Abstract
MicroRNA-203 (miR-203), possessing tumor suppressive or promotive activities, has been found to be downregulated or upregulated in different cancer types. The purpose of this study was to investigate whether the increased expression of miR-203 can be used as a noninvasive diagnostic and prognostic biomarker in epithelial ovarian cancer (EOC). Real-time quantitative PCR was performed to detect the expression levels of miR-203 in EOC tissues. The expression levels of miR-203 were significantly higher in EOC tissues compared to adjacent non-cancerous tissues (p < 0.001). High expression of miR-203 was observed in 65.38 % (102/156) of EOC. In addition, high miR-203 expression was found to be closely correlated with advanced FIGO stage (p < 0.001), higher histological grade (p = 0.02), lymph node involvement (p < 0.001), and positive recurrence (p < 0.001). Moreover, high miR-203 expression was correlated with shorter overall survival (p < 0.001) and shorter progression-free survival (p < 0.001) of EOC patients. Furthermore, multivariate analysis showed that the status of miR-203 expression was an independent predictor for both overall survival and progression-free survival in EOC. These findings provide the convincing evidence for the first time that the upregulation of miR-203 may serve as a novel molecular marker to predict the aggressive tumor progression and unfavorable prognosis of EOC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy and the second leading cause of cancer-related deaths among women worldwide [1]. 5-year survival for EOC depends on the spread of the disease at diagnosis. Since most EOC patients are not diagnosed until the disease is in advanced stages due to mild and diffuse symptoms, the prognosis of EOC patients is very poor [2]. In EOC patients with disease limited to the ovary [International Federation of Gynecology and Obstetrics (FIGO) stage I], survival is close to 80 %, but in cases in which the disease involves the upper abdomen or beyond (FIGO stages III and IV), only about 20 % of patients survive at 5 years [3]. This disparity in survival between early and late stage emphasizes the need to improve early detection of EOC. However, the cellular/molecular mechanisms of tumorigenesis and tumor progression of EOC remain poorly understood, even though extensive clinical and basic research efforts have been undertaken. Therefore, it is still a great challenge to identify novel and efficient molecular markers for early disease diagnosis, clinical outcome prediction, and treatment guidance.
Tumorigenesis and tumor progression are involved in many genetic events. MicroRNAs (miRNAs) are an abundant class of small noncoding RNAs with 18–25 nucleotides in length [4]. These small molecules have been found to regulate gene expression either by translational repression or degradation of mRNA after targeting the 3′UTR [5]. By regulating their target genes, miRNAs are involved in diverse biological processes such as cell proliferation, development, differentiation, apoptosis, and others [6]. In particular, accumulating studies have found the aberrant expression of miRNAs in various cancer types and have described the association of miRNA deregulation with the initiation and progression of human cancers [7]. Some miRNAs are overexpressed in tumors and act as oncogenes, promoting carcinogenesis by targeting tumor suppressors. The other miRNAs are downregulated in tumors and generally participate in oncogene overexpression [8]. Growing evidence has also indicated the possible use of miRNA expression profiles to distinguish between normal and neoplastic tissues, leading to the identification of new diagnostic and/or prognostic markers. Especially, Hong et al. [9] detected miR-221 expression in serum samples from patients with EOC and healthy age-matched controls, and found that serum miR-221 may be associated with clinicopathological features and prognosis in EOC patients, suggesting that this miRNA may have a role as a novel diagnostic and prognostic marker, and may have potential as a therapeutic target for this disease; Chung et al. [10] found specific profiles of serum-derived miRNAs of ovarian cancer based on a comparative study using a miRNA microarray of serum, tissue, and ascites, and identified serum miR-132, miR-26a, let-7b, and miR-145 as potential biomarkers in serous ovarian cancer; Peng et al. [11] indicated that low miR-100 expression may be an independent poor prognostic factor and miR-100 can function as a tumor suppressor by targeting PLK1 in human EOCs. These findings suggest that full understanding of the biological functions and molecular mechanisms of miRNA-mediated processes may provide a great advancement in the diagnosis and treatment for human cancers.
