Abstract
We showed previously that the neuropeptide pituitary adenylyl cyclase-activating polypeptide (PACAP) negatively regulates proliferation of postnatal rat retinal progenitor cells through the downregulation of cyclin D1 in a cAMP/protein kinase A dependent manner. In the present study, we describe by microarray analysis several putative PACAP targets regulated by different transcription factor families. One of these families is the Sp/Klf family of transcriptional factors capable of regulating cyclin D1, and among members, we demonstrate by immunocytochemistry that KLF4 is expressed throughout rat retinal development by retinal progenitor cells and in most differentiated cell types. Using retinal explants preparations, PACAP treatment can transiently increase Klf4 mRNA levels; from electrophoretic mobility shift assays, PACAP is also able to increase the nuclear KLF4 content. From these results, we suggest that KLF4 may be involved in the anti-proliferative effects of PACAP as one mechanism regulating progenitor cell transition from proliferation to differentiation throughout retinal development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vertebrate retina is composed of six types of neurons and one type of glia (Müller glia) organized in alternate cellular and synaptic layers. During development, multipotent retinal progenitor cells (RPCs) proliferate and pass through an overlapping sequence of events in which the cells commit to particular fates, exit the cell cycle, and differentiate to give rise to various retinal cell types in an orderly manner that is conserved in vertebrates (Cepko 1999; Marquardt and Gruss 2002; Hatakeyama and Kageyama 2004).
The proliferation of RPCs is regulated by the coordination of cell-intrinsic and cell-extrinsic factors. Soluble factors such as sonic hedgehog (Shh), fibroblast growth factors (FGFs), epidermal growth factor (EGF), transforming growth factor-α (TGF-α) and transforming growth factor-β (TGF-β), glutamate and pituitary adenylyl cyclase-activating polypeptide (PACAP), among others, affect cell proliferation in the retina (Anchan et al. 1991; Lillien and Cepko 1992; Anchan and Reh 1995; Jensen and Wallace 1997; Young and Cepko 2004; Close et al. 2005; Martins et al. 2006; Martins and Pearson 2008; Njaine et al. 2010). PACAP is a neuroactive peptide of the secretin/glucagon/vasoactive intestinal peptide (VIP) superfamily. It was first isolated for its ability to induce the production of cAMP in the anterior pituitary of rats (Miyata et al. 1989). Its precursor is post-translationally processed into two biologically active products, PACAP38 and PACAP27 (Miyata et al. 1989, 1990), which share high amino acid homology with VIP. In the nervous system, PACAP has been associated to the regulation of cell proliferation (Lu et al. 1998; Suh et al. 2001; Carey et al. 2002; Njaine et al. 2010), differentiation (Grumolato et al. 2003; Borba et al. 2005), and cell survival (Silveira et al. 2002; Kanekar et al. 2010; Baxter et al. 2011; Reglodi et al. 2012). Our group demonstrated that mRNA for both PACAP as well as its receptors is detected throughout retinal development and that PACAP behaves as an antiproliferative agent that negatively regulates cyclin D1 expression in a cAMP/PKA-dependent manner (Njaine et al. 2010).
Several transcription factors directly modulate the expression of cell cycle genes, through interactions with their regulatory regions. KLF4 is both a well-known regulator of cyclin D1 expression (Chen et al. 2003) and a target of PACAP signaling in sympathetic neurons (Braas et al. 2007). KLF4 (originally termed gut Kruppel-like factor (GKLF)) is involved in the control of cell proliferation, tissue homeostasis, inhibition of neuronal fiber outgrowth, and promotion of stemness in coordination with other transcription factors (Ehlermann et al. 2003; Katz et al. 2005; Rowland et al. 2005; Takahashi and Yamanaka 2006; Takahashi et al. 2007; Moore et al. 2009). In controlling cell proliferation, the overexpression of Klf4 in a colon cancer cell line has been shown to decrease DNA synthesis through downregulation of cyclin D1 and upregulation of p21 (Chen et al. 2003). Moreover, expression of KLF4 is decreased in certain tumor cell lines and tumors of the digestive tract, which supports its role as a tumor suppressor (Chen et al. 2001; Yoon et al. 2003; Katz et al. 2005). Notwithstanding, a context-dependent oncogene function has also been proposed for KLF4 (Rowland et al. 2005).
Recent studies showed that sustained expression of Klf4 inhibits neural stem cells (NSCs) self-renewal and yet attenuates neuronal differentiation in the cerebral cortex (Qin et al. 2011; Qin and Zhang 2012), suggesting that the expression of Klf4 must be finely tuned to allow proper cell cycle exit and differentiation. The roles of KLF4 in retinal development have not been well described; however, it was recently shown to act as a transcriptional repressor of axon regeneration in retinal ganglion cells (Moore et al. 2009).
To further establish and characterize the pleiotropic roles of PACAP in retinal development, we examined the abilities for PACAP to regulate the retinal transcriptome. In good agreement with our previous work (Njaine et al. 2010), the data showed that PACAP treatment decreased cyclin D1 transcript levels. In assessing the roles of Klf4, our findings indicate that Klf4 expression may participate in the control of cell proliferation and differentiation throughout retinal development and that its regulation by PACAP signaling may represent a mechanism underlying PACAP-mediated inhibition of retinal progenitor cells proliferation.
