Abstract
In the spite of the impressive results achieved with imatinib in chronic myeloid leukemia (CML) patients, differences in patient’s response are observed, which may be explained by interindividual genetic variability. It is known that cytochrome P450 enzymes play a major role in the metabolism of imatinib. The present study aimed to understand the functional impact of CYP2B6 15631G>T polymorphism on the response of imatinib in CML patients and its relation to CML susceptibility. We have genotyped CYP2B6 G15631T in 48 CML patients and 64 controls by PCR-RFLP. CYP2B6 15631G>T was not found to be a risk factor for CML (OR 95 % CI, 1.12, 0.6–2, p > 0.05). Hematologic response loss was higher in patients with 15631GG/TT genotype when compared with 15631GT (36.8 vs. 13.8 %; X 2 = 3.542, p = 0.063). Complete cytogenetic response was higher in 15631GG/GT genotype groups when compared with 15631TT (X 2 = 3.298, p = 0.024). Primary cytogenetic resistance was higher in patients carrying 15631GG/TT genotype when compared with 15631GT carriers (52.6 vs. 17.2 %; X 2 = 6.692, p = 0.010). Furthermore, side effects were more common for patients carrying 15631GG genotypes when compared with GT/TT carriers (36 vs. 13.8 %; X 2 = 8.3, p = 0.004). In light of our results, identification of 15631G>T polymorphism in CML patients might be helpful to predict therapeutic response to imatinib.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myeloid leukemia (CML) is a malignant proliferation of hematopoietic cells marked by reciprocal translocation t(9; 22)(q34; q11) on the cytogenetic map and the Bcr-Abl gene fusion at the molecular level [1]. CML management was revolutionized by imatinib mesylate (Gleevec), a selective tyrosine kinase inhibitor; imatinib blocks growth of cells expressing Bcr-Abl but also other receptors such as c-kit tyrosine kinases, receptor of stem cell factor and the receptor for platelet-derived growth factor [2, 3]. Imatinib has ensured patients a good hematologic, cytogenetic and molecular response, causing a decrease in the time of response, the toxicity, morbidity and mortality of patients [4, 5]. In spite of these impressive results, certain patients develop resistance and fail to reach expected results or at least do not respond well to imatinib. These differences in terms of treatment can be explained, firstly, by mutation/amplification of Bcr-Abl domain or up-regulation of MDR-1 [6, 7]; secondly, by genetic variability of enzymes involved in imatinib metabolism, including the famous family of cytochrome P450 (CYP). Indeed, it is known that imatinib is largely metabolized by CYP3A4 and in a lesser extent by other isoenzymes such as CYP1A2, CYP2D6, CYP2C9 and CYP2C19 [8]. Inter-patient genetic variability can explain differences in imatinib bioavailability, which influences plasma and intracellular concentrations and finally affects the therapeutic response [8]. In this paper, we have proposed to understand the impact of CYP2B6 G15631T (Gln172His) in exon 4, which represents the most common exonic SNP [9] in CML patients treated with imatinib mesylate. CYP2B6 gene has been located on chromosome 19q13.2 [10], the organs involved in the expression of the gene are predominantly represented by the liver, kidney, intestine and lung [11–13]. It is known that this gene participates in the metabolism of many drugs such as cyclophosphamide, isophamide, tamoxifen, ketamine, propofol, nevirapine, efavirenz and other environmental carcinogens [13, 14]. However, the impact of this polymorphism on the metabolism of imatinib remains controversial [15, 16]. To the best of our knowledge, this is the first pharmacogenetic study investigating CYP2B6 gene on imatinib response in Moroccan patients.
Materials and methods
Patients
A total of 64 controls and 48 patients monitored at the department of Onco-Hematology of the Ibn Rochd University Hospital in Casablanca, Morocco from 2009 to 2012, participated in the present study. The median age at diagnosis was 41.5 years, range (20–59) at diagnosis. Each patient was treated with imatinib (IM) at 400 mg per day; this dose was increased to 600 or 800 mg in the absence of complete hematologic response (CHR) at 3 months, or no major cytogenetic response (MCyR) at 6 months, or no complete cytogenetic response (CCyR) at 12 months. All patients were confirmed with translocation t(9; 22)(q34; q11). Clinical and biological data were obtained from each patient’s data file with the help of hematologist. Hematologic and cytogenetic responses were previously described by [17, 18]. All participants have signed the informed consent.
