Abstract
Autophagy is disturbed in Alzheimer’s disease (AD) and maintaining normal autophagy homeostasis is a new therapeutic strategy for AD treatment. Amyloid β-derived diffusible ligands (ADDLs), the most toxic species of which are oligomeric forms of amyloid β peptide (Aβ) that originate from amyloid β precursor protein (APP) via autophagy; however, whether ADDLs are involved in autophagy-related AD pathogenesis remains unclear. In this study, we primarily defined the specific subsets of ADDLs, A-0, A-12, A-24, and A-48, which were generated from ADDL aggregation mixtures at different time courses of assembly. The secondary structures of ADDL subsets were detected by circular dichroism (CD). Neuronal or non-neuronal cells were exposed to the subsets of ADDLs in vitro, and then, autophagic markers were detected. Our results first showed that exogenous or endogenous LC3 puncta (autophagosomes) were induced in the cytoplasm of cells exposed to ADDLs and that the LC3 puncta were the strongest with A-24 exposure. Then, the CD spectroscopy data also indicated that the proportion of α-helices decreased, whereas the proportion of β-strands and β-turns increased during ADDL assembly from 0 to 24 h. In addition, the quantitative Western blot data demonstrated that the ratio of LC3B-II/I was significantly increased, and SQSTM1/p62 decreased over time. Finally, our results indicated that the level of phosphorylated p70 S6 kinase (p-p70 S6 kinase), which is a substrate protein in the MTOR pathway, and the ratio of p-p70 S6 kinase/p70 S6 kinase significantly decreased following A-24 exposure. Taken together, our data suggest that ADDL-induced abnormal autophagy is correlated with Aβ aggregation degree and the MTOR pathway, which might contribute to ADDL-induced AD pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autophagy is the natural, regulated, destructive mechanism of the cell that disassembles unnecessary or dysfunctional components. It has described as the process of sequestrating cytoplasmic proteins or organelles into the lytic compartment, is an evolutionarily conserved response to various stresses (Sutter et al. 2013). Autophagy homeostasis is a pivotal physiological condition for survival during starvation, differentiation and normal growth control (Sreelatha et al. 2013). Autophagy has become the target of therapy (Steele and Gandy 2013) in many protein misfolding disorders (Metcalf et al. 2010), particularly neurodegenerative diseases (Castillo et al. 2013), including Alzheimer’s disease (AD), which is characterized by extracellular senile plaques (SPs), intracellular neurofibrillary tangles (NTFs), neuron loss, microglial activation and impaired memory (Nassif and Hetz 2012; Nixon 2007). Recent evidence has shown that the amyloid β-peptide (Aβ) is generated from amyloid β precursor protein (APP) supplied by both autophagy and endocytosis through the turnover of APP-rich organelles and then degraded by lysosomes (Nixon 2007). In the AD brain, Aβ accumulates within the autophagic vacuoles (Yu et al. 2005), suggesting that autophagy contributes to AD pathogenesis (Garcia-Arencibia et al. 2010; Tammineni et al. 2017). However, the relationship between Aβ and autophagy remains unclear (Neely et al. 2011; Selfridge et al. 2013; Wolfe et al. 2013). Aβ-related autophagy begins before amyloid deposition (Cecarini et al. 2012; Rubinsztein et al. 2005), which indicates that Aβ may induce the autophagosomes during the early stage of AD (Son et al. 2012); however, the mechanisms of this induction are poorly understood.
In contrast to type I programmed cell death (PCD), namely, apoptosis, autophagy has been recognized as type II PCD, in which early destruction of the cytoplasm occurs either without or prior to nuclear collapse (Paglin et al. 2001). Three different autophagic pathways are known: (1) macroautophagy, (2) microautophagy, and (3) chaperone-mediated autophagy (CMA) (Garcia-Arencibia et al. 2010). Macroautophagy in AD has been a focus of recent study (Yu et al. 2005). Macroautophagy occurs in three steps: (1) the elongation of the phagophore, which is a type of isolated double-membraned structure, to engulf a portion of the cytoplasm, (2) the formation of autophagosomes or initial autophagic vacuoles (AVis) (Kabeya et al. 2000; Morvan et al. 2009), through the fusion of the edges of the phagophore; these vacuoles are the primary morphological marker of macroautophagy, and (3) the formation of autolysosomes or the degradative autophagic vacuoles (AVds) (Kabeya et al. 2000; Morvan et al. 2009), which are the result of autophagosomes fusing with lysosomes, providing the sites where the cytosolic contents are degraded by lysosomal hydrolases (Garcia-Arencibia et al. 2010).
Two types of signaling pathways regulate mammalian autophagy: mammalian target of rapamycin (MTOR)-dependent and mTOR-independent pathways. MTOR is a sensor of nutrient signals. (1) Under nutrient-rich conditions, MTOR acts to suppress autophagy; in contrast, under starvation or stress conditions, MTOR is suppressed and MTOR-mediated phosphorylation of Atg13 and ULK1 is inhibited, ultimately triggering autophagy. One of the well-characterized downstream effectors of MTOR activation is p70 S6 kinase (p70 S6 K) (Hartmann et al. 2013; Jefferies et al. 1997). (2) The MTOR-independent pathway is regulated by several factors, such as intracellular inositol and inositol 1,4,5-trisphosphate (IP3) levels, Ca-calpains, and reactive oxygen species (ROS). No reports regarding which of the pathways mentioned above, if any, would be induced by ADDLs have been published.
The changes of autophagy related marker proteins have been reported within AD brains (Caccamo et al. 2016): (1) Rat microtubule-associated protein 1 light chain 3 (MALP1LC3 or LC3) is the first specific marker protein for autophagic vacuoles in mammalian cells and is the first identified protein localized in the autophagosome membrane (Kabeya et al. 2000; Klionsky et al. 2012; Wang et al. 2009). Similar to the yeast homolog Apg8/Aut7p (Kirisako et al. 1999), LC3 is essential for the formation of autophagosomes (Kabeya et al. 2000). Two forms of LC3, which are called LC3-I and LC3-II, are produced post-translationally in various cells. LC3-I is cytosolic, whereas LC3-II is membrane-bound and exists both inside and outside autophagosomes (Kabeya et al. 2000). Mutational analysis has suggested that LC3-I is formed by the removal of the 22 amino acid C-terminus from newly synthesized LC3, followed by the conversion of a fraction of LC3-I into LC3-II (Kabeya et al. 2000). LC3-II remains membrane-bound even after fusion with lysosomes, after which LC3-II on the cytosolic face of autophagosomes can be recycled (to LC3-I) by Atg4, whereas the LC3-II on the inner face of the membrane is degraded (Kabeya et al. 2000). The human LC3 family has three isoforms: MAP1LC3A (LC3A), MAP1LC3B (LC3B), and MAP1LC3C (LC3C) (He et al. 2003; Tanida et al. 2004). LC3B elevation or LC3B-I/II (Wohlgemuth et al. 2007) shifting and the LC3B-II/I ratio are also used to test autophagy activation in neurodegenerative diseases (Bjorkoy et al. 2009; Congdon et al. 2012; Nakagaki et al. 2013; Solano et al. 2012; Zschocke et al. 2011). (2) Sequestosome 1 (SQSTM1/p62, p62) (Caccamo et al. 2016), one of the selective substrates of autophagy, has been considered involved in Alzheimer’s disease. Since SQSTM1 accumulates when autophagy is inhibited, and decreases when autophagy is induced, SQSTM1 may be considered a marker of autophagic flux (Bjorkoy et al. 2009).