Material and Methods
Material
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum, and Trizol were from Invitrogen (Carlsbad, CA, USA). PACAP38 was obtained from Bachem (Torrance, CA, USA). Primary antibodies are described in Table 1. Sp1 consensus and mutant oligonucleotides (cat# 2502 and cat# 2503) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Secondary antibodies linked to horseradish peroxidase (HRP) (cat# 7074 and cat# 7076) were from Cell Signaling (Beverly, MA, USA). Fluorescent secondary antibodies used were Alexa Fluor 555-conjugated goat anti-rabbit and Alexa Fluor 488-conjugated donkey anti-mouse from Molecular Probes (Eugene, OR, USA). First-strand cDNA kit was from GE Healthcare (Little Chalfont, UK). In addition, recombinant DNase I (cat# AM1906) was from Ambion (Austin, TX, USA), Power SYBR Green PCR Master Mix (cat# 4367659) was from Applied Biosystems (Foster City, CA, USA), Luminata™ Western Chemiluminescent HRP Substrate was from Millipore (cat# WBLUC0100) (Billerica, MA, USA), and GoTaq® DNA Polymerase (cat# M829) was from Promega (Fitchburg, WI, USA).
Tissue Culture and Histology
All experimental procedures with animals were approved by the Ethics Committee on Animal Experimentation of the Health Sciences Center of the Federal University of Rio de Janeiro (CEUA/CCS/UFRJ) under protocol #IBCCF121, based on currently accepted international rules. Retinal explants were maintained in vitro as previously described (Rehen et al. 1996; Varella et al. 1997, 1999; Silveira et al. 2002). In brief, P1 Lister hooded or Sprague Dawley rats were euthanized by instantaneous decapitation, their eyes were dissected, and retinas were cut into 1-mm2 pieces (total of eight explants per P1 retina). For each experimental design, both retinas from at least four rats were used in each of two to five independent experiments. All explants in each experiment were pooled, and at least eight explants were used for each replicate of each experimental group. Retinal explants were maintained in DMEM supplemented with 5 % fetal bovine serum, 2 mM glutamine, 10 U/mL penicillin, and 100 μg/mL streptomycin at pH 7.4 for 24 h (except where noted) in Erlenmeyer flasks with orbital agitation. For histological and immunofluorescence analysis, at the end of each experiment, the explants were fixed by immersion in 4 % paraformaldehyde in sodium phosphate buffer (pH 7.4) for 2 h and cryoprotected in 30 % sucrose in 0.2 M phosphate buffer for cryosectioning (10 μm sections). Except for microarray analysis, most experiments were performed with Lister hooded rats; key experiments were also reproduced with Sprague Dawley rats as described in “Results.” In all experiments, PACAP was used at 10 nM in accordance with our previous studies to allow comparative analyses (Silveira et al. 2002; Njaine et al. 2010).
Microarray
Following retinal explant exposure to vehicle or 10 nM PACAP38 (4 h), total RNA was prepared using RNASTAT-60 total RNA/mRNA isolated reagent (Tel-Test B Inc., Friendswood, TX, USA) and submitted to the Vermont Cancer Center Microarray Facility for target preparation using standard Affymetrix protocols. The Bioinformatics Core Facility at the University of Vermont performed the initial analyses. The data with statistical analysis was included as Supplementary Material. Subsequent gene ontology analyses were performed with GOToolBox (http://genome.crg.es/GOToolBox) using the rat genome as the reference data set. WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/) was used in the enrichment analysis for targets of transcription factors among PACAP-regulated genes.
mRNA Extraction, cDNA Synthesis, and RT-PCR
Total RNA from rat retinas of the following developmental stages: E18, E21, P1, P3, P4, P6, P10, P16, P22, P35, and P45 was extracted with Trizol following manufacturer’s instructions. RNA integrity was confirmed by visualization of Rn18s and Rn28S in 1 % agarose gel electrophoresis. RNA was treated with DNA-free kit following manufacturer’s instructions, quantified with NanoDropTM Spectrophotometer ND-1000 (Thermo Scientific), and stored at −80 °C. Quantity and quality of RNA extracted were assessed to confirm good RNA yields and purity with a mean A260/A280 ratio of 1.9 ± 0.2. DNA contamination was ruled out by standard PCR and agarose gel electrophoresis. cDNA was synthesized from 1 μg of RNA with pd(N)6 random primers, as described in kit manual. For RT-PCR, amplification was performed with a pair of primers that recognize the N-terminal region Klf4 FW 5′-TGATGGGCAAGTTTGTGCTGAAGG-3′, Klf4-REV 5′-ACAGTGGTAAGGTTTCTCGCCTGT-3′; Klf5 FW 5′-TCGCCAAATATCCAACCTGTCCGA-3′, Klf5 REV 5′-TCTTGACAATCTGTACTGGGTCCT-3′; Klf7 FW 5′-AGCAGGACTTTGCACACACAAGG-3′, Klf7 REV 5′-ACCCACACACATACCCACTGTCAT-3′; or Actb FW 5′-TCATGAAGTGTGACGTTGACATCCGT-3′, Actb REV 5′-CCTAGAAGCATTTGCGGTGCACGATG-3′.
