Abstract
The carcinogenic role of Hepatitis B X (HBX) in hepatocellular carcinoma (HCC) remains largely unknown. Histone H3 lysine 4 methyltransferase SMYD3 was found to be over-expressed and have a pro-carcinogenic effect in HCC. The role of HBX in regulating SMYD3 activity and the corresponding C-MYC gene in HCC carcinogenesis was investigated. SMYD3 and C-MYC expression in HBV-negative HepG2 and HBV-positive HepG2.2.15 were detected by real time PCR and Western blot. After transfection of HBX into HepG2, SMYD3 and C-MYC protein expression was detected and the apoptosis and proliferation of hepatoma cells were assayed. After SMYD3 expression in HepG2 with HBX transfection downregulated by siRNA, the corresponding C-MYC expression, cellular apoptosis, and proliferation were assayed by FACS. SMYD3 mRNA and protein and C-MYC protein were significantly higher in HepG2.2.15 than in HepG2. HBX transfection resulted in enhanced SMYD3 and C-MYC expressions, decreased cell apoptosis, and increased cell proliferation in HepG2 cells. Knocking down of SMYD3 in HepG2 with HBX transfection inhibited C-MYC expression and promoted apoptosis. These results suggest that HBX upregulates SMYD3 expression in HepG2, which may promote hepatoma development and progress. C-MYC may act as a down-stream gene in HBX-SMYD3-related hepatocarcinogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) continues to be one of the most common malignancies worldwide, and chronic hepatitis B virus (HBV) infection is the most important risk factor for HCC in East Asia including China [1]. HBV-associated HCC developed directly from chronic hepatitis and cirrhosis due to chronic HBV infection, the integration of HBV DNA sequences in HCC cells is well established [2]. HBV is a partial double-stranded DNA virus with a 3.2 kb genome, containing four known open reading frames, S, C, P, and X. Among these four genes, HBX is integrated into the host genome more frequently and considered as an important factor in HBV-related hepatocarcinogenesis [1, 3]. Although it is known that HBX plays an important role in hepatocarcinogenesis, the exact functions and molecular mechanisms of HBX in HCC are not well understood. Like several other viral oncoproteins, HBX protein is involved in a wide variety of cellular functions, such as trans-activation of transcription, deregulation of cell cycle checkpoints, participation in the cellular signal transduction pathway apoptosis [2, 3]. HBX also plays a role in the regulation of series of cell-signaling cascades, most notably in the Ras- and Raf-induced mitogen-activated protein kinase pathways, and the inactivation of tumor suppressive genes (TSG) such as P21, P16, P15, MGMT, or E-Cadherin [4–6] through promoter hypermethylation.
Histone modification is implicated in the regulation of chromatin structure, as well as transcriptional activation and repression in an epigenetic manner. Multiple modifications that function in a combinational or sequential fashion, in single or multiple tails, dictate ‘histone codes’, which are closely linked to many biological functions [7]. Among these modifications, histone lysine methylations on lysine residues 4, 9, 27, and 36 in H3, and on residue 20 in H4, are considered to be critical for transcriptional regulation [7]. SET and MYND-domain containing 3 (SMYD3), a H3-K4 histone methyltransferase (HMT), was found to be over-expressed in colorectal carcinoma (CRC), breast carcinoma, HCC, and other malignancies [8–10]. It di- or tri-methylates histone H3-K4 in the presence of heat shock 90 kDa protein (HSP90A) and activates the transcription of downstream genes. However, during HBX-related HCC development and progression, whether HBX plays its pro-oncological role by upregulating SMYD3 and correspondingly hypermethylating H3-K4 is not yet understood. Oncogene C-MYC is known to be crucial in various biological processes such as cellular proliferation, growth, apoptosis, metabolism, adhesion, protein synthesis, DNA replication, and angiogenesis [11, 12]. Overexpression of MYC in different cellular lineages of transgenic mice resulted in the formation of lymphomas, osteosarcomas, and HCC [13]. Furthermore, inactivation of mice MYC caused sustained tumor regression and cellular senescence; for example, tumor cells differentiated into normal hepatocytes and biliary cells in hepatocellular carcinoma [14, 15]. However, whether SMYD3 regulates C-MYC activity in hepatoma remains unclear. Hence, in this article, the role of HBX in regulating SMYD3 and whether HBX-SMYD3 regulates HCC carcinogenesis through C-MYC, were investigated.
