Abstract
No study in China has focused on the relationships between germline and somatic hMLH1/hMSH2 gene mutations, hMLH1 promoter methylation, and the prognosis of colorectal cancer (CRC), especially in sporadic CRC. Therefore, we carried out this study with 433 primary sporadic CRC patients to investigate the associations between germline and somatic hMLH1/hMSH2 gene mutations, hMLH1 promoter methylation, and the overall survival (OS) of CRC; to evaluate the effect of interaction between gene mutation and methylation on the risk of CRC prognosis. As a result, the 3-, 5-, and 7-year survival of the sporadic CRC patients was 67, 57, and 50.0 %, respectively. There were no significant associations observed between germline and somatic hMLH1/hMSH2 gene mutations after adjusted (HR = 1.37, 95 % CI 0.70–2.67, p = 0.35; HR = 1.31, 95 % CI 0.69–2.47, p = 0.42, respectively). When the analyses were stratified based on tumor stage, tumor location, and chemotherapy, no significant survival advantage of hMLH1/hMSH2 gene mutation was illustrated. In addition, no significant association between germline and somatic hMLH1 promoter methylation and OS of CRC was observed (HR = 1.46, 95 % CI 0.57–3.74, p = 0.43; HR = 0.70, 95 % CI 0.32–1.53, p = 0.37, respectively). In conclusion, the research did not find the significant association between germline and somatic hMLH1/hMSH2 gene mutations, hMLH1 promoter methylation, and sporadic CRC prognosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most common malignancies, representing the third most common cancer in men and the second in women worldwide [1]. Accounting for 8.5 % (6,94,000) of the total cancer deaths, CRC is the fourth leading cause of cancer death in 2012. [1]. Although the relative 5-year survival of CRC increased in Europe during 1995–2007 [2], it was only about 30–65 % worldwide [3]. Anatomic and pathological stages are still the most accurate predictors of CRC prognosis until now. Therefore, novel molecular prognostic markers for colorectal cancer are needed for the accurate prediction of prognosis.
One of the genetic pathways in the development of CRC is the failure of the DNA mismatch repair (MMR) system [4], which contributes to maintain the genomic stability by recognizing and removing insertion/deletion mutations that occur during DNA replication [5]. The two main mismatch repair genes are hMLH1 [6] and hMSH2 [7]. Germline mutations of them have been identified as the main cause of Lynch syndrome (LS) CRC (traditional HNPCC: meet the Amsterdam I or II criteria, Japanese criteria, or revised Bethesda guidelines). LS CRC patients who carried with germline mutations of hMLH1/hMSH2 gene were reported to have a better prognosis than non-carriers [8] and sporadic CRC patients (there is no obvious CRC family history) [9]. However, few studies have focused on the relationship between germline and somatic mutations of hMLH1/hMSH2 gene and prognosis of sporadic CRC. Only one published paper [10] has not observed statistically significant different survival rates between carriers (74 %) and non-carriers (63 %) of hMLH1/hMSH2/hMSH6 gene mutations in 870 consecutively ascertained sporadic CRC patients under the age of 55 in Scotland.
hMLH1 promoter methylation-induced transcriptional silencing of hMLH1 gene has been proposed as an important mechanism in the development of sporadic CRC [11]. It also has been reported having potential application for treatment response prediction (5-FU-based chemotherapy) [12]. The predominant cause of sporadic CRC has been reported to be microsatellite instability (MSI) [13], which has been hypothesized to be the most promising molecular marker for CRC prognosis [14]. In sporadic CRC, MSI is mainly caused by methylation-induced silencing of the hMLH1 gene [15]. Therefore, we hypothesized that hMLH1 promoter methylation may be associated with the prognosis of sporadic CRC.
Since evidence suggests that rare mutations of severe effect are responsible for a substantial portion of complex human cancer [16]. Therefore, we conducted the study to investigate the associations between germline and somatic hMLH1/hMSH2 gene mutations, hMLH1 promoter methylation, their interactions, and the prognosis of CRC.