Quantitative Real-Time Polymerase Chain Reaction
Quantitative real-time polymerase chain reaction (qRT-PCR) reactions were performed with Power SYBR Green PCR Master Mix using an ABI7500 System (Applied Biosystems). Control without reverse transcriptase was included to ensure that the results were not caused by amplification of genomic DNA. PCR product identity was confirmed by electrophoresis and by melting-point analysis. Primers used for qRT-PCR were: Klf4 FW 5′-ACCCCTCTCTCTTCTTCGG-3′, Klf4 REV 5′-GAGTTCCTCTCGCCAACG-3′; Klf5 FW 5′-TATCTGACACCTCAGCTTCCTCCA-3′, Klf5 REV 5′-TGTGTTACGCATGGTCTCTGGGAT-3′; and Klf7 FW 5′-TGACACCAGCTGGAGCAGTTAAGA-3′, Klf7 REV 5′-ACAGCGATGGACCCTCTTCTTGTT-3′. For endogenous control, Gapdh (FW 5′-GACATGCCGCCTGGAGAAA-3′, Gapdh REV-5′-AGCCCAGGATGCCCTTTAGT-3′) and Mapk1 (FW 5′-TGTTGCAGATCCAGACCATG-3′, Mapk1 REV 5′-CAGCCCACAGACCAAATATCA-3′) were previously validated as reference genes for expression analysis over retinal development (Rocha-Martins et al. 2012). Actb (FW 5′-TCATGAAGTGTGACGTTGACATCCGT-3′ and REV 5′-CCTAGAAGCATTTGCGGTGCACGATG-3′) was also determined to be adequate for comparisons on a specific developmental stage (Njaine et al. 2010). Expression levels of all the samples were determined in the exponential phase by the number of cycles necessary to reach the prior established threshold (Ct). The comparative Ct method (ΔΔCt) was used to determine the target quantity in sample as compared with the mean of different reference genes in combination and relative to a calibrator [(Cttarget gene − Ctreference gene)sample − (Cttarget gene − Ctreference gene)calibrator]. A mathematical correction similar to the one of the software qBase, which is based on the use of the average of ΔCt of all groups, was applied to define the calibrator (Hellemans et al. 2007; Rocha-Martins et al. 2012).
Immunofluorescence
To examine KLF4 immunoreactivity, retinal explants from E18, P1, and P10 rats were acutely fixed as described above; the methodological details for each antibody are described in Table 1. For PACAP-induced changes in protein content, P1 explants were maintained in vitro with culture medium for 3 h. After this period, the explants were treated either with or without 10 nM PACAP38 for 1 h, and the tissue was processed as described above. The sections were immunostained overnight with anti-KLF4, or anti-cyclin D1. 4,6-Diamidino-2-phenylindole (DAPI) was used for nuclear staining. All immunofluorescence reactions were processed using Alexa Fluor conjugate secondary antibodies (Alexa Fluor 555-conjugated goat anti-rabbit or Alexa Fluor 488-conjugated donkey anti-mouse) at room temperature for 2 h.
Image Acquisition
Images were acquired with structural illumination microscopy (Imager. M2 Apotome; Zeiss GmbH, Jena, Germany). Proper filter sets were used for specific detection of each fluorochrome. Apotome images were acquired with a Plan-Neofluar 40/1.3 Oil DIC objective, using Axio Vision 4.8.2 software and AxioCam MRm. Confocal microscope images were acquired with a LSM 510 META (Zeiss GmbH, Jena, Germany) using the 488-nm band of an Ar laser and the 543-nm band of an He/Ne laser for excitation of Alexa Fluor 488 and Alexa Fluor 555, respectively. All paired images were obtained with the same acquisition parameters.
Western-Blotting
Retinal explants were washed with phosphate-buffered saline, and total retinal protein was extracted (10 mM Tris–HCl, 150 mM NaCl, 1 % NP-40, 1 % Triton X-100, 5 mM EDTA, 0.1 % sodium dodecyl sulfate, 1 % sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, 1 μg/mL pepstatin, 1 μg/mL leupeptin, 1 % sodium orthovanadate, and 50 mM sodium fluoride). Protein concentrations in the lysates were determined by the Lowry assay (Lowry et al. 1951). Lysates (30 μg) were electrophoresed in 12 % sodium dodecyl sulfate–polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked with either 5 % bovine serum albumin or 5 % non-fat dry milk and incubated with primary antibodies for KLF4 (1:2,000) or MAPK1/ERK2 (1:4,000) (Rocha-Martins et al. 2012) followed by secondary antibodies (1:2,000 goat HRP-conjugated anti-rabbit and 1:2,000 horse HRP-conjugated anti-mouse) for protein detection using a Luminata system according to the manufacturer’s instructions.
Electrophoretic Mobility Shift Assay
Oligonucleotides (5 pmol) were end-labeled with [γ-32P] ATP, T4 polynucleotide kinase and purified in Promega Biogel 30 column (Fitchburg, WI, USA). For binding reactions, 10-μg nuclear protein extracts were incubated with labeled consensus or mutant probe in 1× binding buffer (10 mM Hepes pH 7.9, 60 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10 % glycerol, and 1 μg of poly(dI-dC)–poly(dI-dC) sodium salt) for 40 min at room temperature. The samples were then fractionated on 4 % native PAGE in 0.5× TBE (1× TBE is 45 mM Tris/borate/1 mM EDTA) at 4 °C. The following oligonucleotides were used: Oligo Sp1 wildtype (wt), 5′-ATT CGA TCG GGG CGG GGC GAG C-3′; and Oligo Sp1 mutated (mut), 5′ATT CGA TCG GTT CGG GGC GAG C-3′. For supershift analysis, protein extracts were incubated for 1 h with 2, 4, or 6 μg of the anti-KLF4 at 4 °C prior to probe addition. The antibody (H-180; Santa Cruz Technology) was directed against an epitope corresponding to amino acids 1–180 mapping to the amino-terminal activation domain of KLF4. The assays were analyzed on an Amersham Phosphorimager (Amersham Pharmacia).
