≥64 vs. <64 years), sex, depth of invasion (T3 vs. T1 + T2), lymph node metastasis (N1 + N2 vs. N0), histopathological criteria (grade 2 + 3 vs. grade 1), SLC31A1 rs10981694 A > C (A/A vs. A/C + C/C genotype), and ATP7B rs9535828 A > G (A/A vs. A/G + G/G genotype and A/A + A/G vs. G/G genotype), are shown in Table 2. The only factor found to be significantly related to survival was the SLC31A1 rs10981694 A > C polymorphism (P = 0.033) (Table 2). The results of the multivariate analysis using the Cox proportional hazard regression model are shown in Table 3. The SLC31A1 rs10981694 A > C polymorphism remained an independent factor influencing the 5-year overall survival after neoadjuvant CRT, and the age, sex, lymph node metastasis and histopathological criteria had no effect on the association between the SLC31A1 rs10981694 A > C polymorphism and 5-year overall survival rate (Table 3).
Discussion
To the best of our knowledge, this is the first report of an effect of the SLC31A1 rs10981694 A > C polymorphism on overall survival and disease-free survival in patients with ESCC undergoing neoadjuvant CRT (cisplatin plus 5-FU or nedaplatin plus 5-FU therapy). The SLC31A1 rs10981694 A > C polymorphism was not associated with the histopathological criteria used to determine the effect of neoadjuvant CRT; however, the 5-year overall and disease-free survival rates were approximately 20% higher in patients with the SLC31A1 rs10981694 C allele than in those with the A/A genotype. In addition, the SLC31A1 rs10981694 A/A genotype was identified as a risk factor for a poorer 5-year overall survival rate after neoadjuvant CRT. We do not have sufficient information to fully explain this phenomenon. The histopathological grade was evaluated in the tissues resected after neoadjuvant CRT. Therefore, similar to the survival rates, the histopathological grade was also considered to be related to the SLC31A1 rs10981694 A > C polymorphism, because patients with the SLC31A1 rs10981694 C allele had a high rate of intracellular platinum drug translocation. In the present study, patients with the SLC31A1 rs10981694 C allele tended to have high histopathologic criteria of grade 2 + 3 compared with A/A genotype, but the difference was not significant. Therefore, we considered the sensitivity for platinum-based anticancer drugs and clinical outcomes separately. In the present study, the clinical outcomes after neoadjuvant CRT were better in patients with the SLC31A1 rs10981694 C allele.
Xu et al. reported no significant difference in the response rate or overall survival rate after cisplatin-based chemotherapy in lung cancer patients with the SLC31A1 rs10981694 A/A genotype compared with the C allele, whereas carrying the SLC31A1 rs10981694 C allele was associated with severe cisplatin-induced ototoxicity [18]. In that report, the major reason for the cisplatin-induced ototoxicity seemed to be cellular accumulation of the platinum-based anticancer drug, as patients with the SLC31A1 rs10981694 C allele had higher levels of intracellular cisplatin. In contrast, Li et al. reported that 12 patients with epithelial ovarian cancer with the SLC31A1 rs10981694 C allele treated with carboplatin had a significantly higher rate of carboplatin resistance, defined as recurrence within 6 months after chemotherapy [19]; however, in that report, this phenomenon was not observed in 94 patients with epithelial ovarian cancer receiving cisplatin therapy [19]. Therefore, further studies evaluating the effect of this polymorphism on survival in patients with epithelial ovarian cancer may be necessary.
Li et al. reported a higher rate of response (complete or partial response) to platinum-based chemotherapy in lung cancer patients with the ATP7B rs9535828 A allele compared with the G/G genotype [20]. In that report, the ATP7B rs9535828 A > G polymorphism showed an ability to predict the clinical outcome after platinum-based chemotherapy in lung cancer patients. However, in the present study, the ATP7B rs9535828 A > G polymorphism affected neither survival nor the histopathological criteria used to determine a treatment effect after neoadjuvant platinum-based CRT in ESCC patients. The clinical outcomes after neoadjuvant platinum-based chemotherapy in ESCC patients in the present study were different from those after platinum-based chemotherapy in lung cancer patients in the study by Li et al. [20]. Thus, differences in patient background characteristics, cancer type, and assessment methods may be attributed to the different results among clinical studies, but further studies are necessary. Both in vivo and in vitro studies have shown CTR1 to be essential for platinum translocation into cells [23, 24]. Moreover, Yu et al. reported lower CTR1 protein expression in a cisplatin-resistant human ESCC cell line compared with the parental cell line [25]. Thus, the SLC31A1 rs10981694 A > C polymorphism may be a more appropriate biomarker of the efficacy of platinum-based anticancer drugs compared with the ATP7B rs9535828 A > G polymorphism. Different genotypes may be related to different levels of drug accumulation in tumor tissues, contributing to different responses to platinum chemotherapy.
