Abstract
Pyrroline-5-carboxylate reductase 1 (PYCR1) is an enzyme involved in cell metabolism, which has been shown to be up-regulated in cancers. However, the functions of PYCR1 in prostate cancers (PCa) are still largely unknown. In the present study, we found that PYCR1 was highly expressed in prostate cancer tissues and then knocked down PYCR1 in PCa cell lines (DU145, PC-3 and LNCap) via lentivirus-mediated gene delivery and analyzed its biological function. Both qRT-PCR and western blotting indicated that PYCR1 was suppressed efficiently after sh-PYCR1 infection. Further analysis indicated knockdown of PYCR1 significantly inhibited PCa cell growth and colony formation ability. The inhibition effects on growth were likely due to G2/M-phase arrest and enhanced cell apoptosis, as determined by flow cytometer analysis. At last, we verified the expression levels of cell cycle regulatory proteins, including CDK1, CDK2, CDK4 and Cyclin B1 were all downregulated and cell apoptotic-related proteins, including cleaved caspase 3 and cleaved PARP were increased in PCa cells after PYCR1 knockdown. Furthermore, PYCR1 has been shown not to be directly regulated by androgen receptor (AR) levels. These results show the functions of PYCR1 in PCa tumorigenesis for the first time and suggest that PYCR1 might be a good potential therapy approach for treating PCa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) is the most common non-cutaneous male cancer as well as the second leading cause of cancer-related death in USA and Europe [1, 2]. And most notably, its incidence and mortality are rapidly increasing in Asia-Pacific region [3]. Currently, several therapy methods have been applied to treat PCa, such as androgen deprivation therapy [4] and docetaxel chemotherapy [5]. However, the survival rate of PCa patients is still low. Therefore, there is an urgent need to identify novel molecular marker to improve the prognosis of PCa patients.
Pyrroline-5-carboxylate reductase 1 (PYCR1) plays a critical role in proline biosynthesis and catalyzes the NAD(P)H-dependent conversion of pyrroline-5-carboxylate (P5C) to proline. Recent studies show that PYCR1 is able to converse Delta (1)-piperideine-6-carboxylate (P6C) into pipecolic acid as well [6]. Besides, PYCR1 is a house-keeping gene which is conserved from yeast to animals. Studies found that mutations of human PYCR1 caused cutis laxa, which is a multisystem disorder characterized by the appearance of premature aging, wrinkled and lax skin, joint laxity and a general developmental delay [7–12]. In addition, PYCR1 is also related with matrix metalloproteinases (MMPs) [9].
Metabolic remodeling is now widely regarded as a hallmark of cancer. The expression of PYCR1 is up-regulated by fourfold in PC at issues compared with normal tissues [13]. Moreover, PYCR1 has been reported to be associated with androgen receptor (AR) signaling [14], which might play a role in PCa progression. However, the detailed functions of PYCR1 and its relation with AR remain largely unclear in PCa.
In this study, we found that PYCR1 was accumulated in human PCa tissues and then knocked down the PYCR1 in PCa cell lines via recombinant lentivirus taking shRNA to analyze its effects on cell proliferation and colony formation of the PCa cells. Furthermore, we tested the cell cycle and apoptosis of PCa cells after RNAi of PYCR1.
Materials and methods
Patient tissues and immunohistochemistry (IHC)
Tumor tissues and its adjacent normal tissues from 142 consecutive patients were prepared for immunohistochemistry assay (IHC) as previously described. PYCR1 protein was determined using rabbit anti-PYCR1 (1:5000, Abgent, Cat No. AP9893b). All specimens were collected from Fuzhou General Hospital of Nanjing Military Command in Fuzhou, China. The use of human tumor tissue was approved by the ethics committee of the Fuzhou General Hospital, Fujian Medical University. Intensity of immunoreactivity was scored visually and defined as negative (−), slightly positive (−+), moderately positive (+) and strongly positive (++), respectively.
Cell lines and culture conditions
Human PCa cell lines, PC-3, DU145 and LNCap cells and human embryonic kidney 293T cells (HEK293T) were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). PC-3 and DU145 cells were cultured in Ham’s/f-12, supplemented with 10% fetal bovine serum (FBS). LNCap cells were cultured in RPMI 1640 medium (Gibco, USA). WPMY-1 and HEK293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS. All cell lines were maintained at 37 °C in a humidified atmosphere containing 5% CO2.