MiR-203, located in a region at chromosome 14, has been demonstrated to be abnormally expressed in various malignant diseases. Previous study of Iorio et al. [12] detected the increased expression levels of miR-203 in ovarian cancer compared with normal tissues. However, its clinical significance in EOC is still unclear. The purpose of this study was to investigate whether the increased expression of miR-203 can be used as a noninvasive diagnostic and prognostic biomarker in EOC.
Materials and methods
Patients and tissue samples
This study was authorized by the Research Ethics Committee of Maternal and Child Health Hospital of Hainan Province, People’s Republic of China. All patients agreed to the procedure and signed consent forms. All specimens were handled and made anonymous according to the ethical and legal standards.
A total of 156 EOC tissues and adjacent non-cancerous tissues from primary EOC patients were surgically obtained between 1998 and 2008 in Maternal and Child Health Hospital of Hainan Province, China. The patients had a mean age of 50.22 ± 6.68 years (range 30–70 years). No patient had received chemotherapy or radiation therapy prior to surgery. The clinicopathological characteristics of all 156 EOC patients were collected, including age at diagnosis, the International Federation of Gynaecologists and Obstetricians (FIGO) stage, histological grade, histological type, serum CA-125, status of lymph node, and status of recurrence. Among these, FIGO stage was performed using FIGO staging system (2009). Histological subtype and tumor grade were classified according to the World Health Organization (WHO) criteria. Tumor grade was subdivided into low (G1) and high grade (G2–G3). The patient characteristics are summarized in Table 1. Resected tissue samples were immediately cut and snap-frozen in liquid nitrogen before stored at −80 °C until RNA extraction.
The survival information from the postoperative follow-up of all 156 EOC patients was received by telephone or mail. The median follow-up time was 36 months (range 6–88 months). During the follow-up period, the patients were performed the examinations including serum CA-125, pelvic MRI, color Doppler ultrasound of the liver and kidney, X-rays every 3 months for 2 years, at 6-month intervals in years 3–5 thereafter. The overall survival (OS) was defined as the interval from the end of treatment to death due to any cause or to the date of last contact. The progression-free survival (PFS) was defined as the period after the conclusion of treatment to proven local recurrence or distant metastasis.
Real-time quantitative RT-PCR for miRNA
Real-time quantitative RT-PCR for miRNA was performed to detect the expression levels of miR-203 in 156 EOC tissues and adjacent non-cancerous tissues. In brief, total RNA was extracted from tissue samples using TRIzol (Invitrogen) according to the manufacturer’s protocol. The quality of RNA was evaluated by calculating the RNA integrity number (RIN). High quality RNA (RIN value was greater than 5.5) was used for reverse transcription of the first-strand cDNA synthesis by reverse transcription. The sequences of the primers used for reverse transcription of mature miR-203 with stem-loop structure, miR-203 RT (5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTAGTGGT-3′), were designed according to the corresponding sequence from miRBase database. After that, the amplification process was performed using the Hairpin-it TM miRNAs qPCR Quantitation Kit (Genepharma, Shanghai, China) according to the manufacturer’s protocol. The reaction mixture was initially incubated at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min, and then held at 4 °C for the reverse transcription step until the preparation for the PCR had been completed. PCR amplification was initiated with one cycle of 95 °C for 10 min, followed by 40 cycles of 94 °C for 15 s and 60 °C for 60 s. The U6 small nuclear RNA was amplified as an internal control. The primers were as follows: for miR-203 F: 5′-ACA CTC CAG CTG GCG TGA AAT GTT TAG GAC CA-3′, R: 5′-CTC AAC TGG TGT CGT GGA-3′; for U6 F: 5′-CTC GCT TCG GCA GCA CA-3′, R: 5′-AAC GCT TCA CGA ATT TGC GT-3′. Relative quantification of target miRNA expression was evaluated using the comparative cycle threshold (CT) method. The raw data were presented as the relative quantity of target miRNA, normalized with respect to U6. Each sample was examined in triplicate. Mean normalized gene expression ± standard deviation (SD) was calculated from independent experiments.