Statistical Analyses
Prism 5.0 software (GraphPad Software, San Diego, CA, USA) was used for all analyses. Statistical comparisons between two experimental groups were made with Student’s two-tailed unpaired t test. For experiments with more than two groups, either one-way or two-way ANOVA tests were used, as appropriate, followed by Tukey or Bonferroni multiple comparisons tests, respectively. Results are reported as means ± standard error of the mean (SEM), and the level of significance was set as p < 0.05.
Results
Microarray Analyses from Neonatal Rat Retina After PACAP Treatment
We and others have shown previously that PACAP signaling in the retina prevents cell death from apoptotic challenges (Silveira et al. 2002; Atlasz et al. 2009, 2011; Wada et al. 2013; Szabadfi et al. 2014) and is antiproliferative (Njaine et al. 2010). To understand which gene targets are regulated by PACAP signaling in developing retina, we performed a microarray analysis after acute (4 h) treatment of newborn Sprague Dawley rat retinal explants with PACAP and performed comparative analyses against untreated controls. The rat strain and treatment time were selected initially to allow comparisons of PACAP-regulated genes in other neuronal populations (Braas et al. 2007). All genes plotted (261 genes) in visual two-dimensional representation of the data showed at least a 1.2-fold difference between control and PACAP-treated groups, where log2-fold change (M) for each gene was expressed as a function of −log10p. Upregulated genes (191 genes) are plotted in the upper half of the graph, whereas the downregulated transcripts (70 genes) are shown in the lower half (Fig. 1a). A survey of the functional annotations of PACAP gene targets by Gene Ontology (http://www.geneontology.org) revealed a wide spectrum of biological activities (Fig. 1b). Biological processes significantly enriched in this data set confirmed, at the expression level, the pleiotropic functions of PACAP signaling in the retina and reinforced the hypothesis of its involvement in the control of cell proliferation and differentiation. Some enriched biological processes match previously reported functions for PACAP signaling in retinal tissue, including its neuroprotective effect in the developing retina (e.g., “regulation of programmed cell death”), antiproliferative effect (e.g., “regulation of cell cycle, cell proliferation, and neurogenesis”), and cAMP/PKA activation (e.g., “response to cAMP”). Other ontology groupings, such as axonal projection and synaptic transmission, have not been well-studied with respect to PACAP function and may be important for retinal differentiation. WebGestalt software (http://bioinfo.vanderbilt.edu/webgestalt) was used to analyze enriched targets of transcription factors in this data set. This approach may indicate possible mediators of PACAP signaling that may account for changes in gene expression upon treatment. Many of the identified genes were targets of cAMP-responsive element binding-protein (CREB). PACAP has been shown to potently stimulate CREB phosphorylation in a variety of cell types, including retinal progenitor cells (Njaine et al. 2010).
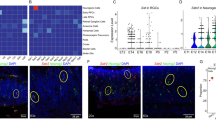
PACAP signaling regulates targets classified in diverse cellular processes. Retinal explants were treated for 4 h with 10 nM PACAP. a PACAP regulated transcripts identified on the Affymetrix 230A microarray are represented as correlation between gene expression ratio (log2) and p value (−log10(p)). All 261 genes plotted showed at least a 1.2-fold variation between control and PACAP treated groups, p < 0.05. Upregulated transcripts are shown in the upper half and downregulated transcripts in the lower half of the graph. b Gene Ontology of biological processes among PACAP target genes. Data represent enrichment and number of target genes in each category. c Enrichment analysis for the targets of different transcription factors among PACAP regulated genes. The number of target genes in each category is shown
Among transcripts, the notable decrease in Sprague Dawley retinal cyclin D1 transcript levels in response to PACAP treatment was also identified in our related PACAP work using Lister hooded rats (Njaine et al. 2010). Several rat strains have been used to examine retina development, and although the PACAP-mediated changes in Sprague Dawley cyclin D1 expression were congruent with those in the well-studied Lister hooded rats, we sought to verify that the PACAP effects in Sprague Dawley retina were meaningful and similar among strains. In cell proliferation assays, PACAP significantly decreased the number of bromo-deoxyuridine (BrdU)-labeled cells in P1 Sprague Dawley retinas more than 60 % compared to untreated controls (control = 100.0 ± 21.9 % vs PACAP = 38.4 ± 3.3 %; p < 0.05). As the antiproliferative effects of PACAP were similar to those observed in Lister hooded rat retinas (Njaine et al. 2010), these results suggested that the PACAP-regulated transcripts identified in the microarray analyses may represent broad and shared retinal targets across rat strains.