Autophagy works as damage and repair roles in AD pathogenesis (Rubinsztein et al. 2005), and may promote either cell death or cell survival (Hung et al. 2009; Pelizzaro-Rocha et al. 2013). Increased numbers of AVis have been detected in the AD brain before amyloid deposition (Rubinsztein et al. 2005); however, whether this phenomenon is the result of elevated autophagic activity or decreased autophagosome-lysosome fusion remains unclear. Aβ is a self-associating peptide produced during the process of autophagy (Nixon 2007). Aβ can induce autophagy (Fonseca et al. 2013), and also can be degraded by autophagy activity (Nogalska et al. 2010). Therefore, specific amyloid species may play distinct roles in autophagy induction in various early stages of AD (Agholme et al. 2012; Lai and McLaurin 2012; Zheng et al. 2011). Amyloid β-derived diffusible ligands (ADDLs), which are the most neurotoxic subset of Aβ42 oligomers, have been recognized as the leading cause of pathological or cognitive impairment in AD (Krafft and Klein 2010; Lambert et al. 1998). ADDLs were found to kill mature neurons in organotypic central nervous system tissues at nanomolar concentrations (Lambert et al. 1998); and to induce tau hyperphosphorylation at residues Ser202 and Thr205 in B103 neuroblastoma cells and at residues Thr231 and Ser404 in hippocampal neurons. Dodecameric structures are the relevant neurotoxins (Lambert et al. 1998). Therefore, we have tested our hypothesis that autophagy may be induced by different types of ADDLs in cells and that conformational changes in ADDLs would contribute to potential autophagic stress in AD.
In this study, we report the first investigation regarding the relationship between ADDL conformational changes and autophagical reactivity. To gain insights into the potential capacities of autophagy induction, four subsets of ADDLs were used to treat mammalian cells, and then autophagy markers were analyzed. To further reveal the mechanism and pathway of ADDL-induced autophagy, the secondary structures of ADDLs were measured by circular dichroism (CD) spectroscopy, and autophagy-related signal molecules were probed by Western blot. To our knowledge, this report is the first to describe the aggregation relationship of ADDL-induced autophagy, and our data provide insight for further studies regarding aggregate-related autophagic stress in AD.
Materials and Methods
Plasmid Construction
Total RNA was extracted from neuroblastoma cells (N2a), and reverse transcriptase polymerase chain reaction (RT-PCR) was used to obtain (microtubule-associated protein 1 light chain 3, MAP1LC3 or LC3) gene. LC3 gene was cloned to pEGFP-C1 (Clontech) vector to construct pEGFP-C1-LC3 plasmid. Primer sequences, designed for RT-PCR to clone LC3 gene were
Forward, 5’-GGAAGATCTATGCCGTCCGAGAAGAC-3’.
Reverse, 5’-CCGGAATTCTTACACAGCCATTGCTGTC-3′.
Chemicals and Antibodies
Rapamycin, 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP), 3-Methyladenine (3-MA), and the rabbit anti-LC3B antibody were purchased from Sigma-Aldrich, Ltd. (Sigma-Aldrich, St. Louis, MO, USA). Mouse monoclonal anti-p62/SQSTM1 was from abcam (Cambridge, MA, USA). Phospho-p70 S6 kinase (Thr389) (1A5) mouse mAb, p70 S6 kinase (49D7) rabbit mAb, anti-Beclin 1, and anti-Atg5 were obtained from Cell Signaling Technology, Inc. (Cell Signaling, Danvers, MA, USA). Mouse anti-Aβ (17–24) (4G8) was purchased from Covance (Covance, Emeryville, CA, USA). Anti-β actin was obtained from Santa Cruz Biotechnology Inc. (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
ADDL Preparation
The Aβ 42 peptides (DAEFRH DSGYEV HHQKLV FFAEDV GSNKGA IIGLMV GGVVIA) obtained from the manufacturer (Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd., Shanghai, China) were > 95% pure. ADDL preparation was performed according to the methods described previously (Lambert et al. 2001; Zhang et al. 2011) with some modifications. Briefly, Aβ42 was dissolved in ice-cold 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) (Sigma-Aldrich,St. Louis, MO, USA) and aliquoted into microcentrifuge tubes. Hexafluoro-2-propanol was removed by evaporation with traces removed under vacuum and the tubes were stored at − 80 °C. An aliquot of Aβ42 was dissolved in anhydrous dimethyl sulfoxide (DMSO) (Sigma- Aldrich, St. Louis, MO, USA) to 5 mM, which was then added to ice-cold DMEM/F12 medium without phenol red (Life Technologies, Carlsbad, CA, USA) to 100 μM. This solution was incubated at 4 °C for 0, 12, 24, and 48 h and then centrifuged at 14,000×g for 10 min. The supernatant was defined as the subsets of ADDLs and was composed of fibril-free solutions of oligomers (ADDLs) and monomers. The ADDL samples which were collected at 0, 12, 24, and 48 h were designated A-0 (ADDLs-0 h), A-12 (ADDLs-12 h), A-24 (ADDLs-24 h), and A-48, respectively, and were evaluated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentrations were determined using a BCA protein assay kit (PIERCE, Rockford, IL, USA).
Cell Culture, Transfection, and Treatment
Human glioblastoma astrocytoma cells U87 MG (U87, ATCC HTB-14), human embryonic kidney cells (HEK293, ATCC CRL-1573), Pheochromocytoma PC12 (ATCC CRL-1721), and mouse neuroblastoma Neuro-2a cells (N2a, ATCC CCL-131) were obtained from the Cell Center of Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences, CAS (Shanghai, China). The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) or Dulbecco’s modified Eagle’s/F12 medium (DMEM/F12) (Life Technologies, Carlsbad, CA, USA), which was supplemented with 10% heat-inactivated fetal bovine serum, 0.03% glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Proliferating cell cultures were maintained in a 5% CO2 humidified incubator at 37 °C. U87 Cells were transfected with the GFP-LC3-expressing plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. HEK293 cells were transfected with pEGFP-LC3 or pEGFP (pEGFP-C1; Clontech, Palo Alto, CA, USA) using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The culture medium was then changed by DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS and 700 μg/mL G418 (Invitrogen, Carlsbad, CA, USA) 24 h after transfection. The culture medium was changed once every 3 days. Two weeks later, stable clones were isolated. Stable clones of HEK293-GFP-LC3 or HEK293-GFP were selected for the following further studies.