To explore the direct link between PYCR1 and AR signaling pathway in PCa cells, the LNCap cells were treated with 10 μM AR inhibitor (Bicalutamide) or AR activator (DHT) for 24 h at 30% cell confluence and then collected for the next detection.
RNA interference of PYCR1 by lentivirus
To knock down the endogenous PYCR1 in PCa cells, we selected two siRNA hairpin sequences against different sites of PYCR1 mRNA, shPYCR1(S1) 5′-CACAGTTTCTGCTCTCAGGAACTCGAGTTCCTGAGAGCAGAAACTGTGTTTTT-3′ and shPYCR1(S2)5′-CCCTTCATCCTGGATGAAATACTCGAGTATTTCATCCAGGATGAAGGGTTTTT-3′. A non-target shRNA, shCon was used as negative control, 5′-GATCCTTCTCCGAACGTGTCACGTCTCGAGACGACGCACTGGCGGAGAATTTTTG-3′. These shRNA sequences were cloned to pFH-L vector (Shanghai Hollybio, China) via restriction enzyme cutting sites NheI and PacI. Then the vectors taking shRNA were transfected into HEK293T cells along with pHelper plasmids pVSVG-I and pCMVΔR8.92 (Shanghai Hollybio, China). The supernatant was collected, and recombinant lentivirus was collected after 96 h through centrifuge for further experiment.
Recombinant lentiviruses shCon, shPYCR1(S1) and shPYCR1(S2) were then used to transfect PCa cells. Briefly, PCa cells were cultured in six-well plate at a density of 50,000 cells/well and infected by recombinant lentivirus with multiplicity of infection (MOI) at 80 and 90, respectively. After 96 h, the plates were observed under microscope to check the morphology and infection efficiency of lentivirus according to GFP signal.
qRT-PCR
Du145 and PC-3 cells were infected with recombinant lentivirus under conditions above mentioned. DU145 cells were collected after 7 days, and PC-3 cells were collected after 5 days. In addition, DU145 cells treated with AR inhibitor were collected for detecting the mRNA levels of PYCR1. RNA was extracted from cells, and cDNA was synthesized using PrimeScript RT reagent kit with cDNA Eraser for RT-PCR with oligodT. Primers of PYCR1 were designed according to its sequence in NCBI (NM_001282279.1). The sequences were listed as below. Forward: 5′-GAAGATGGGGGTGAAGTTGA-3′; Reverse: 5′-CTCAATGGAGCTGATGGTGA-3′. Actin was used as control using primers 5′-GTGGACATCCGCAAAGAC-3′ (Forward) and 5′-AAAGGGTGTAACGCAACTA-3′ (Reverse). PCR reactions were performed on BioRad Connet Real-Time PCR platform with the reaction system of 10 µL 2× SYBR premix ex taq, 0.8 µL 2.5 µM forward and reverse primers, 5 µL cDNA and 4.2 µL ddH2O. The procedure was set as below: initial denaturation at 95 °C for 1 min, 40 cycles of denaturation at 95 °C for 5 s followed by annealing extension at 60 for 20 s. Absorbance values were obtained at the end of every extension step. The statistics method was 2−ΔΔCt.
Western blot analysis
PCa cells were collected and incubated with RIPA buffer. The cell lysis was centrifuged at 15,000 g at 4 °C for 10 min. The supernatant was boiled after adding 2× SDS sample buffer (100 mM Tris–Hcl (pH 6.8), 10 mM EDTA, 4% SDS, 10% Glycine). Proteins were separated by 10% SDS-PAGE gels and electro-transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After blocking with 5% milk, the membranes were probed overnight with primary antibodies, rabbit anti-PYCR1 (1:5000, Abgent, AP9893b), anti-CDK1 (1:1000, Proteintech, 19532-1-AP), anti-CDK2 (1:1000, Cell signaling, #2546), anti-CDK4 (1:1000, Proteintech, 11026-1-AP), anti-Cyclin B1 (1:2000, SAB, #21540), anti-cleaved caspase 3 (1:5000, Cell signaling, #9661), anti-cleaved PARP (1:5000, Cell signaling, #9542) and rabbit anti-GAPDH (1:100,000, Proteintech, 10494-1-AP). The membrane was washed and then incubated with HRP-conjugated secondary goat anti-rabbit antibodies (1:5,000, Santa Cruz, SC-2054) for 1 h. Target proteins were detected with enhanced chemiluminescence reagents (Amersham) and photographed.