Materials and methods
Materials
Hepatoma cell lines HepG2 and HepG2.2.15 were stored in our lab. Plasmid pcDNA3.1-HBX was a gift from the Institute of Virology, 302nd Hospital of China. Liposome Lipofectamine2000TM, Trizol reagent, and primers were bought from Invitrogen Co. USA. G418, the RT-PCR kit was bought from GIBCO Co., USA. Mouse anti-human SMYD3 and HBX monoclonal antibodies were bought from ABCAM Co, USA. The fluorescence quantitative PCR detection kit was bought from Roche Co.
Cell culture and gene transfection
Cells were cultured in a RIMP1640 medium and a DMEM high-glucose culture medium containing 10% fetal calf serum, respectively, and incubated in an atmosphere of 5% CO2 at 37°C, with a wetness fraction of saturation. About 0.25% pancreatin was used to digest and passage the culture every 2–3 days. Negative and positive HBV infection in HepG2 and HepG2.2.15 were verified with immunocytochemistry (data not shown). After plasmids were established and sequenced, Lipofectamine2000TM was applied to the transfect by lipid-mediated transfection; meanwhile, the mock plasmid pEGFP-C2 and the liposome served as controls. About 800 μg/ml G418 was added every 48 h for 2 weeks after transfection to select subclone cells. Once all cells in the control group had died, 400 μg/ml G418 was administered to maintain passage culture. Positive subclones were selected and cultured for 3 months.
The distributions of HBX and SMYD3 proteins were measured by confocal microscopy: 5 × 105/ml HepG2 with HBX transfection was cultured in 6-well culture plates. When the confluence achieved 80–90%, the cells were fixed with 4% paraformaldehyde for 15 min. Then, the mixture of anti-HBX and anti-SMYD3 antibodies was incubated with the cell slides at 4°C overnight. The fluorescence-labeled second antibodies were added and incubated for 60 min, and then the sections were mounted using glycerol and observed by confocal microscopy.
Detection of SMYD3 and C-MYC mRNA expression with real time RT-PCR
A one-step method was used to isolate total RNA from cells, which was reversely transcripted to obtain cDNA, and then real-time PCR was performed. Primers amplifying a 172 bp fragment for SMYD3 were: sense 5′-TGAATGTGACTGTTTC CGTTGC-3′, antisense 5′-ATTGC TGCTTATGATCG CCTGG-3′; primers amplifying a 222 bp fragment for C-MYC were: upstream 5′-GGGGCTTTATC TAACTCGCG-3′, downstream: 5′-CTA TGGGCAAAGTTTCGTG-3′; primers for house-keeping β-actin were: upstream: 5′-GTGGACATCCGCAAAGAC-3′, downstream: 5′-AAAGGG TGTAACGCAACT AA-3′, with a PCR product of 192 bp. The reaction parameters were set up as follows: 2 min at 95°C, 30 s at 94°C, 30 s at 57°C, 30 s at 72°C for 45 cycles, followed by 5 min at 72°C. Fluorescent signals were collected during the extension phase, Ct values of the sample were calculated, and SMYD3 mRNA expression level was analyzed by analysis of variance (ANOVA) method.
Detection of SMYD3 and C-MYC protein expression with Western blot
Western blot was performed to detect SMYD3 and C-MYC protein expression in HepG2 cells. Briefly, cells were collected and extracted with a cell lysis buffer (Promega, Madison, WI). Protein (50 μg) from each sample was subjected to 4–20% pre-cast polyacrylamide gel (Bio-Rad, Hercules, CA) electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). The primary antibody dilution was 1:500, followed by 1:3,000 dilution of goat anti-rabbit HRP-labeled antibody (Bio-Rad). An ECL substrate kit (Amersham, Piscataway, NJ) was used for the chemiluminescent detection of the signals with autoradiography film (Amersham).
