Abstract
The study was aimed to investigate the relationship between hypermethylation of Syk gene and clinicopathological characteristics and long-term outcomes in colorectal cancer. The effect of Syk on cell proliferation and invasion ability was also assessed. Methylation and expression status of Syk were explored in CRC tissues and cell lines by MSP, qRT-PCR and western blot assay. The effects of Syk overexpression on tumorigenesis were studied by in vitro assay. The correlation between Syk methylation and clinical relevance in CRC patients was also analyzed. Syk methylation was found 48.6 % in CRC tissue samples and 57.1 % in cell lines, respectively. The loss of Syk expression could be restored by demethylation agent. Overexpression of Syk in CRC cell inhibited cell proliferation (p < 0.01) and invasion (p < 0.01). The methylation of Syk was significantly associated with histological grade (p = 0.002), lymph node status (p < 0.001) and TNM stage (p < 0.001). Five-year overall survival in methylated Syk group was significantly lower than that in unmethylated Syk group (59 vs. 80 %, p < 0.001). Multivariate analysis demonstrated that Syk methylation was an independent prognostic factor for overall survival. Syk is identified as a potential tumor suppressor in CRC progression. Syk methylation is correlated with poor overall survival, which acts as an independent prognostic indicator of CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most common malignancy and one of the leading causes of cancer-related death in the world [1] which arises as a consequence of the accumulation of genetic and epigenetic alterations in colonic epithelial cells during neoplastic transformation. Epigenetic modifications, particularly DNA methylation in promoter region, are recognized as common molecular alterations in CRC. Hypermethylation of CpG island promoters can lead to a complete block of transcription of tumor suppressor genes [2]. The significance of DNA methylation in the initiation and promotion of CRC has been reported widely [3–7].
Spleen tyrosine kinase (Syk) gene is widely expressed in various types of normal and tumor tissues and cancer cells [8–11]. Emerging evidence indicates that Syk may be a potent modulator of epithelial cell growth and a potential tumor suppressor in human breast carcinoma [12, 13]. Hypermethylation of Syk gene silencing its expression was found in breast cancer [14], gastric cancer [15], melanoma [16] and oral squamous cell carcinoma [17]. However, the function of Syk gene in sporadic CRC remains unknown, so we explored whether methylation of Syk promoter region was associated with the loss of Syk expression, and the relationship between Syk methylation status and clinicopathological factors in CRC. At the same time, we also investigated whether Syk expression could be restored in CRC cell lines with methylated Syk treated with demethylation reagent (5-Aza-2-deoxycytidine, 5-Aza). Additionally, the effect of Syk overexpression on cell proliferation and invasion were also analyzed. In the present study, we found that the Syk gene is frequently hypermethylation and a potential tumor suppressor gene in CRC. Overexpression of Syk inhibited cell proliferation and invasion ability. Methylation of Syk could be act as an independent prognostic factor for CRC patients.
Materials and methods
Tissue samples and cell lines
Tumor and corresponding adjacent non-cancerous tissue samples were collected at the time of surgery from 210 consecutive primary sporadic CRC patients in the Department of Gastrointestinal Surgery of the Sixth Affiliated Hospital, Sun Yat-sen University. Tissues were snap-frozen in liquid nitrogen immediately after resection and stored at −80 °C until analysis. CRC diagnosis was confirmed by histology. Informed written consent was given by all the patients enrolled in the study. This study was approved by Ethics Committee of Sun Yat-sen University in China.
Seven CRC cell lines (Colo320, HCT15, SNU1197, SW116, HCT116, SNU61 and SW-403) were used in the present study. Breast cancer cell lines T-47D and MDA-MB-231 were used as negative and positive control for methylation. The cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in RPMI 1640 medium or Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum and incubated in 5 % CO2 at 37 °C.
In order to test whether 5-Aza-2′-deoxycytidine (5-Aza) restored Syk expression in CRC cell lines, CRC cell lines (Colo320 and HCT15) were treated with 3 μM 5-Aza (Sigma) for 3 consecutive days, with culture medium changing every day, to check for methylation status of Syk gene and restoration of Syk expression. All experiments were conducted in triplicate.