Statistical analysis
All statistical analyses were carried out using the software of SPSS version 13.0 for Windows (SPSS Inc, IL, USA). Data were expressed as mean ± SD. Fisher’s exact test, chi-square test, and two-sample t test were used to evaluate the statistical differences among the groups with different clinicopathological data. Survival curves were made using the Kaplan–Meier method, and the log-rank test was used to analyze the differences between clinicopathological characteristics and survival in EOC patients. Differences were considered statistically significant when p was less than 0.05.
Results
Upregulation of miR-203 in human EOC tissues
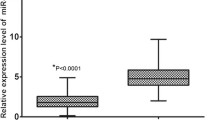
The expression levels of miR-203 in 156 EOC and adjacent non-cancerous tissues were analyzed by real-time quantitative RT-PCR for miRNA. For the EOC tissues, the mean level of miR-203 expression was 8.17 (range 4.04–12.00). For the adjacent non-cancerous tissues, the mean level of miR-203 expression was 3.89 (range 0.20–7.93). The statistical analysis showed that the expression level of miR-203 was significantly higher in EOC tissues than in adjacent non-cancerous tissues (p < 0.001; Fig. 1a). In addition, the expression level of miR-203 was significantly higher in EOC patients with advanced FIGO stage (III/IV) than those with low FIGO stage (I/II) (p < 0.001; Fig. 1b). Moreover, in order to classify all 156 EOC patients into high miR-203 expression and low miR-203 expression groups, a cutoff value for miR-203 expression levels was chosen on the basis of a measure of heterogeneity with the log-rank test statistic with respect to overall survival. An optimal cutoff value (7.60) was identified: the low miR-203 expression group [expression level lower than the cutoff value; mean expression value 5.90, n = 70, 44.87 %] and the high miR-203 expression group [expression level higher than the cutoff value; mean expression value 10.02, n = 86, 55.13 %].
Expression levels of miR-203 in epithelial ovarian cancer (EOC) and adjacent non-cancerous tissues. The expression level of miR-203 was significantly higher in EOC tissues than in adjacent non-cancerous tissues (p < 0.001, a). In addition, the expression level of miR-203 was significantly higher in EOC patients with advanced FIGO stage (III/IV) than those with low FIGO stage (I/II) (p < 0.001, b)
Association of miR-203 expression with clinicopathological parameters of EOC patients
Table 1 summarized the association of miR-203 expression with clinicopathological parameters of EOC patients. As the results, high miR-203 expression was found to be closely correlated with advanced FIGO stage (p < 0.001), higher histological grade (p = 0.02), lymph node involvement (p < 0.001), and positive recurrence (p < 0.001). In details, there were 40 patients in the low miR-203 expression group and 70 patients in the high miR-203 expression group who had high histological grade (G2–G3); Lymph node metastases were present in 81.40 and 18.60 % of patients in the low miR-203 expression and high miR-203 expression groups, respectively; The high miR-203 expression group included 12/22 (54.55 %) FIGO stage II, 36/70 (51.43 %) stage III, and 38/38 (100.00 %) stage IV tumors; in the high miR-203 expression group, 82/116 (70.69 %) EOC patients were present of recurrence. No statistically significant associations of miR-203 expression with age at diagnosis, histological type, tumor size, and serum CA-125 level were found (all p > 0.05, Table 1).
Association of miR-203 expression with prognosis in EOC patients
The association of miR-203 upregulation with prognosis in all 156 EOC patients was also analyzed. Clinical follow-up was performed for all patients. In the univariate analysis, we found a significant difference in patients’ overall survival between the miR-203 low expression and high expression groups (p < 0.001; Fig. 2a; Table 2), which was similar to the prognostic value of miR-203 in progression-free survival of EOC patients (p < 0.001; Fig. 2b; Table 2). In addition to miR-203 expression, histological grade (p = 0.01 and 0.02, respectively), FIGO stage (both p < 0.001), lymph node metastasis (both p < 0.001), and recurrence (both p < 0.001) were also correlated with both overall and progression-free survival of EOC patients (Table 2). In the multivariate analysis using the Cox proportional hazards model with significant parameters in univariate Kaplan–Meier analysis, we further found that miR-203 expression (p = 0.005 and 0.009, respectively), histological grade (p = 0.03 and 0.04, respectively), FIGO stage (p = 0.008 and 0.01, respectively), lymph node metastasis (p = 0.006 and 0.01, respectively), and recurrence (p = 0.01 and 0.01, respectively) were independent prognostic factors for both overall and progression-free survival in EOC patients (Table 2).