Downregulation of SMYD3 expression with RNA interference and detection of SMYD3 and C-MYC expression
In order to clarify whether C-MYC is one of the downstream or target genes of the HBX-SMYD3 pathway, SMYD3 was downregulated with RNA interference and expression of C-MYC was detected with Western blot. Briefly, RNA interference (RNAi) plasmids Pgenesil-1-s1 and Pgenesil-1-s2 were designed to interfere with SMYD3 mRNA targeting the nucleic acids 302–323, 267–288. Primers for Pgenesil-1-s1 were: sense 5′-GATCCATACTGTAGTGCTAAGTGTTTCAAGACG ACACACTTAGCACTACAGTATTTTTTTGTCGACA-3′, antisense 5′-AGCTTGT CGACAAAA AA ATACTGTAGTGCTAAGTGTCGTCTTGAAACACTTAGCA CTACAGTATG-3′; primers for Pgenesil-l-s2 were: sense 5′-GATCCGCTGATGC GATGCTCTCAGTTCAAGACGCTGAGAGCATCGCATCAGCTTTTTTGTCGACA-3′, antisense 5′-AGCTYGTCGACAAAAAAGCTGATGCGATGCTCTCAGCG TCTTGAACTG AGAGCATCGCATCAGCG-3′; plasmid Pgenesil-1-hk was designed as a mock control. The plasmids were transiently transfected into HBX transfected HepG2-HBX cells. Cells were harvested at 24, 48, and 72 h after siRNA transfection. Both SMYD3 and C-MYC mRNA levels were detected with RT-PCR. β-actin mRNA was used as a control.
Detection of cell apoptosis and proliferation with flow cytometry
Cell cycles were examined by flow cytometry. Ratio of S-phase cells/all cells was regarded as proliferating cell ratio. Briefly, 2 × 105 of cells were collected, washed twice with 0.01 mol l−1 PBS, and fixed in 70% ethanol overnight at 4°C. Then, cells were washed once with PBS, digested by 200 μl of RNase (1 mg/ml) at 37°C for 30 min, and stained with 800 μl of propidium iodide (PI, 50 μg/ml, Sigma) at room temperature for 30 min. The DNA histograms were assayed with a flow cytometer (Becton Dickson Co., San Jose, CA), using the CELLQEST software (Becton Dickson Co.).
Apoptotic ratios of cells were determined by annexin V-APC according to the manufacturer’s instructions. Briefly, cells were collected, washed twice with cold PBS, resuspended with 100 μl of binding buffer (10 mmol l−1 HEPES, 140 mmol l−1 NaCl, 2.5 mmol l−1 CaCl2, pH 7.4) into 1 × 106 cells/ml density, and incubated with annexin V-APC and 5 μl of 7-AAD at room temperature for 15 min. After washing with binding buffer, the cells were resuspended with 400 μl of binding buffer. Apoptosis was analyzed by a flow cytometer (Becton Dickson Co) at a wavelength of 488 nm.
Statistical analysis
All tests were repeated three times. Unless otherwise stated, all data are shown as mean ± standard error of the mean (SEM). Statistical significance (P < 0.05) was determined by t test or ANOVA followed by assessment of differences using SPSS 12.0 software (SPSS Inc., Chicago, IL).
Results
Expressions of SMYD3 mRNA and protein in different hepatoma cell lines
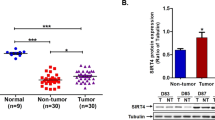
Real-time RT-PCR showed that compared with HBX-negative HepG2, SMYD3 mRNA expression in HBX-positive HepG2.2.15 was higher significantly (HepG2: 0.178 ± 0.053 vs. HepG2.2.15: 0.9233 ± 0.122, P < 0.01). After HBX transfection, SMYD3 mRNA expression increased (HepG2: 0.178 ± 0.053 vs. HepG2-HBX: 0.978 ± 0.154, P < 0.01, Fig. 1a). SMYD3 protein expression was significantly higher in HBX-positive HepG2.2.15 cells than in HepG2 cells (SMYD3/β-actin: HepG2 0.321 ± 0.056 vs. HepG2.2.15 0.565 ± 0.112 P < 0.05). After HBX transfection, SMYD3 protein expression in HepG2 cells was also increased (HepG2-HBX 0.587 ± 0.105 vs. HepG2 0.321 ± 0.089, P < 0.05, Fig. 1b).