DNA isolation, bisulfate sodium treatment and methylation-specific PCR (MSP)
Genomic DNA from tissue samples and cell lines were extracted with Qiagen Tissue and blood Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. DNA was modified with sodium bisulfite as described previously [14]. Briefly, two micrograms of genomic DNA was denatured by incubation with 0.3 mol/l NaOH for 10 min at 37 °C. A total of 30 μl of 0.2 mol/l hydroquinones and 520 μl of 3 mol/l sodium bisulfite were added, and the solution was incubated at 50 °C for 16 h. Treated DNA was purified by Wizard DNA clean-up System (Promega, Madison, WI) according to the manufacturer’s protocol.
The CpG islands within Syk gene promoter have been described previously [14]. MSP was performed to analyze the methylation status in Syk gene promoter region. Primer sequences and PCR conditions were derived from previously published article [14]. Primers designed to amplify methylated and unmethylated Syk promoter sequences: methylated specific: 5′-gattaagatatatttagggaatatg-3′ (forward) and 5′-cacctatattttattcacataatttc-3′ (reverse); unmethylated specific: 5′-attttgtgggttttgtttggtg-3′ (forward) and 5′-acttccttaacacacccaaac-3′ (reverse). The PCR product was 243 bp with 9 CpG sites. The PCR product was 140 bp with 6 CpG sites. Both primers were added to the same reaction and expected to generate 243 or 140 bp products, respectively. PCR conditions were as follows: total reaction volume was 30 μl, 94 °C for 5 min for denaturation, 30 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, with a final elongation step of 10 min at 72 °C. The PCR mixture contained 1× buffer with 1.5 mM MgCl2, 10 pmol of each primer, 0.2 mM dNTPs and 100 ng of bisulfite-modified DNA in a final volume of 50 μl. PCR products were run on 2 % agarose gels stained with ethidium bromide and visualized directly under UV illumination. Samples were considered as methylation or non-methylation, when there was a clearly visible band (243/140 bp) on the gel with methylation or non-methylation primers.
RNA isolation and qRT-PCR
Total RNA was extracted from tissue samples and cell lines using the RNeasy kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Briefly, two micrograms of total RNA from each sample was subjected to cDNA synthesis using the Superscript III first strand synthesis system for RT-PCR (Invitrogen). The mRNA expression level of Syk gene was detected by qRT-PCR. Primer sequences for RT-PCR were as follows: 5′-tgtcaaggataagaacatcaagt-3′ (forward) and 5′-acttccttaacacaacccaaac-3′ (reverse). The PCR conditions were as follows: total reaction volume was 30 μl, 94 °C 5 min for denaturation followed by 30 cycles of 94 °C for 30 s, 57 °C for 30 s, 72 °C for 30 s, with a final extension step of 10 min at 72 °C. β-actin mRNA was used as an internal reference.
Western blotting
Twenty micrograms of protein extracts from each sample was fractionated through SDS-PAGE gels, and the electrophoresed proteins were then transferred to PVDF membranes and subjected to western blotting using specific primary antibody for Syk (N-19) and β-actin (I-19). Primary and secondary antibodies were purchased from Santa Cruz Biotechnology (CA, USA) unless otherwise mentioned.
Immunohistochemistry and immunofluorescence imaging
Formalin-fixed, paraffin-embedded sections of tissues were subjected to immunostaining using a rabbit anti-human Syk antibody. Briefly, tissue sections were deparaffinized, rehydrated and subjected to antigen retrieval by boiling in sodium citrate buffer (10 mmol/l, pH 6.0). The sections were then incubated overnight with the Syk (1:200) antibody at 4 °C, washed, incubated with an anti-rabbit IgG secondary antibody and then streptavidin–biotin complex, and then stained with 3,3-diaminobenzidine. After visualization of immunoreactivity, the sections were counterstained with hematoxylin and mounted.