Primary Culture of Mouse Hippocampal Neurons
Primary neuronal cultures were prepared from the brain of P0 pups of C57BL/6J mice. The mice were purchased from Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) and maintained in Department of Laboratory Animal Science, Peking University Health Science Center. Protocols involving experimentation on animal subjects were developed following the guide for care and use of laboratory animals from Peking University Research Council and approved by the Animal Care and Use Committee in Peking University. Hippocampi of mouse brain were dissected in ice cold HBSS (Gibco) under a dissection microscope (Leica, Zoom 2000, Leica, Wetzler, Germany). Dissociation was performed in ice-cold papain solution (0.01% w/v Papain and 0.01% w/v DNase) and then incubated at 37 °C for 45 min. After mechanically dissociation, cells were strained through cell strainer and were counted and plated (400,000 cells/well) in plating medium at 37 °C for 2 h. When cells attached to the coverslips, all plating medium was aspirated and replaced with neuronal medium. And new medium (30% of starting volume) was added every 3 days until needed. After 15 days, cells were treated with 5 μM ADDLs.
Immunohistochemical Staining for LC3B and Aβ
Cell cultures were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde in PBS for 20 min at 4 °C. After washing in PBS, the cells were blocked for 1 h at room temperature with 2% BSA in PBS, and incubated with rabbit anti-LC3B antibody (Sigma-Aldrich, St. Louis, MO, USA) (1:100) or with mouse anti-Aβ (17–24) (4G8) (SIGNET, Emeryville, CA, USA) in a blocking solution containing 0.1% Tween 20 overnight at 4 °C. Cells were incubated in FITC-labeled goat anti-rabbit IgG (1:100, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature and visualized using a fluorescence microscope (Nikon TE2000-S, Tokyo, Japan) (Tizon et al. 2010).
Western Blot
For the preparation of total cell extracts, U87 cells grown in T25 flask were washed with cold PBS, scraped, re-suspended, and homogenized with RIPA buffer (pH 7.5) (Paitel et al. 2004; Tizon et al. 2010), with the following modifications: 50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 50 mM PIPES/NaOH, 2 mM EDTA, 0.1% CHAPS, 5 mM DTT, 20 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml aprotinin A, 1 mM PMSF, and a commercial protease inhibitor mixture (Complete Protease Inhibitor Mixture; Roche Molecular Biochemicals, Mannheim, Germany) (pH 7.5) on ice for 40 min. Additionally, 1% phosphatase inhibitors (250 mM sodium fluoride, 50 mM sodium orthovanadate, 50 mM sodium pyrophosphate and 50 mM β-glycerophosphate salt) were used for phosphorylated proteins. Then, the samples were centrifuged at 12,000×g for 10 min at 4 °C to remove nuclei and large debris and to collect the supernatant. The protein concentration in the supernatant was measured using a BCA protein assay kit (PIERCE, Rockford, IL, USA). Equal amounts of each protein sample (50 μg) were boiled, loaded, resolved by 10–16% SDS-polyacrylamide gel electrophoresis (16% gel for separating LC3B-I/II), and then electroblotted onto nitrocellulose membranes. After blocking the non-specific binding sites with 5% skim milk in TBS-T, the blots were subsequently probed with appropriate primary antibodies, including rabbit anti-LC3B, anti-p62/SQSTM1, anti-p70 S6 kinase, anti-phospho p70 S6 kinase, anti-Beclin 1, and anti-Atg5 probed further with horseradish peroxidase-conjugated secondary antibodies and then visualized through exposure on X-ray films after the addition of SuperSignal West Pico chemiluminescent substrate (PIERCE, Rockford, IL, USA). The specific bands on the blot were quantified and analyzed using Quantity One software (Bio-Rad).
Determination of Circular Dichroism (CD) Spectroscopy
ADDL assembly in vitro was measured utilizing JASCO-J700 circular dichroism (CD) spectra chromatography (JASCO International Co., Ltd., Tokyo, Japan). The scanning conditions were as follows: range, 190 to 240 nm; 1 mm path length quartz cuvettes; sensitivity, 5 m°/cm; resolution, 0.5 nm; slit, 1 nm; time constant, 8 s; scanning speed, 5 mm/min; and 3 scans for each sample, at room temperature. The blank buffer, which was at the same ion condition, was served as a blank reference control.
Statistical Analyses
Experiments in these studies were analyzed using one-way or two-way ANOVAs and the chi-square test (SPSS19.0). An α level of 0.05 was used for all statistical significance tests.
Results
Autophagy Induction by ADDLs in the Neuronal Cells
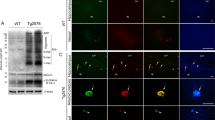
To demonstrate the regulation of autophagosome formation induced by ADDLs, mouse neuroblastoma Neuro-2a (N2a) cells and human glioblastoma astrocytoma U87 MG (U87) cells were treated with ADDLs for 24 h, and with the ADDL assembly buffer (buffer) treatment and non-treatment cells as control groups. Immunofluorescence probed with anti-LC3B antibody (1:200) showed that endogenous LC3 puncta (indicating autophagosomes) accumulated significantly in ADDL group (Fig. 1A, a), compared with buffer (Fig. 1A, b) and cell control (Fig. 1A, c) groups (p < 0.01, Fig. 1C). At the same time, co-treatment with 3-Methyladenine (3-MA), an inhibitor (PI3K) of autophagy, attenuated the accumulation of autophagosomes (p < 0.01, Fig. 1C) (Fig. 1A, d, e, and f), but did not results in complete inhibition in ADDLs group (Fig. 1A, d).