Cell proliferation assay
Briefly, PCa cells (2000 cells/well) were cultured in 96-well plate and infected by recombinant lentivirus. After 96 h, the plates were treated with 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) plus acidic isopropanol (10% SDS, 5% isopropanol and 0.01 mol/L HCl). After incubation, the absorbance was read at a wavelength of 595 nm according to the manufacturer’s protocol.
Colony formation assay
In brief, at 96 h after lentivirus infection, DU145 and PC-3 cells were seeded into 6-well plate with seeding density of 500 cells/well. After culture for 8 days (DU145) and 9 days (PC-3), the cells were stained by crystal purple. The morphology of colonies was observed under microscope. The numbers of colonies were counted.
Cell cycle assay
After lentiviral infection for 5 days, DU145 and PC-3 cells were seeded into 6-cm dish at initial density of 200, 000 cells/dish. The dishes were maintained at 37 °C for 40 h until the confluence of cells reached about 80%. Then DU145 cells were washed and fixed with 70% ethanol overnight at 4 °C. After digestion with RNaseA, the cells were labeled with propidium iodide and went through Cell Lab Quanta Beckman Coulter to measure fluorescence.
Apoptosis detection assay
Apoptotic cells were quantified according to the protocol of the Annexin V-APC/7-AAD Detection kit (Keygen Biotechnology). Briefly, DU145 cells seeded in 6-well culture plates were infected with lentivirus-mediated PYCR1 shRNA for 7 days. After infection, attached cells were trypsinized, washed with PBS and centrifuged. The cells were washed with 1× binding buffer, centrifuged and resuspended in 1 mL 1× staining buffer. Then 5 μL Annexin V-APC was added into 100 μL of cell suspension, followed by a gentle vortex, and 10 min incubation at room temperature in the dark. Data acquisition and analysis were performed by the Becton–Dickinson FACSCalibur flow cytometer plus dedicated software. The experiment was repeated three times.
Statistical analysis
The results were representative of at least three independent experiments and were expressed as the mean ± standard deviation (SD). Associations between PYCR1 expression and clinicopathological variables were analyzed by Pearson Chi-Square test. Student’s t test was used to evaluate the differences between the PYCR1 silenced and non-silenced groups. Statistically significant differences were defined as p < 0.05.
Results
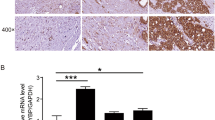
PYCR1 expression in PCa and corresponding normal tissues
To investigate the expression of PYCR1 in PCa, immunohistochemistry was used to evaluate its expression in PCa tissues. As shown in Fig. 1, representative photographs of tumor and adjacent tissues showed that PYCR1 was highly expressed in cancer. We found there was a significant difference between PYCR1 expression and some clinicopathological parameters, such as Gleason score (Table 1, p = 0.0381).
PYCR1 expression is efficiently suppressed in PCa cells
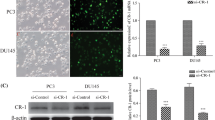
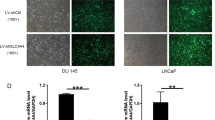
To investigate the role of PYCR1 in PCa, we firstly generated recombinant lentivirus to infect DU145 and PC-3 cells. As shown in Fig. 2a, most of the infected DU145 and PC-3 cells were GFP-positive, suggesting that recombinant lentivirus could deliver shRNAs into PCa cell lines with high efficiency. Then we determined the mRNA and protein levels of PYCR1 in recombinant lentivirus-infected PCa cells using qRT-PCR and western blotting, respectively. The transcription of PYCR1 was suppressed dramatically by shPYCR1(S1) or shPYCR1(S2) compared with shCon in DU145 and PC-3 cells (Fig. 2b, p < 0.01, p < 0.001). No PYCR1 band was detected in the protein extract of shPYCR1-infected DU145 or PC-3 cells (Fig. 2c). Notably, shPYCR1(S1) could more efficiently suppress the expression of PYCR1 than shPYCR1(S2). Furthermore, LNCap cells were transfected with shPYCR1(S1) to determine PYCR1 function as well as its relation with AR level. As shown in Figure S1A and B, shPYCR1(S1) obviously suppressed the expression of PYCR1 protein in LNCap cells.