In assessing mechanisms underlying the anti-proliferative actions of PACAP in retinal progenitor cell development, we reasoned that KLF4 may be one upstream regulator of cyclin D1. In fact, CREB has been described to bind KLF4 gene regulatory regions (Zhang et al. 2005), and transcription factor analyses showed one CREB binding site in the putative rat Klf4 promoter (Supplementary Figure). It is well-established that KLF4 may act on cyclin D1 promoter (Chen et al. 2003), and our previous work in sympathetic neurons demonstrated robust PACAP effects in upregulating Klf4 (Braas et al. 2007). Although an increase in the expression of Sp/Klf family members was not noted under the parameters we defined for the current array study, previously described targets for Sp/Klf transcription factors were enriched among PACAP-regulated genes (Fig. 1c). Based on these findings, we examined the expression pattern and the possible regulation by PACAP of Klf4 and other Sp/Klf family members during retinal development.
Expression of KLFs Throughout Retinal Development
Given the similarities in PACAP responses between strains, the Lister hooded rats were used in the remaining studies for experimental coherency with previous work (Njaine et al. 2010). The expression of Klf4, as well as two related family members Klf5 and Klf7, was studied in several stages of retinal development (from embryonic day (E) 18 up to postnatal day (P) 45) by RT-PCR. We detected Klf4, Klf5, and Klf7 transcripts in all stages analyzed, although the levels for Klf5 transcripts appeared low from P16 to P45 (Fig. 2a). We decided to analyze further the expression of Klf4; notably, Klf4 mRNA levels appeared to increase slightly from P10 to mature stages. This dynamic pattern of Klf4 expression was confirmed when qRT-PCR analysis was performed (Fig. 2b) with endogenous controls more suitable for quantitative analyses among various developmental stages (Rocha-Martins et al. 2012). In addition, we quantified KLF4 protein content by western blotting (Fig. 2c, d). Interestingly, unlike transcript levels, protein content was elevated from P1 until P10 and declined at later stages of retinal development.
Expression of Klf4 during retinal development. a Representative RT-PCR for Klf4, Klf5, and Klf7 from E18 through P45 rat retinas. b qRT-PCR and c western blotting for KLF4 transcript and protein show temporal differences in expression during development. d Densitometric analysis of tissue KLF4 protein levels normalized to ERK2. Results are presented as means ± SEM pooled from three independent experiments. **p < 0.01 and *p < 0.05 in one-way ANOVA followed by Tukey’s multiple comparison’s test
To investigate the expression pattern of KLF4 in specific cell layers, we examined KLF4 staining patterns during development. KLF4 immunoreactivity was present both in nuclei as well as in the cytoplasm, and was not limited to the proliferative neuroblastic layer (NBL) of the retina. In agreement with a previous study (Moore et al. 2009), intense staining was detected in ganglion cells (GCs) in E18 and P1 (Figs. 3 and 4).
Analysis of KLF4 expression in specific cell layers early in retinal development. Sections from E18 rat retinas were stained in green for KLF4 (a and i) and in red for PCNA (b) or βIII-tubulin (j). Sections from P1 rat retinas were stained in green for KLF4 (e) and in red for PCNA (f). DAPI was used for nuclear stain (c, g, and k) and merged images are shown in d, h, and l. Results are representative of two independent experiments, and images were obtained with Imager. M2 Apotome microscope. GCL ganglion cell layer, NBL neuroblastic layer. Calibration bar = 20 μm
Analysis of KLF4 expression in specific cell populations in P10 retinas. Sections from P10 rat retinas were stained in green for KLF4 (a, e, and i) and in red for PKC, a bipolar cell marker (b); glutamine synthetase (GS), a Muller cell marker (f), or rhodopsin, a rod marker (j). DAPI was used for nuclear stain (c, g, and k) and merged images are shown in d, h, and l. Results are representative of two independent experiments, and images were obtained with Imager. M2 Apotome microscope. GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, OPL outer plexiform layer, ONL outer nuclear layer. Calibration bar = 20 μm
To test whether both progenitors and differentiated cells express KLF4, we used a panel of antibodies to label specific retinal cell types. E18 and P1 rat retinas were stained with antibodies for KLF4 and proliferating cell nuclear antigen (PCNA), a marker of retinal progenitor cells (Fig. 3a–d and e–h, respectively). E18 retinas were also processed for both KLF4 and βIII-tubulin to identify early generated neurons, such as ganglion cells (Fig. 3i–l). As expected, we identified KLF4 immunoreactivity both in βIII-tubulin positive cells and in proliferating cells in E18 and P1 (Fig. 3). In P10 retinas, KLF4 was detected in the inner nuclear layer (INL), which in mature retina contains bipolar, horizontal, and amacrine neurons, as well as Müller glial cells (Fig. 4). Double labeling for KLF4 and PKC-alpha (Fig. 4a–d) or KLF4 and glutamine synthetase (GS) (Fig. 4e–h) showed for the first time the expression of KLF4 in rod-bipolar cells and Müller cells, respectively. At this stage, KLF4 immunoreactivity was also detected in βIII-tubulin positive cells, but it was absent in rhodopsin-labeled rod photoreceptors at the outer nuclear layer (ONL) (Fig. 4i–l).