Genotyping CYP2B6 polymorphism
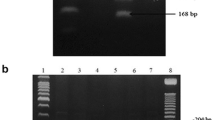
Genomic DNA was isolated from peripheral white blood cells using salting-out method previously described by Miller et al. [19]. DNA quality and quantity were measured by NanoVue Plus spectrophotometer. Polymerase chain reaction followed by restriction fragments length polymorphism method was used to genotype CYP2B6 G15631T; the mixture for PCR amplification consisted of 100 ng of DNA, 1X of 5X GoTaq Flexi Buffer (Promega), 1.25 mM of MgCl2, 0.2 mM of each dNTP, 10 pM of each primer and 0.5 U of GoTaq polymerase (Promega) completed to 20 μl with molecular grade water. Forward and reverse primer used were previously published by Berköz and Yalin [20]. PCR amplification was performed with an initial denaturation at 94 °C for 5 min followed by 35 cycles of three steps: denaturation at 94 °C for 90 s, annealing at 55 °C for 60 s, extension at 72 °C for 90 s and a final extension at 72 °C for 7 min. Amplified product after migration on a 1 % agarose gel shows 570 bp, and then 10 μl of PCR product was digested with 10 units of BsrI enzyme (New England, Biolabs) during 4 h at 65 °C. Digested products were run on a 3 % agarose gel stained with ethidium bromide. Homozygote mutant variant (TT) produces one band at 578 bp, homozygote wild type (GG) produces two bands at 518 and 60 bp, and heterozygote (GT) produces three bands at 570, 518 and 60 bp (Fig. 1).
Statistical analysis
Differences in alleles and genotypes distribution between CML patients and controls as well as between good responders and poor responders to imatinib were calculated by the Chi square (X 2) test. Crude odds ratios (OD) with 95 % confidence intervals (CI) were calculated. p < 0.05 was considered as significant. The statistical package SPSS version 16 (SPSS Inc., Chicago, IL, USA) was used.
Results
The distribution of (CYP2B6 G15631T) polymorphism in patients and controls is summarized in Table 1. No statistical difference was observed in genotype distribution between patients and controls (X 2 = 3.298; p = 0.192). The mutant 15631TT genotype was not associated with the occurrence of chronic myeloid leukemia (OR 95 % CI, 1.35, 0.6–3.1), and the frequency of the T allele in patients was not significantly different from controls (0.58 vs. 0.55; OR 95 % CI, 1.12, 0.6–2). As shown in Fig. 2, 15631GG/TT carriers were more exposed to lose their hematologic response compared with 15631GT carriers (36.8 vs. 13.8 %; X 2 = 3.542, p = 0.063). Surprisingly, in Table 2, the frequency of the homozygous 15631TT was significantly higher in patients who have failed to obtain a CCyR compared with patients who achieved a CCyR (45.8 vs. 12.5 %; p = 0.038). As shown in Table 3, when we consider the recessive model GG/GT vs. TT, a significant difference was observed between poor and good responders (X 2 = 3.298, p = 0.024). The proportion of patients who presented cytogenetic resistance to imatinib was higher in 15631GG/TT carriers compared with 15631GT carriers (52.6 vs. 17.2 %; X 2 = 6.692, p = 0.010) and the risk to develop cytogenetic resistance was 5.33-fold for the 15631GG/TT carriers (Fig. 3). Furthermore, 15631GG genotype carriers were more exposed to have side effects than GT/TT carriers (36 vs. 13.8 %; X 2 = 8.3, p = 0.004) Fig. 4.
Discussion
The CYPs enzymes constitute a large multi-gene family of hemoproteins involved in the endogenous and exogenous metabolism of many xenobiotics. This activity of biotransformation concerns anticancer drugs, environmental carcinogens as well as steroid hormones [13, 14, 21–23]. Thus, CYPs enzymes contribute significantly in drug metabolism. However, the polymorphism of CYPs enzymes leads to a functional change in the expression of the gene that causes variability in patient’s response to drugs. This variability allows the classification of patients into distinct groups (poor, intermediate or extensive metabolizers) [24–27]. Though, numerous studies have shown the importance of CYPs enzymes’ variability in interindividual susceptibility to developing certain diseases [28–30].
In this study, we have found out that the frequency of the 15631TT genotype is higher in patients than in controls (Table 1), but this difference was not significant. It has been shown that the 15631G>T (Gln172His) polymorphism [31, 32] leads to a decrease in enzymatic activity of CYP2B6 in liver [33], which results in a limited ability to eliminate xenobiotics-exposing subjects of developing cancer or not getting an adequate response to treatment [34, 35]. Berköz M and Yalin S indicated that the risk of developing acute leukemia was 2.48-fold for subjects with GT genotype and 1.92-fold for T allele carriers [20]. Other authors have reported that the T allele was associated with acute leukemia [36].