Change of SMYD3 mRNA expression in HepG2, HepG2-HBX, HepG2.2.15 cells. a mRNA level of SMYD3 in HepG2, HepG2 with mock transfection, HepG2 with HBX transfection, and HepG2.2.15 were 0.178 ± 0.053, 0.173 ± 0.014,0.978 ± 0.154, and 0.9233 ± 0.122, respectively. b Protein level of SMYD3 in HepG2, HepG2 with mock transfection, HepG2 with HBX transfection, and HepG2.2.15. Lane 1: HepG2; lane 2: mock transfection; lane 3: HBX transfection; lane 4: HepG2.2.15
HBX and SMYD3 decreased apoptosis and increased cell proliferation in HepG2 cells
Apoptosis ratio of HepG2, mock, and HBX-transfected HepG2 cells were 7.95 ± 0.42%, 7.57 ± 1.71%, 3.27 ± 0.17%; and ratio of cell at S phase in HepG2, mock, and HBX-transfected HepG2 cells were 4.40 ± 1.71%, 2.23 ± 0.14%, 25.19 ± 8.03%, respectively. Compared with HepG2 or mock, apoptosis of HBX-transfected HepG2 cells was significantly lower than that of HepG2 (non-transfected) or mock cells, while cell proliferation at S phase was much higher (P < 0.01).
Knockdown of SMYD3 in HepG2-HBX cells inhibits C-MYC expression and increases cell apoptosis
The results of C-MYC protein assays in HepG2 and HBX-transfected HepG2 (HepG2-HBX) cells showed that C-MYC protein expression was significantly higher in HBX positive HepG2.2.15 (0.775 ± 0.221) and HepG2-HBX (0.924 ± 0.232) than in HepG2 (0.512 ± 0.127 P < 0.05, Fig. 2).
C-MYC protein expression changes in HepG2, HepG2 with mock transfection, HepG2 with HBX transfection, and HBX-positive HepG2.2.15 cells. C-MYC expression level was regarded as ratio of C-MYC/β-actin. As a result, C-MYC expression in HepG2 was 0.512 ± 0.127, which increased to 0.775 ± 0.221 in HBX-positive HepG2.2.15 (P < 0.05). After HBX transfection, expression of C-MYC in HepG2-HBX also increased to 0.924 ± 0.232, which was significantly higher than in HepG2, while no difference was observed with HepG2.2.15. Lane 1: HepG2; lane 2: mock transfection; lane 3: HBX transfection; lane 4: HepG2.2.15
After siRNA transfection into HBX transfected HepG2-HBX for 24, 48, and 72 h, RNA was isolated and mRNA expression was detected by RT-PCR. The results showed that at 24, 48, and 72 h after siRNA transfection, SMYD3 mRNA expressions were inhibited in HepG2-HBX with RNA interference compared with HepG2-HBX without siRNA or mock transfection (P < 0.01). Correspondingly, C-MYC mRNA expressions were also inhibited after transfection (P < 0.01, Fig. 3). Accordingly, at 24 h after SMYD3 siRNA transfection, apoptosis rate of HepG2-HBX cells (27.16 ± 0.57%) was much higher than HepG2-HBX without SMYD3 interference (8.53 ± 0.14%) or mock transfection (13.58 ± 0.50%) (P < 0.05, respectively).
Knockdown of SMYD3 inhibits C-MYC mRNA expression in HBX-transfected HepG2 cells. Pgenesil-1-s1, Pgenesil-1-s2 were siRNAs against SMYD3, with Pgenesil-1-hk as mock. At 24, 48, and 72 h after siRNA transfection, both SMYD3 and C-MYC mRNA expression were inhibited in HepG2-HBX cells. Compared to mock transfection, SMYD3 LSD-t in both Pgenesil-1-s1, Pgenesil-1-s2 groups were 24 h (−0.300, −0.300); 48 h (−0.378, −0.370), 72 h (−0.376, −0.380); P < 0.01 (ANOVA LSD-t test) in all groups. Similarly, compared to mock transfection, C-MYC LSD-t in Pgenesil-1-s1 and Pgenesil-1-s2 was significantly lower than mock transfection (24 h: −0.068, −0.113; 48 h: −0.175, −0.178; 72 h: −0.117, −0.125, P < 0.01 in all groups ANOVA LSD-t test). Lane 1: 24 h after Pgenesil-1-s1 transfection; lane 2: 24 h after Pgenesil-1-s2 transfection; lane 3: 24 h after mock (Pgenesil-1-hk) transfection; lane 4: 48 h after Pgenesil-1-s1 transfection; lane 5: 48 h after Pgenesil-1-s2 transfection; lane 6: 48 h after Pgenesil-1-hk transfection; lane 7: 72 h after Pgenesil-1-s1 transfection; lane 8: 72 h after Pgenesil-1-s2 transfection; lane 9: 72 h after mock transfection
Discussion
Chronic hepatitis B virus (HBV) infection is one of the major causes of hepatocellular carcinoma (HCC) in the world. Integration of HBV genomic DNA into cellular chromosomes occurs during the viral life cycle and is observed in approximately 85% of HBV-associated tumors. HBV has long been suspected to be involved in hepatocarcinogenesis, although its oncogenic effect remains controversial. Hepatitis B virus X protein (HBX), encoded by the smallest open reading frame of HBV, has 154 amino acids and a molecular mass of approximately 17.5 kDa. HBX is a multifunctional regulator that modulates transcription, signal transduction, cell cycle progress, protein degradation pathways, apoptosis, and genetic stability. HBX can moderately stimulate transcription of many different viral and cellular transcription