Autophagy induction by ADDLs in the neuronal cells. ADDLs were added to N2a or U87-GFP-LC3 cells at a concentration of 10 μM in DMEM medium for 24 h. Serum deprivation was used as an autophagy induction control, and 3-MA was used as an autophagy inhibition control. (A) N2a cells were treated with different chemicals for 24 h: (a) ADDLs; (b) ADDL assembly buffer control (buffer); (c) untreated cell control; (d) (e) and (f) respectively represent (a) (b) and (c) with 5 μM of 3-MA trearment. The endogenous LC3B puncta were probed by rabbit anti-LC3B (1:200) and detected by immunofluorescence; the red arrows pointed the LC3B positive puncta. (B) GFP-LC3 transfected U87cells were exposed to ADDLs without (a) or with (b) 3-MA for 24 h. Vector transfected cells were used as control (c). The GFP-LC3 puncta were detected using fluorescence microscopy. (C) and (D) Quantification of the average number of endogenous LC3B puncta and GFP-LC3 puncta per cell. (E) GFP-LC3 transfected U87cells were treated with A-0 (ADDLs at 0 h) (a), A-12 (ADDLs at 12 h) (b), A-24 (ADDLs at 24 h) (c), and A-48 (Aβ aggregates at 48 h) (d) for 24 h, and buffer (e) or serum free (f) was used as control. LC3 puncta were visualized through fluorescence microscopy. (F) LC3B puncta per cell were quantified in each group. (G) U87 cells were treated with ADDLs and Aβ monomers (monAβ) for 24 h, LC3B-I-II and SQSTM1/p62 were detected via Western blot. (H) U87 cells were treated with ADDL species aggregated at 0, 12, 24, and 48 h. The whole cell extracts were analyzed via Western blot. A shift from the one-band to two-band model of LC3B and degradation of SQSTM1/p62 were shown in. The ratio of LC3B-II/LC3B-I (I) and the ratio of LC3B-II/β-actin (J) in (H) were quantified. The experiments were repeated at least three times. (*p < 0.05, **p < 0.01. Scale bar indicates 50 μm in (A) and 20 μm in (B) and (E)
To visually display the LC3 puncta-induced by ADDLs, GFP-LC3B-transfected U87 cells were treated with ADDLs for 24 h, and with the vector-transfected cells as control groups. The LC3B puncta in the U87 cells were directly observed under a fluorescence microscope. As shown in Fig. 1B, the LC3B puncta were induced in the ADDL group (Fig. 1B, a; ADDLs group vs vector control group, p < 0.01, Fig. 1D) but were only weakly induced in 3-MA treatment (Fig. 1B, b; ADDLs group vs ADDLs + 3-MA group, p < 0.01, Fig. 1D) and vector control (Fig. 1B, c) groups. This result confirmed that ADDLs could stimulate the accumulation of autophagy vacuoles in neuronal cells.
To visualize the aggregation-related autophagy induced by ADDLs, we try to investigate whether ADDL-induced autophagosomes were increased along with Aβ aggregation. Equal amounts of ADDL species were collected from aggregate reactions at 0, 12, 24, and 48 h. Correspondingly, these samples were designated as A-0 (ADDLs at 0 h), A-12 (ADDLs at 12 h), A-24 (ADDLs at 24 h), and A-48 (Aβ aggregates at 48 h). Then, the specific ADDL subsets mentioned above (A-0, A-12, A-24, and A-48) were added to GFP-LC3B-transfected U87 cells at a concentration of 10 μM in DMEM medium for 24 h. The LC3B puncta in the cells were directly observed under a fluorescence microscope. As shown in Fig. 1E, the LC3B puncta were induced in the A-0 (Fig. 1EA, a), A-12 (Fig. 1E, b), A-24 (Fig. 1E, c), and serum free groups (Fig. 1E, f), but were only weakly induced in the A-48 (Fig. 1E, d) and ADDL buffer control (Fig. 1E, e) groups. LC3B-positive puncta in each cell were counted, and the quantitative analysis is shown in Fig. 1F. As expected, the appearance of autophagosomes increased in parallel with the formation of distinct oligomer species during ADDL assembly from 0 to 24 h, and this increase was significant in the A-24-treated group (p < 0.01, Fig. 1F).
To investigate how autophagy was regulated by ADDLs aggregation, levels of LC3B and SQSTM1/p62 were analyzed through Western blot in U87 cells treated with ADDLs and Aβ monomers to determine whether LC3B-I/II shifting and p62-degradation occurred. As shown in Fig. 1G, both of the monomers of Aβ and ADDLs produced a marked and significant increase in LC3B-II protein levels compared with buffer control group; cell treatment with ADDLs produced a decrease in SQSTM1/p62 levels after 24 h of incubation, whereas monomeric preparations of Aβ caused a slight, but not significant decrease in SQSTM1/p62 protein levels.
To determine whether specific ADDL species affected autophagy differently, ADDL species aggregated at 0, 12, 24, and 48 h were used to treat the U87 cells. The cells were harvested 24 h later, and then whole cell protein samples were extracted and analyzed by Western blot. A shift from a one-band to two-band model of LC3B and degradation of SQSTM1/p62 occurred, as shown in Fig. 1H. The ratio of LC3B-II/LC3B-I increased (p < 0.05) with ADDLs aggregated from 0 to 24 h but not increased any more with A-48 treatment (Fig. 1I). In addition, SQSTM1/p62 decreased (p < 0.01) especially with A-24 treatment (Fig. 1J). These data showed that the autophagy induced by ADDLs may associate with the aggregate changes of Aβ peptides.
ADDLs Induce Accumulation of Autophagosomes in HEK293 Cells
In order to investigate whether the results can be showed in other cells, the same ADDL subsets mentioned above (A-0, A-12, A-24, and A-48) were added to HEK293-GFP-LC3 stable cells at a concentration of 10 μM in DMEM medium for 24 h. The LC3 dots in the HEK293-GFP-LC3 cell line were directly observed under a fluorescence microscope. As shown in Fig. 2A, the LC3 dots were induced in the A-12 (Fig. 2A, b), A-24 (Fig. 2A, c), A-48 (Fig. 2A, d), and rapamycin groups (Fig. 2A, e) but were only weakly induced in the A-0 (Fig. 2A, a) and ADDL buffer control (Fig. 2A, f) groups. LC3-positive dots in each cell were counted, and the quantitative analysis is shown in Fig. 2B. The results were similar as Fig. 1E, the appearance of autophagosomes increased in parallel with the formation of distinct oligomer species during ADDL assembly, and this increase was significant in the A-24-treated group (p < 0.01, Fig. 2B). The results suggested that there might be a conformational relationship in ADDL-induced autophagy.