Our current results should be interpreted within the context of the study limitations. Specifically, neoadjuvant chemotherapy or CRT is a standard treatment for stage II/III ESCC in Japan. Therefore, the influence of adjuvant therapy on the results of this study is not clear. Hence, additional studies of ESCC patients who have not received neoadjuvant CRT may be necessary. However, in the univariate and multivariate analyses of the present study, only the SLC31A1 rs10981694 A/A genotype was an independent factor associated with a poorer 5-year overall survival rate after neoadjuvant CRT. Therefore, CTR1, which contributes to intracellular translocation, may play a greater role in platinum resistance in ESCC patients than ATP7B, which contributes to extracellular clearance.
Conclusion
In conclusion, the 5-year overall and disease-free survival rates after neoadjuvant CRT were significantly higher in patients with the SLC31A1 rs10981694 C allele than in those with the SLC31A1 rs10981694 A/A genotype. In addition, only the SLC31A1 rs10981694 A/A genotype was an independent factor associated with poorer 5-year overall survival after neoadjuvant CRT. Therefore, patients with the SLC31A1 rs10981694 A/A genotype might require a different treatment strategy, such as a change in the chemotherapy regimen (for example, combination with radiotherapy or addition of docetaxel to cisplatin and 5-FU combination therapy [26,27,28]). However, further studies on the influence of the SLC31A1 rs10981694 A > C polymorphism on clinical outcomes after docetaxel, cisplatin, and 5-FU therapy are necessary. The biomarkers found in this study might be useful for personalization of ESCC treatment. Further prospective studies of the effect of the SLC31A1 rs10981694 A > C polymorphism on platinum resistance are warranted.
References
Chen MF, Yang YH, Lai CH, Chen PC, Chen WC. Outcome of patients with esophageal cancer: a nationwide analysis. Ann Surg Oncol. 2013;20:3023–30.
Kumagai K, Rouvelas I, Tsai JA, Mariosa D, Klevebro F, Lindblad M, Ye W, Lundell L, Nilsson M. Meta-analysis of postoperative morbidity and perioperative mortality in patients receiving neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal and gastro-oesophageal junctional cancers. Br J Surg. 2014;101:321–38.
Motoyama S, Sugiyama T, Ueno Y, Okamoto H, Takasawa S, Nanjo H, Watanabe H, Maruyama K, Okuyama M, Ogawa J. REG I expression predicts long-term survival among locally advanced thoracic squamous cell esophageal cancer patients treated with neoadjuvant chemoradiotherapy followed by esophagectomy. Ann Surg Oncol. 2006;13:1724–31.
Hayashi K, Motoyama S, Sugiyama T, Izumi J, Anbai A, Nanjo H, Watanabe H, Maruyama K, Minamiya Y, Koyota S, Koizumi Y, Takasawa S, Murata K, Ogawa J. REG Ialpha is a reliable marker of chemoradiosensitivity in squamous cell esophageal cancer patients. Ann Surg Oncol. 2008;15:1224–31.
Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U, Digumarti R, Zukin M, Lee JS, Mellemgaard A, Park K, Patil S, Rolski J, Goksel T, de Marinis F, Simms L, Sugarman KP, Gandara D. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51.
von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, Arning M. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–8.
Adelstein DJ, Li Y, Adams GL, Wagner H Jr, Kish JA, Ensley JF, Schuller DE, Forastiere AA. An intergroup phase III comparison of standard radiation therapy and two schedules of concurrent chemoradiotherapy in patients with unresectable squamous cell head and neck cancer. J Clin Oncol. 2003;21:92–8.
Ando N, Kato H, Igaki H, Shinoda M, Ozawa S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, Ikeda K, Kanda T, Tsujinaka T, Nakamura K, Fukuda H. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907). Ann Surg Oncol. 2012;19:68–74.
Kato H, Sato A, Fukuda H, Kagami Y, Udagawa H, Togo A, Ando N, Tanaka O, Shinoda M, Yamana H, Ishikura S. A phase II trial of chemoradiotherapy for stage I esophageal squamous cell carcinoma: Japan Clinical Oncology Group Study (JCOG9708). Jpn J Clin Oncol. 2009;39:638–43.
Ishida K, Ando N, Yamamoto S, Ide H, Shinoda M. Phase II study of cisplatin and 5-fluorouracil with concurrent radiotherapy in advanced squamous cell carcinoma of the esophagus: a Japan Esophageal Oncology Group (JEOG)/Japan Clinical Oncology Group trial (JCOG9516). Jpn J Clin Oncol. 2004;34:615–9.
Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23.
Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–84.
Sprowl JA, Ness RA, Sparreboom A. Polymorphic transporters and platinum pharmacodynamics. Drug Metab Pharmacokinet. 2013;28:19–27.
Kilari D, Guancial E, Kim ES. Role of copper transporters in platinum resistance. World J Clin Oncol. 2016;7:106–13.
Song IS, Savaraj N, Siddik ZH, Liu P, Wei Y, Wu CJ, Kuo MT. Role of human copper transporter Ctr1 in the transport of platinum-based antitumor agents in cisplatin-sensitive and cisplatin-resistant cells. Mol Cancer Ther. 2004;3:1543–9.