Kaplan–Meier analyses for the correlation between miR-203 expression and survival. The overall (a) and the progression-free (b) survival of EOC patients with high and low expression are shown. Log-rank test showed that patients with high miR-203 expression had significantly poorer overall and progression-free survivals versus patients with low miR-203 expression (both p < 0.001)
Discussion
The biological and phenotypic heterogeneity of EOC patients has been demonstrated to be caused by the complex genomic rearrangements and structural variations observed in the ovarian cancer genome, which has until now made it difficult to exploit genome-wide information to stratify patients more precisely for diagnosis and prognosis [13]. Therefore, it is essential to discover effective biomarkers that could predict the prognosis and improve the prognosis of patients with advanced EOC. According to the recent studies, miRNAs may offer a new regulatory model of gene expression, and miRNA expression signatures correlate well with specific clinical characteristics of cancer, so that they can be used to classify normal and cancerous tissues, as well as for prognosis [14–16]. In the present study, we detected the expression of miR-203 in 156 EOC specimens by real-time quantitative RT-PCR for miRNA and found that the expression of miR-203 was an independent factor for both overall and progression-free survivals. These findings reveal that the aberrant expression of miR-203 may play important roles in the tumorigenesis and aggressiveness of EOC patients. To date, the associations between miR-203 expression and prognosis in EOC have not been reported yet. This is the first study to investigate the impact of miR-203 expression on prognosis using a large number of clinical samples.
miR-203 is a keratinocyte-derived miR that not only regulates embryonic epidermal differentiation and the construction of the skin’s protective barrier but also acts as a tumor suppressor or oncogene by regulating proliferation, differentiation, invasion, metastasis, and apoptosis of tumor cells via actions on its target genes [17]. For one thing, miR-203 exhibits significantly downregulated expression in some tumors such as head and neck squamous cell carcinomas, hematopoietic malignancy, and colon cancer [18–20]. It is involved in the apoptotic program observed in head and neck squamous cell carcinoma and hematopoietic malignancies, and functions as a tumor suppressor by targeting ABL1 [18, 19]. Yuan et al. [21] demonstrated that miR-203 could inhibit cell proliferation in human esophageal squamous cell carcinoma through ΔNp63-mediated signal pathway. Zhao et al. [22] indicated that the downregulated miR-203 in cervical cancer may be correlated with lymph node metastasis only, and the overexpression of miR-203 may decrease cancer cell proliferation. Bo et al. [23] verified that the expression of miR-203 was decreased in bladder cancer tissues; the ectopic expression of miR-203 promoted the apoptosis of human bladder cancer cell lines and inhibited cell proliferation, whereas its depletion increased cell growth. Viticchiè et al. [24] also showed that miR-203 may be downregulated in clinical primary prostatic tumors compared to normal prostate tissue and in metastatic prostate cancer cell lines compared to normal epithelial prostatic cells. In contrast, recent studies indicated that it may act as an onco-miRNA. For instance, Ikenaga et al. [25] reported that miR-203 was overexpressed in pancreatic adenocarcinoma samples compared with chronic pancreatitis and normal pancreas samples. The overexpression of miR-203 was an independent predictor of poor prognosis in pancreatic adenocarcinoma patients. Iorio et al. [12] also examined the upregulation of miR-203 in EOC tissues. Consistent with these findings of previous studies, we here verified the increased expression of miR-203 in a large cohort of EOC patients, compared with the normal controls. In addition to its aberrant expression, we also found that the upregulation of miR-203 may be dramatically associated with the advanced tumor progression, such as high FIGO stage and histological grade, the present of lymph node metastasis and recurrence, and unfavorable overall and progression-free survivals in EOC patients.