Full-length Syk was cloned into the vector pCDNA3.1, and pCDNA3.1-Syk was transfected into HCT15 cells to screen stable cells for analysis. Cells were grown on glass coverslips following incubation at 37 °C in 5 % CO2 for 48 h, the cells were fixed in 4 % paraformaldehyde for 1 h, permeabilized in 1 % Triton X-100 for 10 min, blocked in 1 % bovine serum albumin for 1 h and incubated in anti-Syk antibody at 4 °C overnight. The cells were then washed three times in PBST, incubated in goat anti-rabbit Cy3 antibody for 1 h, again washed three times with PBST and then stained with 1 μg/ml Hoechst 33542 for 10 min. The cells were then analyzed by fluorescence microscopy.
Cell proliferation assay and invasion assay
[3H] thymidine incorporation analysis was performed to analyze the cell proliferation. Cells were seeded at a density of 7 × 103 per well in 24-well plates. Media were changed every 24 h during the experiment. [3H] thymidine (2 μCi) was added to fresh medium and incubated with cells for half an hour at 37 °C. Cells were then washed, methanol-fixed and solubilized prior to scintillation counting. Counts from the experiments were performed in triplicate for each group. Meanwhile, cell viability was examined using the Vybrant MTT Cell Proliferation Assay Kit (Invitrogen) according to the manufacturer’s instructions.
The cell invasion ability was determined with BD BioCoat Growth Factor Reduced Matrigel Invasion Chamber according to the manufacturer’s instructions. Complete culture medium (supplemented with 20 % fetal bovine serum) was used as chemoattractant. Cells (1 × 104 cells/well) were washed twice with PBS, re-suspended in 200 μl of serum-free medium and then transferred into the upper chamber. After 24 h of incubation, the filter was gently removed from the chamber, the cells on the upper surface were removed by wiping with a cotton swab, and cells that had invaded to the lower surface areas were fixed, stained with hematoxylin and eosin (H&E) and counted in 5 randomly selected fields on microscope (×100). Experiments were performed in triplicate for each group.
Statistical analysis
Statistical analysis was performed using the SPSS statistical package (version 16.0; SPSS, Chicago, IL, USA). Comparisons were made by chi-square test or Fisher’s exact test and Student’s t test where appropriate. The effect of Syk methylation on overall survival was performed using Kaplan–Meier method and compared using log-rank test. Overall survival (OS) was defined as the time from the date of surgery to the date of death from any causes. Multivariate analysis of the factors that influenced overall survival was carried out using the Cox proportional hazards regression model. All analyses were two-sided, and p < 0.05 was considered statistically significant.
Results
Methylation status and expression of Syk gene in CRC tissues and cell lines
To determine whether methylation of Syk gene in CRC tissues, MSP was performed with sodium bisulfate-treated DNA isolated from 210 primary sporadic colorectal cancers and their corresponding non-cancerous tissues. Methylation of Syk was observed in 102 of 210 tumor tissues (48.6 %), 32 of 210 non-cancerous tissues (15.2 %) from corresponding CRC patients (Fig. 1a) and 4 of 7 CRC cell lines (57.1 %) (Fig. 1c).Hypermethylation of Syk in CRC tissues was more frequent than that in corresponding normal tissues (48.6 vs. 15.2 %, p < 0.001).Western blotting was performed to detect the Syk protein expression level in 7 CRC cell lines. Syk protein was detected in 3 unmethylated CRC cell lines (Colo320, SNU1197 and HCT1116) and not in 4 methylated CRC cell lines (HCT15, SW1116, SW403 and SNU61) (Fig. 1f). Expression level of Syk mRNA by RT-qPCR in CRC tumor samples with unmethylated Syk gene was significantly higher than that in samples with methylated Syk gene (about 2.5-fold, p < 0.01) (Fig. 1b). Meanwhile, to detect the effect of Syk methylation on Syk protein expression, MSP and immunohistochemistry of Syk protein were performed in 210 primary CRC samples. Syk protein expression was either lost or present in more than 50 % of CRC cells per sample, and the patients were divided into Syk-negative and Syk-positive groups. CRC tissues with Syk methylation were significantly more Syk-negative compared to those with Syk unmethylation [83.4 % (120/210) vs. 20 % (18/90), p < 0.01]. Representative data were shown in Fig. 1d.