Klf4 mRNA and Protein Levels Increase Transiently in Response to PACAP
From the results above, we assessed whether changes in the expression of Sp/Klf family members could be detected upon PACAP signaling in P1 rat retinas. As transcription factors of this family are described as early response genes, transcript levels for Klf4, 5, and 7 and KLF4 protein content were measured after stimulation of retinal tissue with PACAP. Klf4 transcripts were slightly increased already at 0.5 h after PACAP treatment although a significant difference was only observed at 1 h after treatment. This upregulation of Klf4 mRNA was transient as the levels declined to baseline after 3–6 h of treatment (Fig. 5a). The same PACAP-induced Klf4 transcription expression pattern was observed with Sprague Dawley retinas (data not shown). Hence, while there can be strain differences in retinal development and function, our studies to date have failed to reveal any differences with respect to the antiproliferative effect of PACAP in the developing retina. The PACAP-induced changes in expression were specific to Klf4, since no effect was observed for Klf5 (Fig. 5b) or Klf7 (Fig. 5c). KLF4 protein levels were also transiently increased after 1 h of PACAP treatment (Fig. 5d, e). Three hours after PACAP treatment, the levels of KLF4 are similar in treated and untreated conditions (Fig. 5d, e).
PACAP induces a transient increase in the expression of Klf4 in neonatal retina. Protein and RNA extracts were obtained from P1 rat retinal explants maintained in vitro for the indicated times and treated with PACAP. qRT-PCR analysis for (a) Klf4 (n = 4), (b) Klf5 (n = 3), and (c) Klf7 (n = 3) after treatment for 0.5, 1, 3, and 6 h. Western-blotting and densitometric analysis for KLF4 after treatment for 1 and 3 h (d, e) (n = 3).***p < 0.001 in two-way ANOVA followed by Bonferroni’s post hoc test (a) **p < 0.01 in two-way ANOVA followed by Bonferroni’s post hoc test (e)
As described above, KLF4 immunoreactivity was detected in both GCL as well as NBL. As we had observed a transient induction of Klf4 expression by qRT-PCR and western blotting, we tested KLF4 immunoreactivity after 1 h PACAP treatment. Interestingly, KLF4 labeling at the NBL was greatly increased after 1 h of PACAP treatment (Fig. 6d), compared with untreated explants (Fig. 6a). Consistent with the idea that retinal progenitor cells directly respond to this peptide, the PACAP-induced increase in KLF4 labeling was clearly observed in cyclin D1-positive cells (Fig. 6).
KLF4 content is increased in the neuroblastic layer upon PACAP treatment. Retinal explants were maintained in vitro and treated with PACAP for 1 h. Images were obtained by confocal microscopy. KLF4 immunoreactivity was examined in control (a) and PACAP-treated (d) P1 rat retinal explants. Cyclin D1 was used as a marker of retinal progenitors in control (b) and PACAP-treated retinal explants (e) and merged images are shown in c and f. GCL ganglion cell layer, NBL neuroblastic layer. Results are representative of three independent experiments
KLF4 Protein Is Detected in the Nucleus of Retinal Cells Upon PACAP Signaling
As described in the microarray analysis, Sp/Klf target genes were enriched among PACAP-regulated genes. Electrophoretic mobility shift assay (EMSA) was used to test whether the nuclear content of KLF4 was increased upon treating P1 retinal explants with PACAP. Probes containing Sp1 motifs found in cyclin D1 promoter were used. A complex was detected in control conditions (Fig. 7, lane 1), which was greatly increased in extracts from PACAP-treated retinas (Fig. 7, lane 2), whereas no complex was observed with a mutant probe (Fig. 7, lanes 3 and 4). Pre-incubation of nuclear extracts with increasing concentrations of an antibody to KLF4 progressively reduced the amount of protein-probe complex (Fig. 7, lanes 5–10), suggesting that KLF4 was a component of the complex with Sp1 probe and that the incubation with the antibody prevented KLF4 binding to the probe. Altogether, these findings indicate that activation of PACAP signaling in early postnatal rat retina leads to a KLF4-mediated inhibition of retinal progenitor cells proliferation.
PACAP increases retinal nuclear KLF4 content in EMSA. Sp1 consensus (C) or mutant (M) probes were radiolabeled with γ32-ATP and incubated with nuclear protein extracts from P1 retinal explants after vehicle (control) or PACAP treatment (1 h). A decrease in DNA–protein complex was observed after nuclear extract pre-incubation with increasing amounts of KLF4 antibody (2, 4, and 6 μg). * indicates DNA–protein complex. Result is representative of three independent experiments
Discussion
The transcription factor KLF4 has been implicated in a wide variety of cellular events including cell proliferation (Wei et al. 2008; Hu et al. 2009; Shimizu et al. 2010), cell differentiation (Dai and Segre 2004; Ghaleb et al. 2005), tumorigenesis (Rowland et al. 2005), maintenance of pluripotency and cell reprogramming (Takahashi and Yamanaka 2006; Takahashi et al. 2007), and axon regeneration (Moore et al. 2009; Moore and Goldberg 2011). In this report, we present data in support of KLF4 roles in retinal development, particularly in the regulation of progenitor cells proliferation after PACAP treatment. We showed that KLF4 is expressed in both retinal progenitor cells as well as postmitotic differentiated cells in various stages of retinal development in the rat. Furthermore, we demonstrate that PACAP treatment leads to a rapid and transient induction of KLF4 levels and DNA-binding activity.