Autophagy induced by various ADDL species in HEK293 cells. (A) The specific ADDL species (a: A-0, b: A-12, c: A-24, and d: A-48) were added to HEK293-GFP-LC3 stable cells at a concentration of 10 μM in DMEM medium for 24 h. Rapamycin (100 nM) treatment was used as an autophagy induction control. The GFP-LC3 puncta were detected using fluorescence microscopy. (B) Quantification of the average number of EGFP-LC3 puncta per cell. The experiments were repeated at least three times. *p < 0.01. Scale bar = 50 μm
ADDLs But Not Aβ Monomers Colocalize with LC3B Puncta
In order to investigate the conformation relationship between ADDLs and autophagy induction, human glioblastoma astrocytoma N2a cells were treated with ADDLs and rhodamine-conjugated Aβ monomers respectively for 24 h. Colocalization of Aβ aggregates (red) and LC3B puncta (green) could be detected well in ADDL-treatment and serum free group but not in Aβ monomer treatment and cell control group (Fig. 3A). LC3B-Aβ co-localized puncta (yellow) in each cell were counted, and the quantitative analysis was shown in Fig. 3B. Compared with control group, the co-localized puncta increased significantly (p < 0.05) in ADDL-treatment group or in serum free group but not in Aβ monomer group (Fig. 3B).
Colocalization of Aβ and LC3B puncta was induced by ADDLs. (A) ADDLs and rhodamine-conjugated Aβ monomers were added to monolayer N2a cells at a concentration of 10 μM in DMEM medium for 24 h. Serum deprivation was used as an autophagy induction control. ADDLs were probed by rabbit anti-Aβ (4G8) (1:500) and LC3B puncta were probed by rabbit anti-LC3B (1:200). Red color indicates Aβ, green color indicates LC3B, blue color indicates the nucleus stained by DAPI, and yellow color indicates the merge of Aβ and LC3B. (B) Quantitative analysis of colocalized Aβ aggregates and LC3B puncta in (A). *p < 0.05. Scale bar = 50 μm
Characterization of Specific ADDL Species
To determine the contribution of Aβ aggregation to autophagy induction, equal amounts of the ADDL species, A-0 (ADDLs at 0 h), A-12 (ADDLs at 12 h), A-24 (ADDLs at 24 h), and A-48 (Aβ aggregates at 48 h) were analyzed by SDS-PAGE. At the same time, equal amounts of non-aggregate Aβ peptide were loaded to show the same amount of Aβ was used. These samples were detected using the Coomassie Brilliant Blue staining with SDS-PAGE (Fig. 4A), and the results showed that the Aβ aggregates in the gel could be divided into three groups based on their molecular weight, as shown in Fig. 4A (marked on the right edge): (1) the aggregates in Group I were about 6.5 kD in size; (2) the aggregates in Group II were approximately 14.3~20 kD; and (3) the aggregates in Group III were higher order of oligomers that smeared about 175 kD in A-48. The ADDLs were the mixtures of low molecular weight oligomers and some higher order aggregates with the molecular weight ranged from higher than 6.5 to 20 kD. In Fig. 4B, C, quantitative analysis showed that the ratio of ADDLs/monomers and the ratio of Group II/Group I increased significantly with aggregation, particularly in A-24 (p < 0.05). The results indicate the dynamical conformation changes in the different time course of ADDLs formation.
Preparation and identification of particular ADDL species. The aggregate products were collected at 0, 12, and 24 h during the ADDL assembly process from 0 to 24 h. Non-boiled samples were loaded, separated by SDS-PAGE and stained by the Coomassie Brilliant Blue. (A) Profiles of gel stained with Coomassie Blue G250. M indicates protein molecular markers. (B) Ratio of ADDLs/Aβ monomers in ADDLs, as analyzed by the band intensities in (A). (C) Quantitative analysis of ratio of Group II/Group I in (A) (*, p < 0.05)
A Relationship Between Conformational Changes and ADDL-Induced Autophagy
To investigate whether ADDL-induced autophagy is conformation-dependent, circular dichroism data were collected for ADDL aggregation. The dynamic change in the secondary structure was calculated and analyzed using the online software from Dichroweb (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml). The results are shown in Table 1, the proportion of α-helices decreased, whereas the proportion of β-strands increased with ADDL aggregation from A-0 to A-24. These data suggested that the ability to induce autophagy may have a reverse correlation with the proportion α-helices but a positive correlation with random coils.
ADDL-Induced Autophagy Was MTOR-Associated
The effect of ADDLs on the level of phosphorylated p70S6 kinase (p-p70S6 kinase), which is a substrate protein in the MTOR pathway, was analyzed to investigate the autophagy signaling pathway induced by ADDLs. U87 cells were incubated in serum-containing medium in the presence of A-0, A-12, and A-24 for 24 h. As a positive control, serum-free condition was used for autophagy induction. Cell lysate proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and blotted with antibodies directed against either p70S6 kinase or p-p70S6 kinase (Fig. 5A). Band intensities were measured, and p-p70S6 kinase levels were calculated relative to total p70S6 kinase values. Although the level of p70S6 kinase did not decreased by ADDLs treatment (Fig. 5A), the degree of its phosphorylation was significantly reduced by ADDL exposure in serum-containing medium (Fig. 5A). Meanwhile, there was a slight increase detected in the expression level of Beclin 1 and Atg5 (Fig. 5A) in ADDL treatment groups.
ADDL-induced autophagy is MTOR-associated. U87 cells were treated with A-0, A-12, A-24 for 24 h, with serum deprivation as autophagy control and with no treatment as the cell control. Autophagy pathway markers were analyzed via Western blot (A). Quantitative analysis of the ratio of p-p70/p70 is shown in (B) (p-p70 indicates p-p70 S6 kinase, **p < 0.01)
The ratio of p-p70 S6 kinase/p70 S6 kinase was calculated and the results indicated that the levels of the molecule decreased with ADDL aggregation (Fig. 5B). The phosphorylation state of p70S6 kinase is an established index of MTOR activity, with the dephosphorylation of p70S6 kinase indicating MTOR inhibition (Erlich et al. 2007; Lavieu et al. 2006). Therefore, these data indicate that ADDLs induce autophagy through inhibiting the MTOR-signaling pathway.
ADDL-Induced Autophagosome Accumulation in Primary Cultures of Mouse Hippocampal Neurons
To further confirm the findings from cell lines, primary culture of mouse hippocampal neurons screened by neuronal medium was treated with ADDLs. Immunofluorescence showed that endogenous LC3 significantly accumulated in ADDLs group (Fig. 6A) compared with buffer control group (p < 0.01, Fig. 6B). This data suggested that the relationship between conformation and ADDL-induced autophagy could be duplicated in neuronal primary cultures.
Primary neuronal cultures treated with 5 μM ADDLs confirm findings from cell lines on autophagy. (A) Immunofluorescence probed with anti-LC3B antibody (1:200) to show autophagy puncta; (B) Quantification of the cells with LC3B puncta. The experiments were repeated at least three times. *p < 0.05. Scale bar = 50 μm
Discussion
To determine the mechanism of Aβ-induced autophagy, we focused on the three types of distinct misfolded forms during ADDL which assembly at 0, 12, and 24 h. We investigated the relationship between the secondary structures of specific ADDL species and autophagy and then detected the autophagy markers to clarify the involved signaling pathway. As expected, we found that ADDL-induced autophagy is conformation-dependent and may develop via the MTOR pathway.