Methods
Study participants
After obtaining informed written consent from study subjects and approval from Ethics Committee of Harbin Medical University, we identified CRC patients who underwent surgery at the Cancer Hospital of Harbin Medical University and they were diagnosed with pathology. Patients with no family history of CRC were categorized as sporadic CRC. Totally, 433 primary sporadic CRC patients were recruited. A total of 418 blood and 329 tumor tissue DNA were collected for molecular genetic analysis.
We followed patients until Mar 2012 or death. After surgery, the clinical data of patients were collected based on the medical records, which included age at diagnosis, pathological diagnosis, and the level of serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) before surgery. During follow-up, chemotherapy and radiotherapy protocols were obtained. Meanwhile, we obtained information about disease progression, recurrence, and the date and cause of death (if deceased).
Screening for germline and somatic mutations of hMLH1 and hMSH2 genes
PCR-SSCP: sequencing analysis
The primers for 20 pairs of all the 19 exons in the hMLH1 gene and 17 pairs of all the 16 exons in the hMSH2 gene, including exon–intron boundaries, were synthesized for genomic PCR, SSCP, and sequencing [17].
Tumor MSI analysis
MSI status was determined using PCR-SSCP for the Bethesda markers: three dinucleotide (D5S346, D2S123, and D17S250) and two mononucleotide (BAT-25, BAT-26) markers. Three levels of MSI were identified as follows: high-level MSI (MSI-H), generally defined as MSI in two or more than two of the standard markers; low-level MSI (MSI-L), one of the standard markers exhibited MSI; and microsatellite stable (MSS) in the absence of microsatellite alterations [18].
Methylation-sensitive high-resolution melting (MS-HRM)
Genomic DNA was sodium bisulfite modified by the EZ Methylation Gold Kit (Zymo Research, Orange, CA). High-resolution melting (HRM) was used to assess the methylation status of the “A” region of hMLH1 promoter [19]. Human methylated and unmethylated DNA set from Zymo Research was used as 100 % methylated and 0 % methylated controls, and the methylation percentage of 0, 1, 5, 25, 50, and 100 % were used as the standard curve. For germline and somatic hMLH1 methylation, 5 % of methylation was used as cutoff value.
Statistical analysis
Calculating from the first diagnosis of colorectal cancer to the death from any cause or Mar 2012, OS was defined as the primary end point in the study. The survival curves were estimated using Kaplan–Meier product-limit method. Cumulative survival probability was calculated at the third, fifth, and seventh year, respectively. Proportional hazards regression models were fitted with computing hazard ratios (HR) and the corresponding 95 % confidence intervals (95 % CI). All statistical tests were 2 sided; p values < 0.05 were considered statistically significant. For multiple tests, α level of 0.05 was adjusted to α′ divided by the number of multiple tests. All statistical analysis was conducted with SAS 9.1 (SAS Institute, Cary, NC, USA).
Results
Characteristics of CRC patients
The mean age of the 433 sporadic CRC patients was 58.61 years (range 24–82 years). Among the 433 CRC patients, 254 (58.66 %) were males and 179 (41.34 %) were females. Of the 432 patients with available information of TNM stage (UICC), 234 (54.17 %) were in early stage (stages I and II) and 198 (45.83 %) were in advanced stage (stages III and IV). Thirty-seven percent (159/432) tumors located at colon cancer; 63 % (271/432) tumors located at rectal cancer.
A total of 188 (43.42 %) CRC patients received 5-FU-based chemotherapy after surgery. A total of 19 (4.39 %) CRC patients received radiotherapy after surgery; 17 of the 19 patients received both chemotherapy and radiotherapy.
The median follow-up time was 52 months (range 1–87 months). During follow-up, 164 (37.88 %) CRC patients died and 29 (6.70 %) CRC patients were lost to follow-up.
Mutation of hMLH1 and hMSH2 genes and hMLH1 promoter methylation
Germline mutations of hMLH1/hMSH2 genes were identified in 12.44 % (52/418) CRC patients; somatic mutation frequencies of hMLH1/hMSH2 genes were 14.59 % (48/329). Synonymous mutations were excluded from the mutation calculation, whereas a polymorphic mutation in exon 7 of hMSH2 gene (4.07 %, 17/418) remained in the analysis.