PYCR1 expression is suppressed efficiently in PCa cells using RNAi mediated by recombinant lentivirus. a The gene delivery efficiency of recombinant lentivirus in DU145 and PC-3 cells. Upper panels, bright field; lower panels, GFP fluorescence (green). Scale bar, 10 μm. b qRT-PCR analysis of PYCR1 mRNA levels in recombinant lentivirus-infected DU145 cells and PC-3 cells. Data are expressed as mean ± standard deviation (SD) of three independent experiments. **p < 0.01***; p < 0.001. c Western blotting analysis of PYCR1 protein levels in recombinant lentivirus-infected DU145 cells and PC-3 cells. GAPDH was used as control protein
Suppression of PYCR1 inhibits cell proliferation and colony formation of PCa cells
To investigate the functions of PYCR1, we analyzed the cell proliferation of PCa cells following shPYCR1(S1) or shPYCR1(S2) infection using MTT assay. As shown in Fig. 3, we found there was an obvious growth inhibition effect of shPYCR1(S1) or shPYCR1 (S2) on DU145 cells (p < 0.001). Similarly, shPYCR1(S1) or shPYCR1 (S2) significantly suppressed cell growth in PC-3 cells (p < 0.05, p < 0.01 and p < 0.001). Obviously, shPYCR1(S1) presented more inhibitory effects than shPYCR1(S2) in cell proliferation. Additionally, we evaluated the effect of PYCR1 knockdown on the cell growth of LNCap cells and found shPYCR1(S1) could suppressed cell growth as well (Figure S1-C, p < 0.001).
What’s more, we tested the colony formation ability of DU145 cells after PYCR1 knockdown. As shown in Fig. 4a, representative photographs of PYCR1 knockdown DU145 cells had much smaller colonies, indicating weaker colony formation ability. Further analysis showed the colony number was significantly reduced in shPYCR1(S1) group, compared with shCon (Fig. 4b, p < 0.001).
Suppression of PYCR1 leads to a decrease in PCa cell colony formation. a Representative images of colonies formed by shCon- and shPYCR1(S1)-infected DU145 cells. Upper panel, crystal violet staining; middle panel, bright field; lower panel, GFP fluorescence field. Scale bar, 25 μm. b Colony numbers of shCon- and shPYCR1(S1)-infected DU145 cells. Data are expressed as mean ± standard deviation (SD) of three independent experiments. ***p < 0.001
Suppression of PYCR1 induces cell cycle arrest at G2/M phase
To investigate whether growth inhibition was related with cell cycle arrest, we performed flow cytometry on shCon- and shPYCR1(S1)-infected DU145 and PC-3 cells. Representative images of cell cycle distribution in DU145 are shown in Fig. 5a. As expected, knockdown of PYCR1 arrested cell cycle in G2/M phase in and accordingly decreased the cell numbers in G0/G1 phase and S phase DU145 cells (Fig. 5b, p < 0.05, p < 0.001) and PC-3 cells (Fig. 5d, p < 0.05, p < 0.01). Additionally, in the absence of PYCR1, more cells were obviously accumulated in the sub-G1 phase representing apoptotic cells in both of these two cell lines (Fig. 5c, e, p < 0.05, p < 0.001). Moreover, knockdown of PYCR1 apparently downregulated the protein expression of CDK1, CDK2, CDK4 and Cyclin B1 in both DU145 (Fig. 5f) and PC-3 cell lines (Fig. 5g).
Suppression of PYCR1 induces cell cycle arrest in PCa cells. a Representative graphs of flow cytometry assay of cell cycle of shCon- and shPYCR1(S1)-infected DU145 cells. Calculation of cells at G0/G1, S and G2/M phase in DU145 (b) and PC-3 (d) cells following shPYCR1(S1) infection. Calculation of cells at sub-G1 phase in DU145 (c) and PC-3 (e) cells following shPYCR1(S1) infection. Data are expressed as mean ± standard deviation (SD) of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. The expression of CDK1, CDK2, CDK4 and Cyclin B1 protein was measured in DU145 (f) and PC-3 (g) cells following shPYCR1(S1) infection. GAPDH was used as an internal control
Suppression of PYCR1 promotes cell apoptosis in PCa cells
Annexin V-APC/7-AAD detection was used to determine whether silencing PYCR1 had an accelerated effect on the apoptosis of DU145 cells (Fig. 6a). The results showed there were more apoptosis cells, including early-stage and late-stage apoptosis, in the shPYCR1(S1)-infected DU145 cells (Fig. 6b, p < 0.001).To further explore the molecular mechanisms underlying PYCR1-mediated PCa cell apoptosis, we detected the protein levels of pro-apoptotic markers, including cleaved caspase-3 and cleaved PARP and found both of them were increased in the shPYCR1(S1)-infected DU145 cells (Fig. 6c).
