Abstract
MicroRNAs (miRNAs) are a type of small noncoding RNAs that are strongly implicated in carcinogenesis. However, the potential diagnostic, prognostic and therapeutic roles of the majority of miRNAs in the pathological processes of tumorigenesis remain largely unknown. Our and others’ data revealed that miR-204-5p was significantly downregulated in gastrointestinal tumor tissues compared with adjacent noncancerous tissues. The downregulation of miR-204-5p was confirmed in our gastric cancer (GC) cohort, and we showed that ectopic expression of miR-204-5p inhibited, whereas silencing miR-204-5p expression promoted GC cell proliferation in vitro. Subsequent mechanistic investigations identified that USP47 and RAB22A are direct functional targets of miR-204-5p in GC. Silencing the expression of USP47 and RAB22A using siRNA phenocopied the proliferation-inhibiting function of miR-204-5p in GC cells. Our results uncovered that miR-204-5p acts as a tumor suppressor in GC through inhibiting USP47 and RAB22A.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a type of noncoding small RNAs approximately 19–24 bp in length [1], and they negatively regulate a wide variety of genes expression mainly through direct interaction with the 3′untranslated regions (3′UTR) of their corresponding mRNA targets [2]. miRNAs play critical roles in many physiological and pathological processes at the posttranscriptional level, including the development and progression of nearly all types of tumors [3]. Although the studies of miRNAs provide new insights into carcinogenesis, the molecular mechanisms underlying remain to be elucidated.
Gastric cancer (GC) is the second cause of cancer-associated death and the fourth most widespread tumor worldwide [4, 5]. The development of GC involves various factors, including oncogenes and tumor suppressors. It has been demonstrated that aberrant miRNAs expression is associated with GC [6–8], and some miRNAs have been shown to be involved in stomach tumorigenesis by targeting tumor-associated genes [9–12]. In addition, miRNAs appear to be promising tumor biomarkers [13–15]. Our and others’ expression profiling data uncovered that miR-204-5p (previously named as miR-204 before miRBase release 19.0) was one of the most significantly downregulated miRNAs in gastrointestinal tumor tissues compared with adjacent noncancerous tissues (NCTs) [9, 11, 16]. Several studies have also shown that miR-204-5p is frequently downregulated in other cancers [16–18], suggesting a common role of miR-204-5p in human tumorigenesis. Recent studies suggest that miR-204-5p may inhibit GC cells invasion through restraining the activity of the SIRT-LKB1 pathway [11] and promote GC cells apoptosis by targeting Bcl-2 [9]. However, the exact mechanism and clinical value remain largely unknown.
In this study, we confirmed that the miR-204-5p expression is significantly decreased in GC tissues and revealed that miR-204-5p could inhibit cell proliferation through directly targeting USP47 and RAB22A in GC.
Materials and methods
Cell lines and clinical samples
Human GC cell lines, including HGC-27, MGC-803, NCI-N87, SGC-7901 and S206 were purchased from the American Type Culture Collection (ATCC). The cells were cultured in high-glucose (4.5 g/L) DMEM (Hyclone, USA) supplemented with 10 % fetal bovine serum (FBS) (Gibco), penicillin and streptomycin in a 5 % CO2 water-saturated atmosphere.
A total of 102 paired GC tissues and NCTs were collected from the Fourth Affiliated Hospital of Soochow University. The patients’ information is listed in Supplementary Table S1. All of the samples were gathered according to the Institutional Review Board approved protocol and the written informed consent from each patient.
Total RNA extraction and real-time qRT-PCR
Total RNA was extracted using TRIzol reagent (TaKaRa, Japan). A NanoDrop 2000 (Thermo, USA) was used to determine the concentrations of RNA.
Complementary cDNA was generated using the PrimeScript RT reagent kit (TaKaRa, Japan). SYBR Premix Ex Taq (TaKaRa, Japan) was used to detect the relative mRNA expression via qRT-PCR analyses, with β-actin as an internal control. TaqMan miRNA probes (Applied Biosystems, USA) were used to detect the levels of the mature miRNAs through tem-loop qRT-PCR assays. The levels of U6B (a ubiquitously expressed small nuclear RNA) was used to normalize the relative levels of miRNAs. All the primer sequences could be obtained from Supplementary Table S2.