Methylation status and expression level of Syk gene in CRC tissues and cell lines. a Representative results for methylation status and mRNA expression of Syk gene in CRC tumor tissues (T) and adjacent normal tissues (N). b RT-qPCR was used to analyze the expression level of Syk gene in methylated and unmethylated CRC tumor tissues. The relative expression of Syk mRNA level was significantly higher in unmethylated CRC tissues than that in methylated samples (p < 0.01) c Methylation status and protein level of Syk gene in CRC cell lines (Colo320, HCT15, SW1116, SUN1197, SW403, SNU61 and HCT116). Breast cancer cell lines T-47D and MDA-MB-231 as negative and positive methylation controls. d Immunohistochemical staining of Syk protein in CRC tissues. Representative results for normal tissues (left), methylation/Syk protein negative (middle) and unmethylation/Syk protein positive (right)
Syk expression was restored by 5-Aza in CRC cell lines
To investigate the effect of DNA methylation on Syk silencing, two CRC cell lines, Colo320 and HCT15, were treated with 5-Aza. As for Colo320 cell with unmethylated Syk, expression level in both protein and mRNA was not significantly different with or without 5-Aza treatment. However, as for HCT15 with methylated Syk gene, Syk expression was restored and significantly upregulated in mRNA level (>3.5-fold, p < 0.01) by 5-Aza (Fig. 2).
Effect of 5-Aza on Syk expression in methylated cell (HCT15) and unmethylated cell line (Colo320). No significant difference of Syk expression level in Colo320 cell treated with 5-Aza or not, however, methylated Syk was transformed to unmethylated Syk by 5-Aza and restored its expression in both mRNA and protein level in HCT15 cells. All experiments were performed in triplicate. Data are mean ± SD
The effect of Syk gene overexpression on CRC cell proliferation and invasion
In order to analyze the function of Syk gene in CRC carcinogenesis, we constructed and transfected a pcDNA3.1-Syk expression vector into HCT15 cell to set stable cells with overexpression of Syk confirmed by cell immunohistochemistry. Cells with empty vector were used as control (Fig. 3a). The effect of Syk overexpression on cell proliferation and viability was evaluated by 3H thymidine incorporation assay and MTT assay. The overexpression of Syk resulted in a significant decrease in proliferation in HCT15-Syk cell compared to the control group (decreased to 65 %, p < 0.01, Fig. 3b). The inhibitory effect was also confirmed by MTT assay. After about 4-day culture, the cell growth was significantly suppressed in HCT15 cell with overexpression of Syk compared to the control (p < 0.01, Fig. 3c).
Effect of Syk overexpression on CRC cell proliferation and invasion. a Overexpression of Syk in CRC cell line HCT15 stably transfected with Syk cDNA was confirmed by cell immunohistochemistry. Effect of Syk overexpression on cell proliferation and viability was evaluated by 3H thymidine incorporation assay (b) and MTT assay (c). d HCT15 cells transfected by pCDNA3.1-Syk or empty vector were seeded in Matrigel-coated transwells and allowed to migrate for 48 h. There were significantly less migrating cells in those transfected with pCDNA3.1-Syk compared to empty vector (p < 0.01). All experiments were performed in triplicate. Data are mean ± SD
To explore the effect of Syk on cancer cell invasion ability, transwell Matrigel invasion assay was performed. HCT15 cells transfected by pCDNA3.1-Syk or empty vector were seeded in Matrigel-coated transwells and allowed to migrate for 48 h. There were significantly less migrating cells in those transfected with pCDNA3.1-Syk compared to that in those cells with empty vector (down to 50 %, p < 0.01).
Correlation of Syk methylation status with clinicopathological characteristics and long-term outcomes of CRC patients
The relationship between the clinicopathological characteristics of CRC and methylation status of Syk gene is given in Table 1. With the exception of the histological grade (p = 0.002), lymph node status (p < 0.001) and TNM stage (p < 0.001), there was no significant difference in other clinicopathological factors such as age, gender, tumor location, and T and M stages.