Suppression of PYCR1 promoted cell apoptosis in PCa cells. a Apoptotic cells were characterized by Annexin V-APC/7-AAD double staining and analyzed by flow cytometry. b Deficiency of PYCR1 in DU145 cells could trigger early (Annexin V-APC+/7-AAD−) and late apoptosis (Annexin V-APC+/7-AAD+). Data are expressed as mean ± standard deviation (SD) of three independent experiments. ***p < 0.001. c Western blotting analysis of cleaved caspase-3 and cleaved PARP proteins levels in shCon- and shPYCR1(S1)-infected DU145 cells
The expression of PYCR1 is not directly regulated by AR signaling pathway in PCa cells
To explore the link between PYCR1 and AR signaling pathway in PCa cells, we treated LNCap cells with AR inhibitor (Bicalutamide) or AR activator (DHT) and detected the expression of PYCR1 by Western blot analysis. The results showed that the expression of PSA was significantly decreased or increased after treated with Bicalutamide or DHT, respectively, compared with the control (Figure S1-D). However, there was no significant decrease in the expression of PYCR1 after AR inhibitor treatment, which suggests PYCR1 might not have a direct relation with AR signaling pathway.
Discussion
We reported here that PYCR1 was accumulated in human PCa tissues and significantly correlated with Gleason score, suggesting the expression of PYCR1 was increased with the tumor progression. To investigate the role of PYCR1 in the PCa progression, we knocked down PYCR1 in PCa cells via lentivirus-mediated gene delivery and analyzed its effects on tumorigenesis. The results showed that the suppression of PYCR1 led to cell proliferation inhibition, colony formation inhibition, cell cycle arrest and cell apoptosis in PCa cells.
PYCR1 has been shown to possess anti-apoptotic functions. Moreover, not only P5C, the substrate of PYCR1, plays a role in the pro-apoptotic and growth inhibition [15], but also its product proline has anti-apoptotic and anti-oxidant activities [16]. Consistent with those results, we also demonstrated that knockdown of PYCR1 led to inhibition of cell proliferation and increased apoptosis in DU145 cell. Yasuda et al. [17] showed that PYCR1 directly bounded with DJ-1 both in vivo and in vitro. PYCR1 is also localized in mitochondria together with DJ-1. The knockdown of PYCR1 resulted in decreased mitochondria membrane potential and lower viability under oxidative stress conditions.
Previous studies found that PYCR1 was a target of oncogene Myc [18], which induces proline biosynthesis from glutamine, in which PYCR1 catalyzes the final step of proline synthesis from glutamate in cancer cells. The reduction of Myc by siRNA resulted in the decrease in PYCR1 along with the increase in other proline enzymes, such as POX/PRODH, P5CDH and GS. The studies of the metabolism profiles about PCa cells are still underway in PYCR1 knockdown PCa cells now.
Androgen ablation therapy has been the standard treatment of PCa for many years [4]. The androgen receptor (AR) plays a pivotal role in PCa progression through regulating cell proliferation, differentiation and apoptosis [19–21]. Jariwala et al. [14] found that PYCR1 was less strongly or hardly stimulated in LNCap cells by AR by DHT, but it was identified as one of AR target genes. To further confirmed the relation between PYCR1 and AR signaling, we treated LNCap cells with DHT or Bicalutamide and determined the expression of PYCR1. Our results showed the expression of PYCR1 is not obviously altered under changed PSA levels in LNCap cells, which might be regulated other signaling pathways in PCa progression.
In conclusion, we have proved that PYCR1 plays an important role in PCa tumorigenesis for the first time. The suppression of PYCR1 by lentivirus could induce cell apoptosis and cell cycle arrest indicating that PYCR1 might be a good target gene for human PCa therapy.
References
Cancer statistics. JAMA. 2013;310:982. doi:10.1001/jama.2013.5289.
Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403.
Baade PD, Youlden DR, Cramb SM, Dunn J, Gardiner RA. Epidemiology of prostate cancer in the Asia-Pacific region. Prostate Int. 2013;1:47–58.
Scher HI, Buchanan G, Gerald W, Butler LM, Tilley WD. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocr Relat Cancer. 2004;11:459–76.
Gomella LG, Petrylak DP, Shayegan B. Current management of advanced and castration resistant prostate cancer. Can J Urol. 2014;21:1–6.