Plasmid and siRNA
miR-204-5p mimic, miR-204-5p inhibitor (anti-miR-204-5p), negative control were purchased from RiboBio (Guangzhou, China). Duplex siRNAs were purchased from GenePharma (Shanghai, China) (si-USP47: 5′-GCUGUCGCCUUGUUAAAUAT TUAUUUAACAAGGCGACAGCTT-3′; si-RAB22A: 5′-GCAUACAGGUGUAGG UAAATTUUUACCUACACCUGUAUCCTT-3′). The amplified 3′UTRs were then cloned into the region directly downstream of a CMV promoter—driven firefly luciferase cassette in a pcDNA3.0 vector (p-Luc). The mutant 3′UTR of USP47 and RAB22A, which carried the mutated sequence in the complementary site for the seed region of miR-204-5p, was constructed on the basis of the p-Luc-USP47 and p-Luc-RAB22A UTR-WT plasmid by overlap-extension PCR [16]. Plasmid, mimic and siRNA were transfection into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Cell proliferation assay
Cells (2000) were plated in 96-well plates, detected with Cell Counting Kit-8 (CCK-8, Dojindo) at the indicated time points according to the manufacturer’s instruction.
Colony formation assay
Cells (1000) were placed in 6-well plates and cultured in media containing 10 % FBS for 10 days. The colonies were stained with 1.0 % crystal violet in 20 % methanol for 30 min.
Luciferase assay
S206 cells were seeded in 96-well plates. A total of 50 nM of miR-204-5p mimic (or NC), 10 ng of pRL-CMV Renilla luciferase reporter and 50 ng of luciferase reporter were contransfected into the cells using Lipofectamine 2000 (Invitrogen). Using a luciferase assay kit (Promega, USA) assayed the luciferase activities, 48 h after transfection.
Western blotting
The protein levels of USP47 and RAB22A were analyzed by Western blots using anti-RAB22A, anti-β-actin (Abcam, USA) and anti-USP47 (Santa Cruz, Daly City, CA).
Immunohistochemistry (IHC)
Immunohistochemical staining was executed on 4-μm sections of paraffin-embedded GC tissue samples to test the protein expression levels of RAB22A. In brief, the slides were hatched with RAB22A antibody at a dilution of 1:100 at 4 °C overnight.
Statistical analyses
All experiments were performed in triplicate, and all results are presented as the mean values ± SEM. Differences/correlations between groups were calculated using Student’s t test unless otherwise specified (χ 2 test, Pearson’s correlation). A P value of <0.05 was considered statistically significant. SPSS 16.0 package (IBM, USA) and GraphPad Prism 5.0 software (GraphPad Software, USA) were used for statistical analyses and scientific graphing, respectively.
Results
miR-204-5p was downregulated in human GC tissues
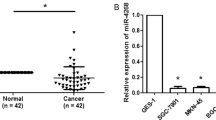
miR-204 emerged as one of the most prominently downregulated miRNAs in GC in previous studies [9, 11]. We examined the miR-204-5p levels in an expanded GC tissues (n = 63) using qRT-PCR and found that miR-204-5p expression was obviously downregulated in human GC tissues compared with their corresponding NCTs (P = 0.0010, Fig. 1a, b). The miR-204-5p expression was negatively related with the tumor TNM stage, and higher miR-204-5p expression was observed in tumor with earlier TNM stage (P = 0.0054, Fig. 1c).