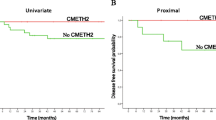
Moreover, the correlation between Syk methylation status and overall survival (OS) of CRC patients was also evaluated. In the present study, the 5-year OS of all patients was 67 % with median survival time of 8.5 years. When stratified by Syk methylation status, the 5-year OS of patients with methylated Syk was 59 % with median survival time of 7.8 years, compared with 80 % with median survival time of 9.3 years for patients with unmethylated Syk gene in the tumor tissues (Fig. 4). On Cox univariate regression analysis for OS in this study, we observed that tumor location, N stage, M stage, TNM stage and Syk methylation status were significant risk factors. However, only TNM stage and Syk methylation status were independent prognostic indicators by multivariate analysis (Table 2). Additionally, multivariate analysis indicated that patients with methylated Syk had an increased risk of death by a factor of 1.766 compared to unmethylated Syk in tumor tissues (Table 2).
Discussion
Hypermethylation of Syk in the promoter region has been reported in various cancers [14–17]. In the present study, Syk was found to be methylated and inactivated in CRC. The methylation of Syk was found 48.6 % in CRC tissue samples and 57.1 % in CRC cell lines, respectively. Hypermethylation of Syk led to silencing of Syk in CRC cell lines, which was restored by treatment with demethylation agent (5-Aza). The methylation of Syk was associated with the decrease in the protein expression in CRC tissue samples. These results demonstrated that DNA methylation in the promoter region played a key role in Syk transcription and led to Syk inactivation. Furthermore, Syk can act as a potential tumor suppress gene in the development of CRC, which was consistent with other reports [15, 18, 19].
To explore the effect of Syk methylation on expression, cell proliferation and invasion ability, we introduced the Syk full-length cDNA into CRC cell line HCT15 to establish the stable cells overexpressing Syk. Then, we examined the proliferation and invasiveness ability of Syk-transfected and mock-transfected cells using MTT assay, [3H] thymidine incorporation assay and in vitro invasion assay. Significant differences were found in proliferation rate and invasion ability compared with empty vector controls. These results indicated that ectopic expression of Syk inhibited cell proliferation and invasion, which are in agreement with previously published data [17, 20].
Analysis of clinicopathological factors and Syk methylation in CRC indicated a significant association of Syk methylation status with poorly differentiation (p = 0.002), lymph metastasis (p < 0.001) and advanced stages (p < 0.001). These results showed that loss of Syk involved in metastasis and progression of CRC. Similar results were found in breast cancer [14], lung cancer [21] and melanoma [20].
Survival analysis of patients in this study indicated that hypermethylation of Syk significantly correlated with worse 5-year OS by multivariate regression analysis, which showed that methylation of Syk could act as an independent prognostic factor for CRC. These results are consistent with those reported in liver cancer [18] and gastric cancer [15], and contrast with head and neck cancer [22].
Based on the present data, we concluded that hypermethylation of Syk is involved in CRC and that an epigenetic mechanism may regulate loss of its expression in CRC tissues and cell lines. Syk contributes to the suppression of tumorigenesis by inhibiting cell proliferation and cell migration and invasion ability. Methylation status of Syk in CRC tissues is correlated with poor OS of CRC patients, which acts as an epigenetic marker to predict prognosis for patients with CRC.
References
Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics. CA Cancer J Clin. 2005;55(1):10–30.
Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1(7):686–92.
1997 update of recommendations for the use of tumor markers in breast and colorectal cancer. Adopted on November 7, 1997 by the American Society of Clinical Oncology. J Clin Oncol. 1998; 16(2):793–5.
Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57(5):808–11.
Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA. 1998;95(15):8698–702.
Shin SK, Nagasaka T, Jung BH, Matsubara N, Kim WH, Carethers JM, Boland CR, Goel A. Epigenetic and genetic alterations in Netrin-1 receptors UNC5C and DCC in human colon cancer. Gastroenterology. 2007;133(6):1849–57.