Struys EA, Jansen EE, Salomons GS. Human pyrroline-5-carboxylate reductase (PYCR1) acts on Delta(1)-piperideine-6-carboxylate generating L-pipecolic acid. J Inherit Metab Dis. 2014;37:327–32.
Guernsey DL, Jiang H, Evans SC, Ferguson M, Matsuoka M, Nightingale M, et al. Mutation in pyrroline-5-carboxylate reductase 1 gene in families with cutis laxa type 2. Am J Hum Genet. 2009;85:120–9.
Reversade B, Escande-Beillard N, Dimopoulou A, Fischer B, Chng SC, Li Y, et al. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat Genet. 2009;41:1016–21.
Kretz R, Bozorgmehr B, Kariminejad MH, Rohrbach M, Hausser I, Baumer A, et al. Defect in proline synthesis: pyrroline-5-carboxylate reductase 1 deficiency leads to a complex clinical phenotype with collagen and elastin abnormalities. J Inherit Metab Dis. 2011;34:731–9.
Lin DS, Yeung CY, Liu HL, Ho CS, Shu CH, Chuang CK, et al. A novel mutation in PYCR1 causes an autosomal recessive cutis laxa with premature aging features in a family. Am J Med Genet Part A. 2011;155A:1285–9.
Dimopoulou A, Fischer B, Gardeitchik T, Schroter P, Kayserili H, Schlack C, et al. Genotype-phenotype spectrum of PYCR1-related autosomal recessive cutis laxa. Mol Genet Metab. 2013;110:352–61.
Scherrer DZ, Baptista MB, Matos AH, Maurer-Morelli CV, Steiner CE. Mutations in PYCR1 gene in three families with autosomal recessive cutis laxa, type 2. Eur J Med Genet. 2013;56:336–9.
Ernst T, Hergenhahn M, Kenzelmann M, Cohen CD, Bonrouhi M, Weninger A, et al. Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: a gene expression analysis on total and microdissected prostate tissue. Am J Pathol. 2002;160:2169–80.
Jariwala U, Prescott J, Jia L, Barski A, Pregizer S, Cogan JP, et al. Identification of novel androgen receptor target genes in prostate cancer. Mol Cancer. 2007;6:39.
Maxwell SA, Davis GE. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA. 2000;97:13009–14.
Krishnan N, Dickman MB, Becker DF. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med. 2008;44:671–81.
Yasuda T, Kaji Y, Agatsuma T, Niki T, Arisawa M, Shuto S, et al. DJ-1 cooperates with PYCR1 in cell protection against oxidative stress. Biochem Biophys Res Commun. 2013;436:289–94.
Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA. 2012;109:8983–8.
Marker PC, Donjacour AA, Dahiya R, Cunha GR. Hormonal, cellular, and molecular control of prostatic development. Dev Biol. 2003;253:165–74.
Geck P, Szelei J, Jimenez J, Lin TM, Sonnenschein C, Soto AM. Expression of novel genes linked to the androgen-induced, proliferative shutoff in prostate cancer cells. J Steroid Biochem Mol Biol. 1997;63:211–8.
Bonkhoff H, Remberger K. Differentiation pathways and histogenetic aspects of normal and abnormal prostatic growth: a stem cell model. Prostate. 1996;28:98–106.
Acknowledgements
The authors are thankful for the financial support from the National Natural Science Foundation of China (81272247 and 81372751).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Conflict of interest relevant to this article was not reported.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional of Fuzhou General Hospital Affiliated to Fujian Medical University.
Additional information
Tengyue Zeng, Libing Zhu and Min Liao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12032_2016_870_MOESM1_ESM.tif
Figure S1. (A) The gene delivery efficiency of shPYCR1(S1) in LNCap cells. Upper panels, bright field; lower panels, GFP fluorescence (green). Scale bar, 10 μm. (B) Western blotting analysis of PYCR1 protein levels in shPYCR1(S1)-infected LNCap cells. (C) The proliferation levels of LNCap cells after shPYCR1(S1) infection analyzed by the MTT assay. Data are expressed as mean ± standard deviation (SD) of three independent experiments. ***p < 0.001. (D) Western blot analysis of PYCR1 and PSA protein levels in Con, AR inhibitor (Bicalutamide) or AR activator (DHT) treated LNCap cells. (TIFF 1329 kb)
Rights and permissions
About this article
Cite this article
Zeng, T., Zhu, L., Liao, M. et al. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol 34, 27 (2017). https://doi.org/10.1007/s12032-016-0870-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-016-0870-5