PACAP has been previously described to play a role in various processes in both developing and mature nervous system (Vaudry et al. 2009). Our microarray analyses identified various genes associated with processes previously described to be regulated by PACAP in the retina as well as other areas of the nervous system. Regulation of Klf4 and Cdkn1a mRNA levels in response to PACAP was shown in sympathetic neurons (Braas et al. 2007). Although the current retinal microarray study did not identify changes in Klf4 transcript levels, our temporal studies suggested that this likely reflected the transient nature of Klf4 expression. The retinal array study examining PACAP-regulated transcripts after several hours of peptide treatment was defined based on previous work on sympathetic neurons. But unlike sympathetic neurons, our current studies demonstrated that the PACAP-induced Klf4 expression in retinal tissues was rapid with maximal changes approximately 1 h after treatment, which attenuated to control levels after prolonged PACAP treatments. While the transient nature of PACAP-regulated Klf4 obviated the detection of its transcripts in the array, we could detect the regulation of targets of Sp/Klf transcription factors, notably cyclin D1, which we had shown previously to be downregulated in response to PACAP treatment (Njaine et al. 2010). Similarly, the microarray data also showed target regulation by other transcription factors, such as CREB, which was expected from our previous study (Njaine et al. 2010).
KLF4 has been previously described to regulate downstream targets, which are involved in cell cycle control in various cell types, acting both as a positive or negative regulator of cell proliferation (Wei et al. 2008; Hu et al. 2009; Shimizu et al. 2010). In targeting and downregulating cyclin D1 expression (Chen et al. 2003), we propose that the induction of Klf4 by PACAP signaling is one potential mechanism for the antiproliferative effect of PACAP in retinal progenitors (Njaine et al. 2010). Recently, Qin et al. (2011) showed that Klf4 overexpression impacts cortical development by reducing proliferation of NSCs in vitro and in subventricular zone in transgenic mice. Moreover, the artificial regulation of Klf4 levels in the developing cortex interfered with both neuronal differentiation as well as migration (Qin and Zhang 2012). Interestingly, Klf4 overexpression also led to hydrocephalus in mice (Qin et al. 2011), the same phenotype observed after overexpression of PACAP-specific G-protein coupled receptor, PAC1 (Lang et al. 2006), which reinforces the putative link between PACAP signaling and Klf4 regulation.
EMSA results also indicate that KLF4 is present in the nucleus and binds to the Sp1 probe alone or as a complex with other transcription factors. The absence of supershift on EMSA assay may suggest the presence of protein complexes associated with Sp1 DNA motif, which were dissociated when KLF4 antibody was added to nuclear extract. For example, in EMSA assays using TM4 cells as a model of Sertoli cell, Sze et al. (2008) described a ternary protein complex composed by GATA-1/GATA-6/KLF4, which was completely abolished with the use of specific antibodies. Similar data was obtained before for various transcription factors (Leone et al. 1995; Grueneberg et al. 1997; Feng et al. 1998; Husmann et al. 2000; Hurgin et al. 2002). However, the precise mechanism of how KLF4 represses expression of PACAP-dependent genes, which contain Sp1 motifs such as cyclin D1, remains unclear.
As proliferation is one of the processes influenced by PACAP in the nervous system both in the retina (Njaine et al. 2010) and in cortical (Suh et al. 2001; Carey et al. 2002) and cerebellar progenitors (Nicot et al. 2002; Lelievre et al. 2008), our current findings together with data from the literature suggest that PACAP regulation of Klf4 may be one molecular mechanism underlying PACAP control of progenitor cell proliferation in different tissues.
In conclusion, we propose that PACAP induces a transient increase in Klf4 expression, which could result in a reduction in cyclin D1 levels in a subpopulation of retinal progenitors (Njaine et al. 2010), leading to cell cycle withdrawal and perhaps affecting the specification and/or differentiation of retinal cell types.
Abbreviations
- KLF:
-
Kruppel-like factor
- BrdU:
-
5-Bromo-2-deoxyuridine
- CREB:
-
cAMP response element-binding protein
- RPCs:
-
Retinal progenitor cells
- DPM:
-
Disintegrations per minute
- GCL:
-
Ganglion cell layer
- HRP:
-
Horseradish peroxidase
- INL:
-
Inner cell layer
- PAC1:
-
Pituitary adenylyl cyclase activating polypeptide-specific receptor
- PACAP:
-
Pituitary adenylyl cyclase-activating polypeptide
- PCNA:
-
Proliferating cell nuclear antigen
- pCREB:
-
Phosphorylated cAMP response element-binding protein
- PKA:
-
cAMP-dependent protein kinase (protein kinase A)
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- Shh:
-
Sonic hedgehog
- VIP:
-
Vasoactive intestinal peptide
- VPAC:
-
Receptor activated by both pituitary adenylyl cyclase-activating polypeptide and vasoactive intestinal peptide
References
Anchan RM, Reh TA (1995) Transforming growth factor-beta-3 is mitogenic for rat retinal progenitor cells in vitro. J Neurobiol 28(2):133–145
Anchan RM, Reh TA, Angello J, Balliet A, Walker M (1991) EGF and TGF-alpha stimulate retinal neuroepithelial cell proliferation in vitro. Neuron 6(6):923–936