elements; promoters and enhancers stimulated by HBX typically contain DNA binding sites for NF-κB, AP-1, AP-2, c-EBP, ATF/CREB, or the calcium-activated factor NF-AT [12, 13,]. HBX also stimulates RNA pol I- and pol III-dependent promoter [14]. It has been established that HBX can stimulate various cytoplasmic signal transduction pathways. HBX was shown to activate the extracellular signal-regulated kinases (ERKs), the stress-activated protein kinases/NH2-terminal-Junkinases (SAPK/JNKs), the p38 kinase [15], telomerase, and VEGF [16, 17]. Cellular factors reported to interact with HBX include a number of transcriptional components, such as proteosome subunit protein C7, p53, the UV light-damaged DNA binding protein complex UVDDB, and the mitochondrial voltage-dependent anion channel-3 protein (HVDAC3), a component of the mitochondrial transition pore that regulates calcium flux [18]. HBX has also been shown to downregulate the proteasomal functions [19]. Some studies have proven that HBX could induce arrested cells to exit G0 and reenter the cell cycle, through activation of signal transduction factors such as Src kinases, MAPK, and JNK [20, 21], increasing HCC multidrug resistance and metastasis [22]. Transgenic mice models also confirmed that liver tumor development in HBX-transgenic animals was directly related to HBX expression [23–25]. Epigenetic modulation, such as DNA promoter hypermethylation of many tumor suppressive genes (TSG) by HBX, is also involved in HBV-associated carcinogenesis [5, 6]. However, the relationship of histone methylation, another major epigenetic regulation means, with HBX in HCC development needs investigation.
HBX exerts its pro-oncologic function through upregulating SMYD3 in hepatoma
Chromatin is composed of genomic DNA and nuclear proteins including histones, and serves as the template for processing genetic information. The dynamic DNA–protein structure of chromatin is influenced by epigenetic modifications on both the DNA and nucleosomal histones. Recent advances have shown that covalent histone modifications play critical roles in chromatin structure. One of the best-characterized modifications is acetylation, which is controlled by both histone acetyltransferases and deacetylases. Additionally, histone methylation has emerged as another modification that significantly impacts chromatin structure [7]. SMYD3, a histone H3-K4-specific methyltransferase, was shown to lead to transcriptional activation of downstream genes and be involved in the development of human colorectal carcinoma (CRC), breast cancer, and hepatocellular carcinoma (HCC) cells [8, 9]. In previous studies, we found that SMYD3 was highly expressed in many HCC cell lines and resected samples. Knockdown of SMYD3 inhibited HepG2 cell proliferation and migration, and increased apoptosis. The pro-oncogenesis of SMYD3 may be carried out through its hypermethylation of RIZ1 promoter CpG islands, thus abrogating the tumor suppressive activity of RIZ1 [10]. In order to clarify whether HBV regulates SMYD3 in hepatoma development, we detected SMYD3 expression in HBV-negative HepG2 and HBV-positive HepG2.2.15 cells, and found that SMYD3 expression was markedly higher in HBX-positive HepG2.2.15 than in HBX-negative HepG2 cells. To make sure that SMYD3 expression was regulated by HBV infection, we transfected HBX into HepG2 cells. SMYD3 was also increased after HBX transfection. After HBX transfection and increased SMYD3 expression in HepG2 cells, the number of HepG2 cells that progressed into S phase increased, and apoptosis decreased, accordingly. In accordance with our previous results that knockdown of SMYD3 inhibits HepG2 cell proliferation and migration, and promotes apoptosis, these results indicate that in HBV-induced HCC carcinogenesis, HBX may upregulate SMYD3 expression, which, in turn, transactivates oncogenes and/or hypermethylate promoter CpG islands of TSG such as RIZ1, and promotes HCC development or progression. Whether HBX directly regulates SMYD3 or not, and what that exact regulatory pathway is, needs to be further investigated. Recent data showed that polymorphism of variable number of tandem-repeat sequence (VNTR) of the transcriptional factor E2F-1 binding site in the 5′-flanking region of SMYD3 was a high risk for HCC and other malignancies [26–28]. Combined with the fact that HBX activation of E2F1 is an important regulatory pathway in gene transcription [29–31], these findings suggest that HBX activating nuclear factor E2F-1, and in turn combining with VNTR of E2F-1 binding site in the promoter of SMYD3, is one means of SMYD3 transactivation by HBX.