We have noticed that the response to imatinib was influenced by CYP2B6 15631G>T polymorphism. It is noteworthy that hematologic response loss (HRL) was more common in patients harboring 15631GG/TT genotypes compared with patients carrying 15631GT (Fig. 2). This finding might be explained by the fact that homozygous 15631GG regarded as extensive metabolizers eliminate drugs before they reach plasma half-life. Regarding, the homozygous 15631TT mutant considered to be poor metabolizers fail to reach plasma half-life because of the lack or reduction in the expression of CYP2B6. However, 15631GT carriers categorized as intermediate metabolizers have responded better to the treatment with less HRL. Thus, the determination of 15631G>T polymorphism might help to predict hematologic response and classify patients into distinct groups.
Surprisingly, we found that the 15631TT genotype was significantly associated with a low CCyR rate (Table 1); which might confirm that 15631TT homozygous mutant are poor metabolizers. The highest rate of CCyR was observed among patients with 15631GT genotype (Table 2). Considerably, the 15631GG/GT genotype was associated with a high rate of CCyR (Table 3). Subsequently, we have noted that patients harboring 15631GG/TT were more frequently exposed to develop cytogenetic resistance to imatinib when compared with 15631GT carriers (Fig. 3). Villuendas et al. [16] have reported that CYP2B6 was associated with primary cytogenetic resistance to imatinib and was considered to be an independent resistance marker, in the absence of the BCR-ABL domain mutation. In contrast, Lewis [15] have shown that CYP2B6 is not involved in imatinib metabolism. Based on our results, determination of the CYP2B6 15631G>T polymorphism in our patients undergoing imatinib treatment might help in the prediction of therapeutic response. However, 15631GG carriers were more frequently exposed to have side effects compared with 15631GT/TT carriers (p = 0.004) (Fig. 4). These side effects might be explained by the clonal evolution of hematopoietic cell in extensive metabolizers despite the respect of imatinib dose.
Conclusion
We have explored the functional impact of the CYP2B6 15631G>T polymorphism in CML patients treated with imatinib mesylate. We have shown that the CYP2B6 15631G>T is not associated with the development of CML, but interestingly, it influences significantly the clinical and cytogenetical outcome of our CML patients and seems to be a good therapeutic predictor. Therefore, taking into account the number of our sample, other studies are needed to confirm our findings.
References
Deininger MWN, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia Review article. Blood. 2000;96:3343–56.
Buchdunger E, Zimmermann J, Mett H, Lydon NB, Meyer T, Muller M, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–4.
Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–53. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15618470.
O’Brien SG, Guilhot F, Larson R a, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12637609.
Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11287972.
Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102:276–83. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12623848.
Werner B, Lutz D, Brümmendorf TH, Traulsen A, Balabanov S. Dynamics of resistance development to imatinib under increasing selection pressure: a combination of mathematical models and in vitro data. PloS One. 2011;6:1–8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3245228&tool=pmcentrez&rendertype=abstract.
Peng B, Hayes M, Resta D, Racine-Poon A, Druker BJ, Talpaz M, et al. Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J Clin Oncol Off J Am Soc Clin Oncol. 2004;22:935–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14990650.
Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, et al. Hepatic CYP2B6 expression : gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther. 2003;307:906–22.
Saarikoski ST, Rivera SP, Hankinson O, Husgafvel-Pursiainen K. CYP2S1: a short review. Toxicol Appl pharmacol. 2005;207:62–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16054184.
Gonzalez F, Crespi C, Czerwinski M, Gelbon V. Tohoku J Exp Med. 1992;168:67–72.
Gervot L, Rochat B, Gautier JC, Bohnenstengel F, Kroemer H, de Berardinis V, et al. Human CYP2B6: expression, inducibility and catalytic activities. Pharmacogenetics. 1999;9:295–306. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10471061.
Ekins S, Wrighton S a. The role of CYP2B6 in human xenobiotic metabolism. Drug Metab Rev. 1999;31:719–54. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10461547.
Lang T, Klein K, Fischer J, Nüssler a K, Neuhaus P, Hofmann U, et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001;11:399–415. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11470993.
Lewis DF V. 57 varieties: the human cytochromes P450. Pharmacogenomics. 2004;5:305–18. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15102545.
Villuendas R, Steegmann JL, Pollán M, Tracey L, Granda a, Fernández-Ruiz E, et al. Identification of genes involved in imatinib resistance in CML: a gene-expression profiling approach. Leukemia. 2006;20:1047–54. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16598311.
Deininger MW, O’Brien SG, Ford JM, Druker BJ. Practical management of patients with chronic myeloid leukemia receiving imatinib. J Clin Oncol Off J Am Soc Clin Oncol. 2003;21:1637–47. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12668652.
Kantarjian HM, O’Brien S, Cortes J, Giles FJ, Rios MB, Shan J, et al. Imatinib mesylate therapy improves survival in patients with newly diagnosed Philadelphia chromosome-positive chronic myelogenous leukemia in the chronic phase: comparison with historic data. Cancer. 2003;98:2636–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14669283.
Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215.
Berköz M, Yalin S. Association of CYP2B6 G15631T polymorphism with acute leukemia susceptibility. Leuk Res. 2009;33:919–23. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19144407.
Waxman DJ, Ko A, Walsh C. Regioselectivity and stereoselectivity of androgen hydroxylations catalyzed by cytochrome P-450 isozymes purified from phenobarbital-induced rat liver. J Biol Chem. 1983;258:11937–47.
Seree EJ, Pisano PJ, Placidi M, Rahmani R, Barra YA. Identification of the human and animal hepatic cytochromes P450 involved in clonazepam metabolism. Fundam Clin Pharmacol. 1993;7:69–75. Available from: http://www.ncbi.nlm.nih.gov/pubmed/8486332.
Dehal SS, Kupfer D. Metabolism of the proestrogenic pesticide methoxychlor by hepatic P450 monooxygenases in rats and humans. Dual pathways involving novel ortho ring-hydroxylation by CYP2B. Drug Metab Dispos Biol Fate Chem. 1994;22:937–46. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7895613.
Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–49. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12571262.
Scripture CD, Sparreboom A, Figg WD. Modulation of cytochrome P450 activity: implications for cancer therapy. Lancet Oncol. 2005;6:780–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16198984.
Smires F, Moreau C, Habbal R, Siguret V, Fadili S, Golmard J, et al. Influence of genetics and non-genetic factors on acenocoumarol maintenance dose requirement in Moroccan patients. J Clin Pharm Ther. 2012;37:594–8.
Elmachad M, Elkabbaj D, Elkerch F, Laarabi F, Barkat A, Oualim Z, et al. Frequencies of CYP3A5 * 1/* 3 variants in a Moroccan population and effect on tacrolimus daily dose requirements in renal transplant. Genet Test Mol Biomarkers. 2012;16:644–7.
Belmouden A, Melki R, Hamdani M, Zaghloul K, Amraoui A, Nadifi S, et al. A novel frameshift founder mutation in the cytochrome P450 1B1 (CYP1B1) gene is associated with primary congenital glaucoma in Morocco. Clin Genet. 2002;62:334–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12372064.
Gan CQ, Wang XY, Cao YD, Ye WX, Liu H, Sun YY. Association of CYP2C19*3 gene polymorphism with breast cancer in Chinese women. Genet Mol Res GMR. 2011;10:3514–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22180071.
Levkovich NN, Gorovenko NG, Myasoedov D V. Association of polymorphic G1934A variant (allele *4) of CYP2D6 gene with increased risk of breast cancer development in Ukrainian women. Exp Oncol. 2011;33:136–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21956465.
Turpeinen M, Raunio H, Pelkonen O. The functional role of CYP2B6 in human drug metabolism: substrates and inhibitors in vitro, in vivo and in silico. Curr Drug Metab. 2006;7:705–14. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17073575.
Waris M, White LJ. Seasonality of respiratory syncytial virus infection. Clin Infect Dis Off Publ Infect Dis Soc Am. 2006;43:541–2. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19713363.
Hofmann MH, Blievernicht JK, Klein K, Saussele T, Schaeffeler E, Schwab M, et al. Aberrant splicing caused by single nucleotide polymorphism c. 516G T [Q172H], a marker of CYP2B6 * 6, is responsible for decreased expression and activity of CYP2B6 in Liver. J Pharmacol Exp Ther. 2008;325:284–92.
Rakhmanina NY, van den Anker JN, Soldin SJ, van Schaik RH, Mordwinkin N, Neely MN. Can TDM improve pharmacotherapy of HIV infection in adolescents? Ther Drug Monit. 2010;32:273–81.
Xu C, Ogburn ET, Guo Y, Desta Z. Effects of the CYP2B6 * 6 allele on catalytic properties and inhibition of CYP2B6 in vitro: implication for the mechanism of reduced efavirenz metabolism and other CYP2B6 substrates in vivo (Abstract). 2012;40:717–25.
Yuan Z, Liu Q, Zhang Y, Liu H, Zhao J, Zhu P. CYP2B6 gene single nucleotide polymorphisms and leukemia susceptibility. Ann Hematol. 2011;90:293–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20878158.
Acknowledgments
We authors thank Hassan II academy of Science and Technology for providing us with financial support for this study.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kassogue, Y., Quachouh, M., Dehbi, H. et al. Functional polymorphism of CYP2B6 G15631T is associated with hematologic and cytogenetic response in chronic myeloid leukemia patients treated with imatinib. Med Oncol 31, 782 (2014). https://doi.org/10.1007/s12032-013-0782-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-013-0782-6