Conformational change is the primary characteristic of Aβ self-associating misfolding (Fawver et al. 2012; Ringman et al. 2012); however, the precise proportions of various structural components in the most toxic Aβ aggregates and even in individual ADDL species remain unclear. This study is the first to measure and analyze the secondary structure components of the specific ADDL species A-0, A-12, A-24, and A-48. As shown in Table 1, the proportion of α-helices were 12.66, 12.86, and 9.00% in the ADDL species A-0, A-12, and A-24, respectively, which indicated the decrease in the proportion of α-helices during aggregate development. In contrast, the proportion of β-strands and β-turns elevated rapidly at 24 h. These results are partially consistent with the previous reports (Guglielmotto et al. 2014; Liu et al. 2011): the oligomeric interfaces in Aβ display a variety of sheet-to-sheet pairing angles, offering a structural explanation for the heterogeneity of amyloid oligomers.
We found that autophagic activation is highly related to the secondary structures of ADDLs. As shown in Fig. 1, the number of LC3 puncta paralleled the degree of aggregation of the ADDLs. A-24, which had the lowest proportion of α-helices, induces the highest level of autophagy among the subsets of ADDLs. Thus, the decrease in the proportion of α-helical components might initiate autophagy during cell survival. Additionally, among the three groups of ADDLs (shown in Fig. 4), Group II is the most common component in A-0, A-12, and A-24, and the ratio of Group II/I (Fig. 4) even increased with aggregate formation. Our data showed that, there were Aβ aggregates with different molecular weight in ADDLs samples. The bands of Group II showed changes with time course, but that of Group I maintained constant density. The ratio of Group II/I (Fig. 4) was used to indicate that Group II contributes much more to autophagy induction than the other two groups of oligomers. The aggregates in Group II have molecular weights similar to those of the toxic oligomers reported in our previous work (Zhang et al. 2011), which contribute to cognitive decline in SAMP8 mice. Further experiments should be performed to purify aggregates from these groups to differentiate their distinct functions. In addition, the total amount of LC3B in Fig. 1G is also elevated. However, LC3B-II/I ratio can be used to test autophagy activation if it increased significantly compared to control.
We found that ADDLs induced autophagy in several cell lines including N2a, U87MG, PC12 (Fig. S1), HEK293, and Saos-2 (data not shown), which suggested that ADDLs induced autophagy may be a ubiquitous phenotype. However, the brightness of the LC3 puncta is much less in HEK293 or Saos-2 than it in neuroblastoma cells. Therefore, our data showed that neurons are more prone to ADDLs induced autophagy.
Autophagy is activated via either MTOR-dependent or MTOR-independent pathway (Garcia-Arencibia et al. 2010). The dephosphorylation of p70S6 kinase, as shown in Fig. 5, suggested that ADDL-induced autophagy is MTOR-dependent. Our data showed that after treatment with Aβ aggregates in the course of ADDL assembly (A-0, A-12, and A-24), the elevated LC3BII/I ratio was consistent with autophagy induction and with MTOR inhibition.
Recently, autophagy, which is responsible for the degradation of long-lived proteins, protein aggregates and organelles, has become a popular target for AD therapy (Zhu et al. 2013). Moreover, autophagy can be induced as a cellular response to various physiological and pathological stimuli. Thus, autophagy dysfunction has been implicated in the pathogenesis of neurodegenerative disorders, which includes impairing autophagosome clearance, reducing autophagosome synthesis, and blocking autophagosome removal processes (Garcia-Arencibia et al. 2010; Salminen et al. 2013). Serving as a site for Aβ generation, the abnormal pool of accumulated autophagic vesicles may additionally enhance AD pathology (Garcia-Arencibia et al. 2010; Salminen et al. 2013).
Most large aggregate-prone proteins are precluded from entering the narrow pore of the proteasome barrel after oligomerization; these proteins are thus highly dependent on autophagy rather than on the ubiquitin-proteasome system for their clearance (Harris and Rubinsztein 2011). Therefore, controlling of autophagy may be used as a therapy for Alzheimer’s disease. However, the prospect of upregulating autophagy as a therapy for certain neurodegenerative diseases raises some challenges. For example, earlier treatment with rapamycin would be desired to delay the onset of disease. However, the resultant long-term inhibition of MTOR is extremely troublesome because MTOR regulates many other cellular processes independent of autophagy (Brown et al. 1994; Brunn et al. 1996). As such, a detailed understanding of the pathways regulating autophagy will be of paramount importance in AD therapy (Garcia-Arencibia et al. 2010).
We can draw the following conclusions from the data in the present study: (i) the secondary structure in ADDL assembly contributes to autophagy initiation; (ii) ADDL-induced autophagy is MTOR-associated. Precisely characterization of autophagy-related species in ADDLs is a further interesting investigation which will help to understand the mechanisms in development of autophagy targeted therapeutic strategies for AD.
Abbreviations
- AD:
-
Alzheimer’s disease
- Aβ:
-
beta amyloid
- APP:
-
amyloid β precursor protein
- ADDLs:
-
amyloid β-derived diffusible ligands
- MAb:
-
monoclonal antibody
- SPs:
-
senile plaques
- NFTs:
-
neurofibrillary tangles
- PCD:
-
programmed cell death
- CMA:
-
chaperone-mediated autophagy
- AVis:
-
initial autophagic vacuoles
- AVds:
-
degradative autophagic vacuoles
- LC3:
-
microtubule-associated protein 1 light chain 3
- MTOR:
-
mammalian target of rapamycin
- SQSTM1/p62:
-
sequestosome 1
- TMB:
-
3,3′,5,5′-tetramethylbenzidine.