Somatic mutation frequencies of hMLH1/hMSH2 genes were 23.44 % (15/64) in proximal colon cancer, 18.03 % (11/61) in distal colon cancer, and 10.40 % (21/202) in rectal cancer (p = 0.02). There was no significant difference between germline mutation frequency of hMLH1/hMSH2 genes and tumor location (proximal colon cancer, distal colon cancer, and rectal cancer) (p = 0.07). Both the germline and somatic mutation frequencies in MSI CRC were significantly higher than in MSS CRC (26.92 vs. 10.85 % for germline mutation; 35.19 vs. 10.57 % for somatic mutation) (p = 0.0005). Germline and somatic mutation frequencies of hMLH1/hMSH2 genes were not significantly different in other clinicopathological characteristics (age, gender, BMI, tumor stage, tumor differentiation, histotypes, and pathological types) of CRC (Table 1). The details of the hMLH1/hMSH2 gene mutations detected in the study have been described before [17].
The prevalence of germline and somatic hMLH1 promoter methylation was 8.29 % (30/362) and 14.62 % (44/301), respectively.
Somatic hMLH1 promoter methylation frequency was 26.22 % (16/61) in proximal colon cancer, 10.91 % (6/55) in distal colon cancer, and 11.96 % (22/184) in rectal cancer (p = 0.02). Somatic hMLH1 promoter methylation frequency in female and male CRC was 20.00 % (25/125) and 10.80 % (19/176) (p = 0.03), respectively. Somatic hMLH1 promoter methylation frequency was 11.38 % (28/246) in MSS CRC and 30.61 % (15/49) in MSI CRC (p = 0.00). Neither germline nor somatic hMLH1 promoter methylation was significantly different in other clinicopathological characteristics (age, gender, BMI, tumor stage, tumor differentiation, histotypes, and pathological types) of CRC (Table 1).
Survival analysis
Overall survival analysis on clinical and pathological status
The 3-, 5-, and 7-year survival of the 433 CRC patients was 67, 57, and 50.0 %, respectively; the mean survival time was 62.56 months. In multivariate Cox regression analysis, tumor stage and CA19-9 level before surgery were significantly associated with the prognosis of colorectal cancer (data not shown).
Overall survival analysis on hMLH1/hMSH2 gene mutations, MSI, and hMLH1 promoter methylation
Neither germline nor somatic hMLH1/hMSH2 gene mutation was associated with overall survival of CRC in the multivariate Cox regression analysis after adjusting by age, gender, tumor location, tumor differentiation, tumor stage, and CA19-9 level before surgery (HRadj = 1.37, 95 % CI 0.70–2.67, p = 0.35; HRadj = 1.31, 95 % CI 0.69–2.47, p = 0.42, respectively). For germline and somatic hMLH1 promoter methylation, no significant association was observed between germline and somatic hMLH1 promoter methylation and overall survival of CRC (Table 2).
When analyses stratified by tumor stage, there was no significant association between germline mutation of hMLH1/hMSH2 gene and overall survival of stage I + II CRC (HRadj = 2.32, 95 % CI 0.97–5.59, p = 0.06) (Fig. 1). No significant association was observed between germline mutation of hMLH1/hMSH2 gene and overall survival of stage III + IV CRC (HRadj = 0.68, 95 % CI 0.33–1.40, p = 0.29). For the somatic mutation of hMLH1/hMSH2 gene, non-significant associations were reported (Table 3).
When analyses by tumor location, MSI status, and chemotherapy after surgery, no significant association was observed between the germline and somatic hMLH1/hMSH2 gene mutation, hMLH1 promoter methylation, and the prognosis of CRC (data not shown).
The study has not found significant interaction between hMLH1/hMSH2 gene mutation and hMLH1 promoter methylation on the prognosis of CRC (data not shown).