Expression of miR-204-5p was usually downregulated in GC. a, b The expression of miR-204-5p was tested by qRT-PCR in 63 paired GC and adjacent noncancerous tissues (NCTs). The level of miR-204-5p was obviously downregulated in tumor tissues contrast with the corresponding NCTs. c The relation of miR-204-5p expression and TNM stage. The levels of miR-204-5p were divided into two groups(TNM stage: I–II; III–IV)
miR-204-5p inhibits GC cell proliferation in vitro
The consistently low expression of miR-204-5p in GC suggests its potential contribution to the stomach tumorigenesis. We tested the levels of miR-204-5p in five GC cell lines (Fig. 2a) and selected S206 and SGC-7901 for the further studies. Cell proliferation assay and colony formation assay showed the proliferation-repressing function of miR-204-5p in S206 cell (Fig. 2b, c). To the contrary, silencing miR-204-5p expression in SGC-7901 cell significantly promoted the cell growth (Fig. 2b, c). Collectively, these data demonstrate that miR-204-5p acts as a tumor suppressor gene in GC cells in vitro.
miR-204-5p inhibits GC cell proliferation in vitro. a The expression of miR-204-5p in five gastric cancer cell lines. b The cell growth rate was determined by the CCK-8 assay. c The colony formation assay was used to verify the cell proliferation, again. Significant differences: *P < 0.05; **P < 0.01
Identification of USP47 and RAB22A as the targets of miR-204-5p
To explore the potential mechanism by which miR-204-5p represses the proliferation of GC cells, we searched for genes regulated by miR-204-5p. Our team had previously performed a microarray analysis of the targets of miR-204-5p in colorectal cancer cells and found that USP47 and RAB22A were observably downregulated in miR-204-5p-overexpressed cells (GSE59897) [16] and both of them were predicted to be the potential targets of miR-204-5p by TargetScan algorithm. We have identified RAB22A as the functional target of miR-204-5p in colorectal cancer [16]. To test whether USP47 and RAB22A are the direct targets of miR-204-5p in GC, the wild type and mutant of USP47 3′UTRs and RAB22A 3′UTRs were independently cloned into p-Luc (Fig. 3a). As shown in Fig. 3b, miR-204-5p could inhibit the expression of the reporter gene in recombinant plasmids containing the 3′UTRs of RAB22A and USP47, whereas the mutant USP47 3′UTR and RAB22A 3′UTR were completely refractory to the miR-204-5p-mediated luciferase reporter repression in S206 cell. In concordance with these results, USP47 and RAB22A protein expression was significantly decreased in miR-204-5p-overexpressed GC cells and enhanced in miR-204-5p-depleted cells (Fig. 3c). In conclusion, our results confirmed that USP47 and RAB22A are the targets of miR-204-5p in GC.
Identification of USP47 and RAB22A as the targets of miR-204-5p. a The putative miR-204-5p-binding sequence in the USP47 3′UTR and RAB22A 3′UTR. A mutation was generated in the site complementary to the miR-204-5p seed region of the USP47 3′UTR and RAB22A 3′UTR, as indicated. b Analyses of the luciferase activity of the luciferase reporter plasmids containing either wild-type (WT) or mutant-type (MT) USP47 3′UTR and RAB22A 3′UTRs in S206 cell. c The protein levels of USP47 and RAB22A were determined by Western blotting in S206 cell transfected with miR-204-5p mimic, and in SGC-7901 cell transfected with miR-204-5p inhibitor or the corresponding NC. Beta-actin served as an internal control
Silence the expression of USP47 and RAB22A could repress GC cell proliferation in vitro
To further clarify whether targeting of USP47 and RAB22A might mediate the inhibition of cell proliferation in miR-204-5p-overexpressing GC cells, we performed functional restoration assays using S206 and SGC-7901 cells. We used siRNA to knockdown USP47 and RAB22A expression (Fig. 4a). As for the cell proliferation assay, siRNA-mediated USP47 and RAB22A silencing could phenocopy the proliferation-repressing effect of miR-204-5p (Fig. 4b), and colony formation assay also confirmed the promoting effects of USP47 and RAB22A in GC cell growth (Fig. 4c). Overall, our data demonstrated that miR-204-5p exerts tumor suppressive effect in GC via directly targeting USP47 and RAB22A.