References
Agholme L, Hallbeck M, Benedikz E, Marcusson J, Kagedal K (2012) Amyloid-beta secretion, generation, and lysosomal sequestration in response to proteasome inhibition: involvement of autophagy. J Alzheimers Dis 31(2):343–358. https://doi.org/10.3233/JAD-2012-120001
Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T (2009) Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452:181–197. https://doi.org/10.1016/S0076-6879(08)03612-4
Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL (1994) A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369(6483):756–758. https://doi.org/10.1038/369756a0
Brunn GJ, Williams J, Sabers C, Wiederrecht G, Lawrence JC Jr, Abraham RT (1996) Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J 15(19):5256–5267
Caccamo A, Ferreira E, Branca C, Oddo S (2016) p62 improves AD-like pathology by increasing autophagy. Mol Psychiatry. https://doi.org/10.1038/mp.2016.139
Castillo K et al (2013) Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 9:1308–1320. https://doi.org/10.4161/auto.25188
Cecarini V et al (2012) Crosstalk between the ubiquitin-proteasome system and autophagy in a human cellular model of Alzheimer's disease. Biochim Biophys Acta 1822:1741–1751. https://doi.org/10.1016/j.bbadis.2012.07.015
Congdon EE et al (2012) Methylthioninium chloride (methylene blue) induces autophagy and attenuates tauopathy in vitro and in vivo. Autophagy 8:609–622. https://doi.org/10.4161/auto.19048
Erlich S, Alexandrovich A, Shohami E, Pinkas-Kramarski R (2007) Rapamycin is a neuroprotective treatment for traumatic brain injury. Neurobiol Dis 26(1):86–93. https://doi.org/10.1016/j.nbd.2006.12.003
Fawver JN et al (2012) Probing and trapping a sensitive conformation: amyloid-beta fibrils, oligomers, and dimers. J Alzheimers Dis 32:197–215. https://doi.org/10.3233/JAD-2012-120880
Fonseca MB, Sola S, Xavier JM, Dionisio PA, Rodrigues CM (2013) Amyloid beta peptides promote autophagy-dependent differentiation of mouse neural stem cells : Abeta-mediated neural differentiation. Mol Neurobiol. https://doi.org/10.1007/s12035-013-8471-1
Garcia-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC (2010) Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol 21:691–698. https://doi.org/10.1016/j.semcdb.2010.02.008
Guglielmotto M et al (2014) Abeta1-42 monomers or oligomers have different effects on autophagy and apoptosis. Autophagy 10:1827–1843. https://doi.org/10.4161/auto.30001
Harris H, Rubinsztein DC (2011) Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol 8(2):108–117. https://doi.org/10.1038/nrneurol.2011.200
Hartmann B, He X, Keller F, Fischereder M, Guba M, Schmid H (2013) Development of a sensitive phospho-p70 S6 kinase ELISA to quantify mTOR proliferation signal inhibition. Ther Drug Monit 35(2):233–239. https://doi.org/10.1097/FTD.0b013e3182804c9b
He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JO, Yu L (2003) Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 278(31):29278–29287. https://doi.org/10.1074/jbc.M303800200
Hung SY, Huang WP, Liou HC, Fu WM (2009) Autophagy protects neuron from Abeta-induced cytotoxicity. Autophagy 5(4):502–510
Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G (1997) Rapamycin suppresses 5'TOP mRNA translation through inhibition of p70s6k. EMBO J 16(12):3693–3704. https://doi.org/10.1093/emboj/16.12.3693
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19(21):5720–5728. https://doi.org/10.1093/emboj/19.21.5720
Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147(2):435–446. https://doi.org/10.1083/jcb.147.2.435
Klionsky DJ, Abdalla FC, Abeliovich H et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8(4):445–544. https://doi.org/10.4161/auto.19496
Krafft GA, Klein WL (2010) ADDLs and the signaling web that leads to Alzheimer's disease. Neuropharmacology 59(4-5):230–242. https://doi.org/10.1016/j.neuropharm.2010.07.012
Lai AY, McLaurin J (2012) Inhibition of amyloid-beta peptide aggregation rescues the autophagic deficits in the TgCRND8 mouse model of Alzheimer disease. Biochim Biophys Acta 1822:1629–1637. https://doi.org/10.1016/j.bbadis.2012.07.003
Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL (1998) Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci U S A 95(11):6448–6453. https://doi.org/10.1073/pnas.95.11.6448
Lambert MP, Viola KL, Chromy BA, Chang L, Morgan TE, Yu J, Venton DL, Krafft GA, Finch CE, Klein WL (2001) Vaccination with soluble Abeta oligomers generates toxicity-neutralizing antibodies. J Neurochem 79(3):595–605
Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P (2006) Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem 281(13):8518–8527. https://doi.org/10.1074/jbc.M506182200
Liu C, Sawaya MR, Cheng PN, Zheng J, Nowick JS, Eisenberg D (2011) Characteristics of amyloid-related oligomers revealed by crystal structures of macrocyclic beta-sheet mimics. J Am Chem Soc 133(17):6736–6744. https://doi.org/10.1021/ja200222n
Metcalf DJ, Garcia-Arencibia M, Hochfeld WE, Rubinsztein DC (2010) Autophagy and misfolded proteins in neurodegeneration. Exp Neurol 238(1):22–28. https://doi.org/10.1016/j.expneurol.2010.11.003
Morvan J, Kochl R, Watson R, Collinson LM, Jefferies HB, Tooze SA (2009) In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy 5(5):676–689
Nakagaki T et al (2013) FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy 9:1386–1394. https://doi.org/10.4161/auto.25381
Nassif M, Hetz C (2012) Autophagy impairment: a crossroad between neurodegeneration and tauopathies. BMC Biol 10(1):78. https://doi.org/10.1186/1741-7007-10-78
Neely KM, Green KN, LaFerla FM (2011) Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a gamma-secretase-independent manner. J Neurosci 31:2781–2791. https://doi.org/10.1523/JNEUROSCI.5156-10.2010
Nixon RA (2007) Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci 120:4081–4091. https://doi.org/10.1242/jcs.019265
Nogalska A, D'Agostino C, Engel WK, Klein WL, Askanas V (2010) Novel demonstration of amyloid-beta oligomers in sporadic inclusion-body myositis muscle fibers. Acta Neuropathol 120(5):661–666. https://doi.org/10.1007/s00401-010-0737-3
Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, Domingo D, Yahalom J (2001) A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res 61(2):439–444
Paitel E, Sunyach C, Alves da Costa C, Bourdon JC, Vincent B, Checler F (2004) Primary cultured neurons devoid of cellular prion display lower responsiveness to staurosporine through the control of p53 at both transcriptional and post-transcriptional levels. J Biol Chem 279(1):612–618. https://doi.org/10.1074/jbc.M310453200