C-MYC may be one downstream gene of the HBX-SMYD3 pathway in HCC development
The overexpression of the C-MYC (MYC) oncogene is associated with tumorigenesis in a wide range of human cancers by causing inappropriate gene expression, resulting in autonomous cellular proliferation, while blocking cellular differentiation [32, 33]. Transgenic mice [34] overexpressed MYC in different cellular lineages, resulting in the formation of lymphomas, osteosarcomas, and HCC, while MYC inactivation caused sustained tumor regression. In hepatocellular carcinoma, MYC inactivation results in differentiation of tumor cells into normal hepatocytes and biliary cells; however, a subpopulation of the cells retains its neoplastic properties upon MYC inactivation, hence exhibiting tumor dormancy [15]. HBX can enhance the intracellular stability of C-MYC by blocking its ubiquitination degradation [35], increasing the rate of C-MYC transcription [36, 37] and facilitating its proliferative action, leading to HCC in transgenic mice [38–40]. Since SMYD3 is a H3-K4 HMT that transactivates oncogenes in HCC and other malignancies, we investigated whether C-MYC expression is regulated by SMYD3 in hepatoma. We found that HBX increased not only SMYD3 but also C-MYC expression in HepG2. After knockdown of SMYD3 in HepG2-HBX cells, C-MYC expression was downregulated, with cell apoptosis increasing correspondingly, indicating that SMYD3 upregulates C-MYC expression in HepG2. This upregulation may play an important pro-oncological role in HCC development and progression. Taken together, our results suggest that HBX may upregulate SMYD3 expression, which in turn increases C-MYC activity in hepatoma during HCC progression and/or development, and that C-MYC may act as a downstream or target gene of SMYD3. SMYD3 modulates transcription by various distinct mechanisms, such as chromatin opening through the histone modification that enables the transcriptional machinery to become accessible to the promoter region, and transcription elongation by the recruitment of RNA polymerase II; SMYD3 also directly regulates gene transcriptional activity through its zf-MYND domain, interacting with SMYD3 binding element (SBE) in the promoter region of target genes [8]. SBE sequences of CCCTCC and GGAGGG in the promoter region of C-MYC [41] suggest that SMYD3 may upregulate C-MYC gene expression through its zf-MYND interacting with the SBE in the promoter of C-MYC.
Combining the above with our findings that HBX increases SMYD3 expression and SMYD3 upregulates C-MYC in hepatoma, we suggest that SMYD3-C-MYC might be one of most important pathways in HBX-related HCC development and progression. Since SMYD3 is abundantly expressed in HepG2 cells, HBX may not initiate HCC development, but facilitates HCC progression through a SMYD3-C-MYC pathway.
References
Feitelson MA, Duan LX. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141–57.
Feitelson MA. Hepatitis B virus in hepatocarcinogenesis. J Cell Physiol. 1999;181:188–202. doi:10.1002/(SICI)1097-4652(199911)181:2<188::AID-JCP2>3.0.CO;2-7.
Su Q, et al. Expression of hepatitis B virus X protein in HBV infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–20. doi:10.1002/hep.510270428.
Su PF, et al. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int J Cancer. 2007;121:1257–64.
Park IY, et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–94.
Zhu R, et al. Association of p16INK4A hypermethylation with hepatitis B virus X protein expression in the early stage of HBV-associated hepatocarcinogenesis. Pathol Int. 2007;57:328–36.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi:10.1126/science.1063127.
Hamamoto R, et al. SMYD3 encodes a histone methyltransferase involved in proliferation of cancer cells. Nat Cell Bio1. 2004;6:731–40.
Hamamoto R, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–8.
Chen LB, Xu JY, Yang Z, Wang GB. Silencing SMYD3 in hepatoma demethylates RIZI promoter, induces apoptosis and inhibits cell proliferation and migration. World J Gastroenterol. 2007;13:5718–24.