Silence the expression of USP47 and RAB22A could repression GC cell proliferation in vitro. a The protein levels of USP47 and RAB22A in siRNA-mediated USP47 and RAB22A silencing cells. b, c By the CCK-8 assay and the colony formation assay confirm that knockdown of RAB22A by siRNA significantly repressed GC cell proliferation. Significant differences: *P < 0.05; **P < 0.01; ***P < 0.001
Upregulation of RAB22A is inversely correlated with miR-204-5p expression in GC
To further study the relationship between miR-204-5p and RAB22A and USP47 in human GC, we measured the protein expressions in 102 paired GC and NCTs using immunohistochemistry (IHC; Fig. s1). As indicated in Fig. 5a, b, 66 of 102 (64.7 %) tumors showed increased RAB22A expression compared with paired NCTs. Furthermore, the enhanced immunoreactivity of RAB22A in GC tissues were inversely correlated with the miR-204-5p expression levels (P = 0.0062, Fig. 5c), suggesting that miR-204-5p acts as a regulator of RAB22A expression in clinical GC tumors (R = −0.263, P = 0.047). We did not get reliable staining results of USP47 in GC tissues for the poor dyeing effect of the USP47 antibody.
Upregulation of RAB22A is inversely correlated with miR-204-5p expression in GC. a Immunohistochemical staining of RAB22A in 102 tumor tissues and adjacent noncancerous tissues (NCTs). Brown cytoplasmic RAB22A staining was observed in GC cells but was nearly absent in normal mucosal epithelia. b RAB22A protein expression was frequently increased in the tumor tissues compared with the matched NCTs (64.7 %). c The expression levels of RAB22A were negatively correlated with the miR-204-5p expression levels in the GC tissues (P = 0.0062)
Discussion
GC is among the most malignant tumors, and the median survival time for GC patients is only 6–9 months [19, 20]. This is mainly attributed to the following reasons: the early diagnosis is difficult, the effect of clinical therapy is very poor and the molecular markers utilized for targeted therapy is lack. It is necessary to make a better understanding of gastric carcinogenesis and identify novel molecular targets to improve diagnosis and therapy of GC.
miRNAs have been studied most intensively in the field of oncological research, and emerging evidence suggests that altered miRNA regulation is involved in the pathogenesis of cancers, including GC. Among these miRNAs, miR-204 has been reported to function as a tumor suppressor in a variety of human cancers [9, 11, 16–18, 21–26]. Recent reports showed that miR-204-5p could inhibit GC cell invasion and promote GC cell apoptosis [9, 11]. In this study, we confirmed that miR-204-5p is downregulated in clinical GC tissues, and ectopic expression of miR-204-5p inhibited GC cell proliferation. In the subsequent mechanistic study, we demonstrated that miR-204-5p directly targets USP47 and RAB22A to inhibit cell proliferation in GC. RNAi-mediated knockdown of USP47 and RAB22A inhibit GC cells proliferation, which phenocopied the proliferation-inhibiting effect of miR-204-5p.
Ubiquitin-specific proteases (USPs) are a subfamily of cysteine proteases that catalyze the removal of ubiquitin from substrates, thus counteracting the activity of E3 ubiquitin ligases [27, 28]. Although the importance of some members of the USP family in a variety of biological processes [29], such as regulation of DNA damage checkpoint response, epigenetic regulation and protein stabilization is well established, very little is known about the biological function of the majority of the USPs. Here, we revealed that USP47 is a direct target of miR-204-5p in GC and knockdown USP47 expression could significantly decrease GC cell growth. These data suggest, for the first time, that USP47 is a potential new oncogene for GC.
RAB22A is another target gene of miR-204-5p identified in this study. It belongs to a Ras superfamily of GTPases, which are usually activated by binding GTP in the transport vesicles, and then hydrolyzed to generate GDP-bound RABs after membrane fusion [30, 31]. However, our knowledge on its role in human tumorigenesis is quite limited. Our previous research showed that RAB22A is a potential new prognostic factor for CRC and is the direct target of miR-204-5p in colorectal cancer [16]. In line with the conclusion, we found that upregulated expression of RAB22A is also frequently observed in GC tissues. Furthermore, RNAi-mediated knockdown of RAB22A inhibit GC cell proliferation. These data suggest the common oncogenic role of RAB22A in gastrointestinal tumors.