Hibi K, Mizukami H, Shirahata A, Goto T, Sakata M, Sanada Y. Aberrant methylation of the netrin-1 receptor genes UNC5C and DCC detected in advanced colorectal cancer. World J Surg. 2009;33(5):1053–7.
Flück M, Zürcher G, Andres AC, Ziemiecki A. Molecular characterization of the murine syk protein tyrosine kinase cDNA, transcripts and protein. Biochem Biophys Res Commun. 1995;213(1):273–81.
Tsuchida S, Yanagi S, Inatome R, Ding J, Hermann P, Tsujimura T, Matsui N, Yamamura H. Purification of a 72-kDa protein-tyrosine kinase from rat liver and its identification as Syk: involvement of Syk in signaling events of hepatocytes. J Biochem. 2000;127(2):321–7.
Dejmek J, Leandersson K, Manjer J, Bjartell A, Emdin SO, Vogel WF, Landberg G, Andersson T. Expression and signalling of Wnt05a/discoidin domain receptor-1 and Syk plays distinct but decisive roles in breast cancer patient survival. Clin Cancer Res. 2005;11:520–8.
Okamura S, Ng CC, Koyama K, Takei Y, Arakawa H, Monden M, Nakamura Y. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res. 1999;11(6):281–5.
Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, Basyuk E, Blancato JK, Vezza PR, McLeskey SW, Mangeat PH, Mueller SC. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–7.
Stewart Z, Pietenpol J. Syk: a new player in the field of breast cancer. Breast Cancer Res. 2001;3:5–7.
Yuan Y, Mendez R, Sahin A, Dai JL. Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res. 2001;61(14):5558–61.
Wang S, Ding YB, Chen GY, Xia JG, Wu ZY. Hypermethylation of Syk gene in promoter region associated with oncogenesis and metastasis of gastric carcinoma. World J Gastroenterol. 2004;10(12):1815–8.
Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, Nelson B, Bosenberg M. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66(23):11187–93.
Ogane S, Onda T, Takano N, Yajima T, Uchiyama T, Shibahara T. Spleen tyrosine kinase as a novel candidate tumor suppressor gene for human oral squamous cell carcinoma. Int J Cancer. 2009;124(11):2651–7.
Yuan Y, Wang J, Li J, Wang L, Li M, Yang Z, Zhang C, Dai JL. Frequent epigenetic inactivation of spleen tyrosine kinase gene in human hepatocellular carcinoma. Clin Cancer Res. 2006;12(22):6687–95.
Li J, Sidell N. Growth-related oncogene produced in human breast cancer cells and regulated by Syk protein-tyrosine kinase. Int J Cancer. 2005;117(1):14–20.
Bailet O, Fenouille N, Abbe P, Robert G, Rocchi S, Gonthier N, Denoyelle C, Ticchioni M, Ortonne JP, Ballotti R, Deckert M, Tartare-Deckert S. Spleen tyrosine kinase functions as a tumor suppressor in melanoma cells by inducing senescence-like growth arrest. Cancer Res. 2009;69(7):2748–56.
Dong SW, Ma L, Xu N, Yan HQ, Liu HY, Li YW, Zhang P. Research on the reactivation of Syk expression caused by the inhibition of DNA promoter methylation in the lung cancer. Neoplasma. 2011;58(1):89–95.
Luangdilok S, Box C, Patterson L, Court W, Harrington K, Pitkin L, Rhŷs-Evans P, O-charoenrat P, Eccles S. Syk tyrosine kinase is linked to cell motility and progression in squamous cell carcinomas of the head and neck. Cancer Res. 2007;67:7907–16.
Acknowledgments
This study was supported by a grant (no. 81072042) from National Nature Scientific Funds, China.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Additional information
Zuli Yang and Lijun Huo contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, Z., Huo, L., Chen, H. et al. Hypermethylation and prognostic implication of Syk gene in human colorectal cancer. Med Oncol 30, 586 (2013). https://doi.org/10.1007/s12032-013-0586-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-013-0586-8