Discussion
The aim of our study was to examine the prognostic significance of germline and somatic mutations of hMLH1/hMSH2 genes and hMLH1 promoter methylation in 433 sporadic CRC patients.
Germline mutations of hMLH1/hMSH2 genes were reported to be associated with about one-third of LS CRC and 3–5 % of sporadic CRC patients [20]. Moreover, carriers of germline mutations in hMLH1/hMSH2 reached up to 70–80 % probabilities of developing colorectal cancer [21, 22]. Whether germline and somatic hMLH1/hMSH2 gene mutation carriers are associated with the prognosis of CRC patients, especially sporadic CRC patients, remain unclear. In our cohort, neither germline nor somatic mutation of hMLH1/hMSH2 genes were associated with the prognosis of CRC, which was in concordance with the non-significant result by Barnetson et al. [10] in 870 consecutively ascertained sporadic CRC patients under 55 years old in Scotland. LS CRC patients who carried with MMR gene mutations were reported to have a better prognosis than non-carriers [8], the different survival advantage of mutations of hMLH1/hMSH2 genes in LS and sporadic CRC patients may be due to the different cause of MSI in LS and sporadic CRC: in sporadic CRC, MSI was mainly caused by hMLH1 promoter methylation [23, 24]; whereas, in LS CRC, MSI was mainly caused by MMR inactivation because of germline mutation [25]. In addition, according to the report in the InSiGHT database and the function prediction by any two of the PolyPhen/SIFT/MMP-MMR software, most of the mutations detected in our cohort were non-pathological or uncertain, and only two mutations of hMLH1 gene (c.1845_1847 del GAA and c.1742 CCG > CTG) in two patients were pathological [17]. This may also explain the non-significant survival advantage of hMLH1/hMSH2 gene mutation carriers.
In our study, germline mutation of hMLH1/hMSH2 genes marginally increased the hazard risk by 2.32-folds in stages I + II CRC patients (p = 0.06). In stages III + IV CRC patients, neither germline nor somatic mutation of hMLH1/hMSH2 gene had significant effect on the prognosis of CRC. The association between germline mutation of hMLH1/hMSH2 gene and CRC survival may be affected by tumor stage. However, the underlying mechanisms need to be investigated.
Until now, six published studies have concerned the association between hMLH1 promoter methylation in tumor DNA and CRC prognosis [26–28] or CpG island methylator phenotype (CIMP) and CRC prognosis [29–31], which provided the information on the association between hMLH1 promoter methylation and CRC prognosis. Four of the six studies suggested that there was no significant association between hMLH1 promoter methylation in tumor tissue and CRC prognosis (one paper provided information without sample size in sporadic MSS CRC patients [29], one paper concerned 199 sporadic CRC patients with MSI/BRAF alteration [26], one focused on 188 advanced CRC patients treated with 5-FU-based chemotherapy [30], and the other paper related 35 CRC patients treated with 5-FU-based chemotherapy [27]). Nevertheless, significant association between hMLH1 promoter methylation in tumor tissue and CRC prognosis was reported in two studies with 72 sporadic CRC patients [28] and 130 sporadic CRC patients [31], respectively. In addition, the study by Ogino et al. [32] also reported a significant association in 30 metastatic MSS CRC patients treated with combination of 5-Fu and other chemotherapy (with only one methylated tumors). Since the inconsistent study subjects, no sufficient data to pool the HR, and 95 % CI, we could not evaluate the association between hMLH1 promoter methylation and CRC prognosis upon meta-analysis. No study has investigated the association between hMLH1 promoter methylation in blood DNA and CRC prognosis. Therefore, we evaluated the association between hMLH1 promoter methylation in blood sample and CRC prognosis. However, no significant association was observed.