In conclusion, these findings provide the convincing evidence for the first time that the upregulation of miR-203 may serve as a novel molecular marker to predict the aggressive tumor progression and unfavorable prognosis of EOC patients. However, there are some limitations. At first, the sample size is small. To solve this problem, further studies and more samples will be required to confirm the prognostic value of miR-203 in EOC. Secondly, the current study has not elucidated the exact molecular mechanisms of miR-203 acting on EOC, which is also worth to be further investigated.
References
Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–29.
Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer Statistics, 2001. CA Cancer J Clin. 2001;51:15–36.
Marchini S, Mariani P, Chiorino G, Marrazzo E, Bonomi R, Fruscio R, Clivio L, Garbi A, Torri V, Cinquini M, Dell’Anna T, Apolone G, Broggini M, D’Incalci M. Analysis of gene expression in early-stage ovarian cancer. Clin Cancer Res. 2008;14:7850–60.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Cho WC. MicroRNAs in cancer-from research to therapy. Biochim Biophys Acta. 2010;1805:209–17.
Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6.
Hong F, Li Y, Xu Y, Zhu L. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J Int Med Res. 2013;41:64–71.
Chung YW, Bae HS, Song JY, Lee JK, Lee NW, Kim T, Lee KW. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient. Int J Gynecol Cancer. 2013;23:673–9.
Peng DX, Luo M, Qiu LW, He YL, Wang XF. Prognostic implications of microRNA-100 and its functional roles in human epithelial ovarian cancer. Oncol Rep. 2012;27:1238–44.
Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707.
Huang Y, Shen XJ, Zou Q, Zhao QL. Biological functions of microRNAs. Bioorg Khim. 2010;36:747–52.
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–39.
Perera RJ, Ray A. MicroRNAs in the search for understanding human diseases. BioDrugs. 2007;21:97–104.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
He JH, Li YM, Li YG, Xie XY, Wang L, Chun SY, Cheng WJ. hsa-miR-203 enhances the sensitivity of leukemia cells to arsenic trioxide. Exp Ther Med. 2013;5:1315–21.
Boldrup L, Coates PJ, Wahlgren M, Laurell G, Nylander K. Subsite-based alterations in miR-21, miR-125b, and miR-203 in squamous cell carcinoma of the oral cavity and correlation to important target proteins. J Carcinog. 2012;11:18.
Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z, Dong JT. Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes Cancer. 2011;2:782–91.
Shah MS, Schwartz SL, Zhao C, Davidson LA, Zhou B, Lupton JR, Ivanov I, Chapkin RS. Integrated microRNA and mRNA expression profiling in a rat colon carcinogenesis model: effect of a chemo-protective diet. Physiol Genomics. 2011;43:640–54.
Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J, Cheng HZ, Huang SD. MicroRNA-203 inhibits cell proliferation by repressing ΔNp63 expression in human esophageal squamous cell carcinoma. BMC Cancer. 2011;11:57.
Zhao S, Yao DS, Chen JY, Ding N. Aberrant expression of miR-20a and miR-203 in cervical cancer. Asian Pac J Cancer Prev. 2013;14:2289–93.
Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu D, Huang Y. microRNA-203 suppresses bladder cancer development by repressing bcl-w expression. FEBS J. 2011;278:786–92.
Viticchiè G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli LG, Knight RA, Candi E, Melino G. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–31.
Ikenaga N, Ohuchida K, Mizumoto K, Yu J, Kayashima T, Sakai H, Fujita H, Nakata K, Tanaka M. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17:3120–8.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shaosheng Wang and Xiaohong Zhao have contributed equally to this article.
Rights and permissions
About this article
Cite this article
Wang, S., Zhao, X., Wang, J. et al. Upregulation of microRNA-203 is associated with advanced tumor progression and poor prognosis in epithelial ovarian cancer. Med Oncol 30, 681 (2013). https://doi.org/10.1007/s12032-013-0681-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-013-0681-x