Secombe J, Pierce SB, Eisenman RN. Myc: a weapon of mass destruction. Cell. 2004;117:153–6.
Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–45.
Felsher DW. Cancer revoked: oncogenes as therapeutic targets. Nat Rev Cancer. 2003;3:375–80.
Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–7.
Jain M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–4.
Su JM, et al. X protein of hepatitis B virus functions as a transcriptional co-repressor on the human telomerase promoter. Hepatology. 2007;46:402–13.
Kunizaki M, et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–65.
Huang J, Kwong J, Sun ECY, Liang TJ. Proteosome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–91.
Rahmani Z, Huh KW, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J Virol. 2002;74:2840–6.
Bouchard M, Giannakopoulos S, Wang E, Tanese E, Schneider RJ. Hepatitis B virus HBx protein activation of cyclin A-cyclin dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol. 2001;75:4247–57.
Chin R, et al. Modulation of MAPK pathways and cell cycle by replicating hepatitis B virus: factors contributing to hepatocarcinogenesis. J Hepatol. 2007;47:325–37.
Ou DP, Tao YM, Tang FQ, Yang LY. The hepatitis B virus X protein promotes hepatocellular carcinoma metastasis by upregulation of matrix metalloproteinases. Int J Cancer. 2007;120:1208–14.
Kim CM, Koike K, Saito I, Miramura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;353:317–20.
Yu DY, et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123–32.
Zheng Y, Chen WL, Louie SG, Yen TS, Ou JH. Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology. 2007;45:16–21.
Tsuge M, et al. A variable number of tandem repeats polymorphism in an E2F–1 binding element in the 5’ flanking region of SMYD3 is a risk factor for human cancers. Nat Genet. 2005;37:1104–7.
Wang H, et al. Association of the variable number of tandem repeats polymorphism in the promoter region of the SMYD3 gene with risk of esophageal squamous cell carcinoma in relation to tobacco smoking. Cancer Sci. 2008;99:787–91.
Frank B, et al. Variable number of tandem repeats polymorphism in the SMYD3 promoter region and the risk of familial breast cancer. Int J Cancer. 2006;118:2917–8.
Choi BH, Choi M, Jeon HY, Rho HM. Hepatitis B viral X protein overcomes inhibition of E2F1 activity by pRb on the human Rb gene promoter. DNA Cell Biol. 2001;20:75–80.
Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:771–8.
Wang WH, Hullinger RL, Andrisani OM. Hepatitis B virus X protein via the p38MAPK pathway induces E2F1 release and ATR kinase activation mediating p53 apoptosis. J Biol Chem. 2008;283:25455–67.
Lara-Pezzi E, Armessila A, Majano P, Redondo J, Lopez-Cabrera M. The hepatitis B virus X protein activates nuclear factor of activated T cells (NF-AT) by a cyclosporin A-sensitive pathway. EMBO J. 1999;17:7066–77.
Maguire HF, Hoeffler JP, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–4.
Wang HD, Trivedi A, Jonson DL. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol Cell Biol. 1997;17:6838–46.
Kalra N, Kumar V. The X protein of hepatitis B virus binds to the F box protein Skp2 and inhibits the ubiquitination and proteasomal degradation of c-Myc. FEBS Lett. 2006;580:431–6.
Balsano C, et al. Full-length and truncated versions of the hepatitis B virus (HBV) X protein (pX) transactivate the c-myc protooncogene at the transcriptional level. Biochem Biophys Res Commun. 1991;176:985–92.
Tu H, et al. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803–10.
Terradillos O, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404.
Madden C, Finegold M, Slagle B. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75:3851–8.
Lakhtakia R, et al. Hepatocellular carcinoma in a hepatitis B ‘X’ transgenic mouse model: a sequential pathological evaluation. J Gastroenterol Hepatol. 2003;18:80–91.
Desjardins E, Hay N. Repeated CT elements bound by zinc finger proteins control the absolute and relative activities of the two principal human c-myc promoters. Mol Cell Biol. 1993;13:5710–24.
Acknowledgement
The paper was supported by Natural Science Foundation of China (NSF) No. 30672067 and 30700190.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, L., He, J., Chen, L. et al. Hepatitis B virus X protein upregulates expression of SMYD3 and C-MYC in HepG2 cells. Med Oncol 26, 445–451 (2009). https://doi.org/10.1007/s12032-008-9144-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12032-008-9144-1