Although hMLH1 promoter methylation-induced transcriptional silencing of hMLH1 gene has been proposed as an important mechanism in the development of colorectal cancer [11], its prognostic significance needs to be further studied and confirmed. hMLH1 promoter methylation has been reported having resistance to 5-FU-based chemotherapy [12]. We conducted subgroup analysis based on chemotherapy; however, no significant survival advantage of hMLH1 methylation was observed in patients treated with chemotherapy. The small sample size may explain the non-significant result. Furthermore, we determined methylation by the percentage of hMLH1 promoter methylation. However, it is unclear to what extent promoter methylation of hMLH1 might affect hMLH1 expression and function. Further studies are needed to confirm which percentage of hMLH1 promoter methylation was pathogenic methylation.
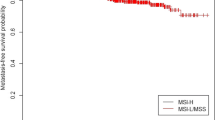
Tumor MSI has been hypothesized to be the most promising molecular marker for CRC prognosis; the two published meta-analyses confirmed the significant association between MSI and favorable prognosis [14, 33]. MSI was reported to be mainly (about 67 %) caused by hMLH1 promoter methylation in sporadic CRC [13]. Whereas, only 30.6 % (15/49) MSI patients presented with somatic hMLH1 promoter methylation and 8.3 % (4/44) MSI patients presented with germline hMLH1 promoter methylation in our study. Neither MSI patients with germline hMLH1 promoter methylation nor MSI patients with somatic hMLH1 promoter methylation had significant survival advantage compared with MSS patients without hMLH1 promoter methylation (HR = 0.95, 95 % CI 0.13–6.93, p = 0.96; HR = 0.18, 95 % CI 0.03–1.32, p = 0.09, respectively). Small sample size of MSI and germline hMLH1 promoter methylation carriers (n = 4) and MSI and somatic hMLH1 promoter methylation carriers (n = 15) limited the statistical power. Retrospective analysis was another limitation of the research, and larger number of sporadic CRC patients may needed to confirm the relationship between MSI, hMLH1 promoter methylation, and CRC prognosis.
Our study could provide references in searching molecular markers to predict the prognosis of colorectal cancer. However, limitations such as different time to initiation of adjuvant chemotherapy and no unified period of chemotherapy should be considered in deriving conclusion.
In conclusion, no significant association was observed between hMLH1/hMSH2 gene mutations, hMLH1 promoter methylation, and CRC prognosis.
References
El-Serafi MM, Bahnassy AA, Ali NM, Eid SM, Kamel MM, Abdel-Hamid NA, et al. The prognostic value of c-Kit, K-ras codon 12, and p53 codon 72 mutations in Egyptian patients with stage II colorectal cancer. Cancer. 2010;116(21):4954–64. doi:10.1002/cncr.25417.
Coleman MP, Forman D, Bryant H, Butler J, Rachet B, Maringe C, et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (the International Cancer Benchmarking Partnership): an analysis of population-based cancer registry data. Lancet. 2011;377(9760):127–38. doi:10.1016/S0140-6736(10)62231-3.
Coleman MP, Quaresma M, Berrino F, Lutz JM, De Angelis R, Capocaccia R, et al. Cancer survival in five continents: a worldwide population-based study (CONCORD). Lancet Oncol. 2008;9(8):730–56. doi:10.1016/S1470-2045(08)70179-7.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–67.
Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med. 2003;138(7):560–70.
Aaltonen LA, Peltomaki P. Genes involved in hereditary nonpolyposis colorectal carcinoma. Anticancer Res. 1994;14(4B):1657–60.
Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75(5):1027–38.
Russo A, Sala P, Alberici P, Gazzoli I, Radice P, Montefusco C, et al. Prognostic relevance of MLH1 and MSH2 mutations in hereditary non-polyposis colorectal cancer patients. Tumori. 2009;95(6):731–8.
Elsakov P, Kurtinaitis J. Survival from colorectal carcinoma in HNPCC families as compared to the general population in Lithuania-initial results. Fam Cancer. 2006;5(4):369–71. doi:10.1007/s10689-006-0007-7.
Barnetson RA, Tenesa A, Farrington SM, Nicholl ID, Cetnarskyj R, Porteous ME, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354(26):2751–63.
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96(15):8681–6.