Konishi M, Imai A, Fujii M, Sugimoto K, Katakami N, Imai Y, Kamoshida S. Correlation of expression levels of copper transporter 1 and thymidylate synthase with treatment outcomes in patients with advanced non-small cell lung cancer treated with S-1/carboplatin doublet chemotherapy. Asian Pac J Cancer Prev. 2018;19:435–41.
Nakayama K, Kanzaki A, Ogawa K, Miyazaki K, Neamati N, Takebayashi Y. Copper-transporting P-type adenosine triphosphatase (ATP7B) as a cisplatin based chemoresistance marker in ovarian carcinoma: comparative analysis with expression of MDR1, MRP1, MRP2. LRP BCRP Int J Cancer. 2002;101:488–95.
Xu X, Ren H, Zhou B, Zhao Y, Yuan R, Ma R, Zhou H, Liu Z. Prediction of copper transport protein 1 (CTR1) genotype on severe cisplatin induced toxicity in non-small cell lung cancer (NSCLC) patients. Lung Cancer. 2012;77:438–42.
Li T, Peng J, Zeng F, Zhang K, Liu J, Li X, Ouyang Q, Wang G, Wang L, Liu Z, Liu Y. Association between polymorphisms in CTR1, CTR2, ATP7A, and ATP7B and platinum resistance in epithelial ovarian cancer. Int J Clin Pharmacol Ther. 2017;55:774–80.
Li XP, Yin JY, Wang Y, He H, Li X, Gong WJ, Chen J, Qian CY, Zheng Y, Li F, Yin T, Gong ZC, Zhou BT, Zhang Y, Xiao L, Zhou HH, Liu ZQ. The ATP7B genetic polymorphisms predict clinical outcome to platinum-based chemotherapy in lung cancer patients. Tumour Biol. 2014;35:8259–65.
Japan Esophageal Society. Japanese classification of esophageal cancer, 11th edition: part I. Esophagus. 2017;14:1–36.
Japan Esophageal Society. Japanese classification of esophageal cancer, 11th edition: part II and III. Esophagus. 2017;14:37–65.
Rabik CA, Maryon EB, Kasza K, Shafer JT, Bartnik CM, Dolan ME. Role of copper transporters in resistance to platinating agents. Cancer Chemother Pharmacol. 2009;64:133–42.
Holzer AK, Samimi G, Katano K, Naerdemann W, Lin X, Safaei R, Howell SB. The copper influx transporter human copper transport protein 1 regulates the uptake of cisplatin in human ovarian carcinoma cells. Mol Pharmacol. 2004;66:817–23.
Yu L, Chen MH, Gu CP, Li YL, Wen J, Fu JH, Cho CH, Liu SW. Cisplatin induces drug resistance in human esophageal squamous carcinoma cell line EC109 by decreasing CTR1 protein expression. Nan Fang Yi Ke Da Xue Xue Bao. 2011;31:801–4.
Watanabe M, Baba Y, Yoshida N, Ishimoto T, Nagai Y, Iwatsuki M, Iwagami S, Baba H. Outcomes of preoperative chemotherapy with docetaxel, cisplatin, and 5-fluorouracil followed by esophagectomy in patients with resectable node-positive esophageal cancer. Ann Surg Oncol. 2014;21:2838–44.
Ui T, Fujii H, Hosoya Y, Nagase M, Mieno MN, Mori M, Zuiki T, Saito S, Kurashina K, Haruta H, Matsumoto S, Niki T, Lefor A, Yasuda Y. Comparison of preoperative chemotherapy using docetaxel, cisplatin and fluorouracil with cisplatin and fluorouracil in patients with advanced carcinoma of the thoracic esophagus. Dis Esophagus. 2015;28:180–7.
Hara H, Tahara M, Daiko H, Kato K, Igaki H, Kadowaki S, Tanaka Y, Hamamoto Y, Matsushita H, Nagase M, Hosoya M. Phase II feasibility study of preoperative chemotherapy with docetaxel, cisplatin, and fluorouracil for esophageal squamous cell carcinoma. Cancer Sci. 2013;104:1455–60.
Acknowledgements
This work was supported by a Grant (No. 20H01096) from the Japan Society for the Promotion of Science, Tokyo, Japan.
Disclosure
All authors report that they have no relevant relationships to disclose.
Author information
Authors and Affiliations
Contributions
KF, SM, YS, AW, YN, YM, and MM participated in the design of the study and reviewed the results. SM, YS, AW, YN, and YM were responsible for the patient collection and involved in data acquisition. KF performed the genotyping. KF, SM, and MM were responsible for the statistical analyses. YS, AW, YN, and YM drafted the manuscript. AS, KS, TN, and KI helped draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No author has any conflict of interest to declare.
Research involving human participants
The study was performed in accordance with the ethical standards of the Declaration of Helsinki and its subsequent amendments.
Informed consent
Signed informed consent was obtained from all patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fujita, K., Motoyama, S., Sato, Y. et al. Effects of SLC31A1 and ATP7B polymorphisms on platinum resistance in patients with esophageal squamous cell carcinoma receiving neoadjuvant chemoradiotherapy. Med Oncol 38, 6 (2021). https://doi.org/10.1007/s12032-020-01450-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-020-01450-1