In conclusion, we determined that miR-204-5p is downregulated in GC, and miR-204-5p acts as a tumor suppressor by inhibiting GC cell proliferation via targeting USP47 and RAB22A. These data suggest that restoration of miR-204-5p may be a promising therapeutic strategy for treatment of human GC.
References
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31.
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Hou Z, Xie L, Yu L, Qian X, Liu B. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Med Oncol. 2012;29(2):886–92.
Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, et al. MiRNA-199a-3p: a potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108(2):89–92.
Chen Y, Zuo J, Liu Y, Gao H, Liu W. Inhibitory effects of miRNA-200c on chemotherapy-resistance and cell proliferation of gastric cancer SGC7901/DDP cells. Chin J Cancer. 2010;29(12):1006–11.
Liu D, Xia P, Diao D, Cheng Y, Zhang H, Yuan D, et al. MiRNA-429 suppresses the growth of gastric cancer cells in vitro. J Biomed Res. 2012;26(5):389–93.
Sacconi A, Biagioni F, Canu V, Mori F, Di Benedetto A, Lorenzon L, et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012;3:e423.
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–33.
Zhang L, Wang X, Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial-mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 2013;13:290.
Zhou X, Li L, Su J, Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS One. 2014;9(7):e101457.
Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20(30):10432–9.
Zhang R, Wang W, Li F, Zhang H, Liu J. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med Oncol. 2014;31(10):243.
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–26.
Yin Y, Zhang B, Wang W, Fei B, Quan C, Zhang J, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by down-regulating RAB22A. Clin Cancer Res. 2014. doi:10.1158/1078-0432.CCR-14-1030.
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, et al. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73(2):990–9.
Li W, Jin X, Zhang Q, Zhang G, Deng X, Ma L. Decreased expression of miR-204 is associated with poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2014;7(6):3287–92.
Kaneko S, Yoshimura T. Time trend analysis of gastric cancer incidence in Japan by histological types, 1975–1989. Br J Cancer. 2001;84(3):400–5.
Verdecchia A, Corazziari I, Gatta G, Lisi D, Faivre J, Forman D, et al. Explaining gastric cancer survival differences among European countries. Int J Cancer. 2004;109(5):737–41.
Chen Z, Sangwan V, Banerjee S, Mackenzie T, Dudeja V, Li X, et al. miR-204 mediated loss of Myeloid cell leukemia-1 results in pancreatic cancer cell death. Mol Cancer. 2013;12(1):105.
Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6(4):e1000730.
Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21(4):532–46.
Bao W, Wang HH, Tian FJ, He XY, Qiu MT, Wang JY, et al. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol Cancer. 2013;12:155.
Chung TK, Lau TS, Cheung TH, Yim SF, Lo KW, Siu NS, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130(5):1036–45.
Gong M, Ma J, Li M, Zhou M, Hock JM, Yu X. MicroRNA-204 critically regulates carcinogenesis in malignant peripheral nerve sheath tumors. Neuro Oncol. 2012;14(8):1007–17.
Parsons JL, Dianova II, Khoronenkova SV, Edelmann MJ, Kessler BM, Dianov GL. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase beta. Mol Cell. 2011;41(5):609–15.
Peschiaroli A, Skaar JR, Pagano M, Melino G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene. 2010;29(9):1384–93.
Huang Z, Wen P, Kong R, Cheng H, Zhang B, Quan C, et al. USP33 mediates Slit-Robo signaling in inhibiting colorectal cancer cell migration. Int J Cancer. 2014;. doi:10.1002/ijc.29226.
Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313(4):889–901.
Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349(6305):117–27.
Acknowledgments
This study was partially supported by grants from the National Natural 39 Science Foundation of China (Nos. 81000867, 81272299, and 81301784) and Medical Key Professionals Program of Jiangsu Province (RC2011031).
Conflict of interest
The authors report no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, B., Yin, Y., Hu, Y. et al. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol 32, 331 (2015). https://doi.org/10.1007/s12032-014-0331-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0331-y