Maier S, Dahlstroem C, Haefliger C, Plum A, Piepenbrock C. Identifying DNA methylation biomarkers of cancer drug response. Am J Pharmacogenomics. 2005;5(4):223–32.
Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69(4):780–90. doi:10.1086/323658.
Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46(15):2788–98. doi:10.1016/j.ejca.2010.05.009.
Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57(5):808–11.
McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–7. doi:10.1016/j.cell.2010.03.032.
Hu F, Li D, Wang Y, Yao X, Zhang W, Liang J, et al. Novel DNA variants and mutation frequencies of hMLH1 and hMSH2 genes in colorectal cancer in the Northeast China population. PLoS ONE. 2013;8(4):e60233. doi:10.1371/journal.pone.0060233.
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–57.
Gausachs M, Mur P, Corral J, Pineda M, Gonzalez S, Benito L, et al. MLH1 promoter hypermethylation in the analytical algorithm of Lynch syndrome: a cost-effectiveness study. Eur J Hum Genet EJHG. 2012;20(7):762–8. doi:10.1038/ejhg.2011.277.
Tomlinson IP, Beck NE, Homfray T, Harocopos CJ, Bodmer WF. Germline HNPCC gene variants have little influence on the risk for sporadic colorectal cancer. J Med Genet. 1997;34(1):39–42.
Aarnio M, Mecklin JP, Aaltonen LA, Nystrom-Lahti M, Jarvinen HJ. Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer. 1995;64(6):430–3.
Aarnio M, Sankila R, Pukkala E, Salovaara R, Aaltonen LA, de la Chapelle A, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81(2):214–8.
Raedle J, Trojan J, Brieger A, Weber N, Schafer D, Plotz G, et al. Bethesda guidelines: relation to microsatellite instability and MLH1 promoter methylation in patients with colorectal cancer. Ann Intern Med. 2001;135(8 Pt 1):566–76.
Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58(15):3455–60.
Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113(4):1146–58.
Maestro ML, Vidaurreta M, Sanz-Casla MT, Rafael S, Veganzones S, Martinez A, et al. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol. 2007;14(3):1229–36. doi:10.1245/s10434-006-9111-z.
Ide T, Kitajima Y, Ohtaka K, Mitsuno M, Nakafusa Y, Miyazaki K. Expression of the hMLH1 gene is a possible predictor for the clinical response to 5-fluorouracil after a surgical resection in colorectal cancer. Oncol Rep. 2008;19(6):1571–6.
Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Frikha F, Kallel L, et al. Aberrant methylation of hMLH1 and p16INK4a in Tunisian patients with sporadic colorectal adenocarcinoma. Biosci Rep. 2011;31(4):257–64. doi:10.1042/BSR20100023.
Ward RL, Cheong K, Ku SL, Meagher A, O’Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21(20):3729–36. doi:10.1200/JCO.2003.03.123.
Shen L, Catalano PJ, Benson AB III, O’Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007;13(20):6093–8. doi:10.1158/1078-0432.CCR-07-1011.
Lee S, Cho NY, Yoo EJ, Kim JH, Kang GH. CpG island methylator phenotype in colorectal cancers: comparison of the new and classic CpG island methylator phenotype marker panels. Arch Pathol Lab Med. 2008;132(10):1657–65. doi:10.1043/1543-2165(2008)132[1657:CIMPIC]2.0.CO;2.
Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450(5):529–37. doi:10.1007/s00428-007-0398-3.
Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–18.
Acknowledgments
Thanks for the Grants from the National Natural Science Foundation of China (Nos. 30671801, 30371243, and 30972538), Harbin Technological Innovation Talent Research Special Foundation (2011RFXXS045), and the Foundation for the Returned Scholars of Harbin (No. 2005AFLXJ017).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
All the subjects included in the study have been approved by the Ethics Committee of Harbin Medical University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Wang, Y., Li, D., Li, X. et al. Prognostic significance of hMLH1/hMSH2 gene mutations and hMLH1 promoter methylation in sporadic colorectal cancer. Med Oncol 31, 39 (2014). https://doi.org/10.1007/s12032-014-0039-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-014-0039-z