Pelizzaro-Rocha KJ, de Jesus MB, Ruela-de-Sousa RR, Nakamura CV, Reis FS, de Fatima A, Ferreira-Halder CV (2013) Calix[6]arene bypasses human pancreatic cancer aggressiveness: downregulation of receptor tyrosine kinases and induction of cell death by reticulum stress and autophagy. Biochim Biophys Acta. https://doi.org/10.1016/j.bbamcr.2013.07.010
Ringman JM, Tomic JL, Coppola G, Elashoff D, Gylys KH, Glabe CG (2012) Conformation-dependent oligomers in cerebrospinal fluid of presymptomatic familial Alzheimer's disease mutation carriers. Dement Geriatr Cogn Dis Extra 2(1):652–657. https://doi.org/10.1159/000345771
Rubinsztein DC et al (2005) Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 1:11–22
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M (2013) Impaired autophagy and APP processing in Alzheimer's disease: the potential role of Beclin 1 interactome. Prog Neurobiol. https://doi.org/10.1016/j.pneurobio.2013.06.002
Selfridge JE, L E, Lu J, Swerdlow RH (2013) Role of mitochondrial homeostasis and dynamics in Alzheimer's disease. Neurobiol Dis 51:3–12. https://doi.org/10.1016/j.nbd.2011.12.057
Solano RM, Casarejos MJ, Gomez A, Perucho J, de Yebenes JG, Mena MA (2012) Parkin null cortical neuronal/glial cultures are resistant to amyloid-beta1-42 toxicity: a role for autophagy? J Alzheimers Dis 32(1):57–76. https://doi.org/10.3233/JAD-2012-120406
Son SM, Jung ES, Shin HJ, Byun J, Mook-Jung I (2012) Abeta-induced formation of autophagosomes is mediated by RAGE-CaMKKbeta-AMPK signaling. Neurobiol Aging 33:1006 e1011–1006 e1023. https://doi.org/10.1016/j.neurobiolaging.2011.09.039
Sreelatha A, Bennett TL, Zheng H, Jiang QX, Orth K, Starai VJ (2013) Vibrio effector protein, VopQ, forms a lysosomal gated channel that disrupts host ion homeostasis and autophagic flux. Proc Natl Acad Sci U S A 110(28):11559–11564. https://doi.org/10.1073/pnas.1307032110
Steele JW, Gandy S (2013) Latrepirdine (Dimebon(R)), a potential Alzheimer therapeutic, regulates autophagy and neuropathology in an Alzheimer mouse model. Autophagy 9:617–618. https://doi.org/10.4161/auto.23487
Sutter BM, Wu X, Laxman S, Tu BP (2013) Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 154:403–415. https://doi.org/10.1016/j.cell.2013.06.041
Tammineni P, Ye X, Feng T, Aikal D, Cai Q (2017) Impaired retrograde transport of axonal autophagosomes contributes to autophagic stress in Alzheimer's disease neurons. Elife 6. https://doi.org/10.7554/eLife.21776
Tanida I, Ueno T, Kominami E (2004) Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem 279(46):47704–47710. https://doi.org/10.1074/jbc.M407016200
Tizon B, Sahoo S, Yu H, Gauthier S, Kumar AR, Mohan P, Figliola M, Pawlik M, Grubb A, Uchiyama Y, Bandyopadhyay U, Cuervo AM, Nixon RA, Levy E (2010) Induction of autophagy by cystatin C: a mechanism that protects murine primary cortical neurons and neuronal cell lines. PLoS One 5(3):e9819. https://doi.org/10.1371/journal.pone.0009819
Wang AL, Boulton ME, Dunn WA Jr, Rao HV, Cai J, Lukas TJ, Neufeld AH (2009) Using LC3 to monitor autophagy flux in the retinal pigment epithelium. Autophagy 5:1190–1193
Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA Jr (2007) Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res 10(3):281–292. https://doi.org/10.1089/rej.2006.0535
Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA (2013) Autophagy failure in Alzheimer's disease and the role of defective lysosomal acidification. Eur J Neurosci 37(12):1949–1961. https://doi.org/10.1111/ejn.12169
Yu WH et al (2005) Macroautophagy—a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol 171:87–98. https://doi.org/10.1083/jcb.200505082
Zhang Y et al (2011) Administration of amyloid-beta42 oligomer-specific monoclonal antibody improved memory performance in SAMP8 mice. J Alzheimers Dis 23:551–561. https://doi.org/10.3233/JAD-2010-091195
Zheng L et al (2011) Macroautophagy-generated increase of lysosomal amyloid beta-protein mediates oxidant-induced apoptosis of cultured neuroblastoma cells. Autophagy 7:1528–1545
Zhu XC, Yu JT, Jiang T, Tan L (2013) Autophagy modulation for Alzheimer's disease therapy. Mol Neurobiol 48(3):702–714. https://doi.org/10.1007/s12035-013-8457-z
Zschocke J, Zimmermann N, Berning B, Ganal V, Holsboer F, Rein T (2011) Antidepressant drugs diversely affect autophagy pathways in astrocytes and neurons—dissociation from cholesterol homeostasis. Neuropsychopharmacology 36(8):1754–1768. https://doi.org/10.1038/npp.2011.57
Acknowledgements
We thank Dr. Cong-Cong He (Northwestern University) for her advice on autophagy detection and critical reading of this manuscript. We thank National Students’ Platform for Innovation and Entrepreneurship Training Program (160170002 and 170170005) to support X.W. in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81100809 and 81271417), the Beijing Natural Science Foundation (7152090), and the Fundamental Research Funds for the Central University of China (2015JBM096) to Y.Z.
Author information
Authors and Affiliations
Contributions
JW, FF, and YZ performed cell culture, treatment, and LC3 puncta detection via IF or fluorescence microscopy, as well as analysis and figure artwork. FF, SG, and YZ contributed to preparation and identification of particular ADDL species via SDS-PAGE and CD, as well as analysis and figure artwork. JW and SG carried out Western blot to detect LC3BII/I, SQSTM1/p62, p70 S6 kinase, and p-p70 S6 kinase, as well as analysis and figure artwork. XP and WS contributed to some experiments in colocalization of LC3B and Aβ. XW performed some cell culture experiments. HT and JH contributed to part work in experiment design. YZ supervised the study and writing of the manuscript. All authors revised the article critically for intellectual content and have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Protocols involving experimentation on animal subjects were developed following the guide for care and use of laboratory animals from Peking University Research Council and approved by the Animal Care and Use Committee in Peking University.
Consent for Publication
All authors agree to publication of this study.
Competing Interests
The authors declare that they have no competing interests.
Electronic supplementary material
Figure S1
Autophagy induction by ADDLs in Pheochromocytoma PC12 cells. ADDLs were added to GFP-LC3 transfected PC12 cells at a concentration of 10 μM in DMEM medium for 24 h. The GFP-LC3 puncta were detected using fluorescence microscopy. Scale bar indicates 50 μm. (JPEG 36 kb)
Rights and permissions
About this article
Cite this article
Wen, J., Fang, F., Guo, SH. et al. Amyloid β-Derived Diffusible Ligands (ADDLs) Induce Abnormal Autophagy Associated with Aβ Aggregation Degree. J Mol Neurosci 64, 162–174 (2018). https://doi.org/10.1007/s12031-017-1015-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-017-1015-9