Atlasz T, Szabadfi K, Kiss P, Marton Z, Griecs M, Hamza L, Gaal V, Biro Z, Tamas A, Hild G, Nyitrai M, Toth G, Reglodi D, Gabriel R (2011) Effects of PACAP in UV-A radiation-induced retinal degeneration models in rats. J Mol Neurosci 43(1):51–57
Atlasz T, Szabadfi K, Reglodi D, Kiss P, Tamas A, Toth G, Molnar A, Szabo K, Gabriel R (2009) Effects of pituitary adenylate cyclase activating polypeptide and its fragments on retinal degeneration induced by neonatal monosodium glutamate treatment. Ann N Y Acad Sci 1163:348–352
Baxter PS, Martel MA, McMahon A, Kind PC, Hardingham GE (2011) Pituitary adenylate cyclase-activating peptide induces long-lasting neuroprotection through the induction of activity-dependent signaling via the cyclic AMP response element-binding protein-regulated transcription co-activator 1. J Neurochem 118(3):365–378
Borba JC, Henze IP, Silveira MS, Kubrusly RC, Gardino PF, de Mello MC, Hokoc JN, de Mello FG (2005) Pituitary adenylate cyclase-activating polypeptide (PACAP) can act as determinant of the tyrosine hydroxylase phenotype of dopaminergic cells during retina development. Brain Res Dev Brain Res 156(2):193–201
Braas KM, Schutz KC, Bond JP, Vizzard MA, Girard BM, May V (2007) Microarray analyses of pituitary adenylate cyclase activating polypeptide (PACAP)-regulated gene targets in sympathetic neurons. Peptides 28(9):1856–1870
Carey RG, Li B, DiCicco-Bloom E (2002) Pituitary adenylate cyclase activating polypeptide anti-mitogenic signaling in cerebral cortical progenitors is regulated by p57Kip2-dependent CDK2 activity. J Neurosci 22(5):1583–1591
Cepko CL (1999) The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol 9(1):37–46
Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW (2001) Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem 276(32):30423–30428
Chen X, Whitney EM, Gao SY, Yang VW (2003) Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol 326(3):665–677
Close JL, Gumuscu B, Reh TA (2005) Retinal neurons regulate proliferation of postnatal progenitors and Muller glia in the rat retina via TGF beta signaling. Development 132(13):3015–3026
Dai X, Segre JA (2004) Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev 14(5):485–491
Ehlermann J, Pfisterer P, Schorle H (2003) Dynamic expression of Kruppel-like factor 4 (Klf4), a target of transcription factor AP-2alpha during murine mid-embryogenesis. Anat Rec A: Discov Mol Cell Evol Biol 273(2):677–680
Feng XH, Zhang Y, Wu RY, Derynck R (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev 12(14):2153–2163
Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW (2005) Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 15(2):92–96
Grueneberg DA, Henry RW, Brauer A, Novina CD, Cheriyath V, Roy AL, Gilman M (1997) A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev 11(19):2482–2493
Grumolato L, Louiset E, Alexandre D, Ait-Ali D, Turquier V, Fournier A, Fasolo A, Vaudry H, Anouar Y (2003) PACAP and NGF regulate common and distinct traits of the sympathoadrenal lineage: effects on electrical properties, gene markers and transcription factors in differentiating PC12 cells. Eur J Neurosci 17(1):71–82
Hatakeyama J, Kageyama R (2004) Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol 15(1):83–89
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8(2):R19
Hu W, Hofstetter WL, Li H, Zhou Y, He Y, Pataer A, Wang L, Xie K, Swisher SG, Fang B (2009) Putative tumor-suppressive function of Kruppel-like factor 4 in primary lung carcinoma. Clin Cancer Res 15(18):5688–5695
Hurgin V, Novick D, Rubinstein M (2002) The promoter of IL-18 binding protein: activation by an IFN-gamma-induced complex of IFN regulatory factor 1 and CCAAT/enhancer binding protein beta. Proc Natl Acad Sci U S A 99(26):16957–16962
Husmann M, Dragneva Y, Romahn E, Jehnichen P (2000) Nuclear receptors modulate the interaction of Sp1 and GC-rich DNA via ternary complex formation. Biochem J 352(Pt 3):763–772
Jensen AM, Wallace VA (1997) Expression of Sonic hedgehog and its putative role as a precursor cell mitogen in the developing mouse retina. Development 124(2):363–371
Kanekar S, Gandham M, Lucero MT (2010) PACAP protects against TNFalpha-induced cell death in olfactory epithelium and olfactory placodal cell lines. Mol Cell Neurosci 45(4):345–354
Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH (2005) Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology 128(4):935–945
Lang B, Song B, Davidson W, MacKenzie A, Smith N, McCaig CD, Harmar AJ, Shen S (2006) Expression of the human PAC1 receptor leads to dose-dependent hydrocephalus-related abnormalities in mice. J Clin Invest 116(7):1924–1934
Lelievre V, Seksenyan A, Nobuta H, Yong WH, Chhith S, Niewiadomski P, Cohen JR, Dong H, Flores A, Liau LM, Kornblum HI, Scott MP, Waschek JA (2008) Disruption of the PACAP gene promotes medulloblastoma in ptc1 mutant mice. Dev Biol 313(1):359–370
Leone TC, Cresci S, Carter ME, Zhang Z, Lala DS, Strauss AW, Kelly DP (1995) The human medium chain acyl-CoA dehydrogenase gene promoter consists of a complex arrangement of nuclear receptor response elements and Sp1 binding sites. J Biol Chem 270(41):24622
Lillien L, Cepko C (1992) Control of proliferation in the retina: temporal changes in responsiveness to FGF and TGF alpha. Development 115(1):253–266
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Lu N, Zhou R, DiCicco-Bloom E (1998) Opposing mitogenic regulation by PACAP in sympathetic and cerebral cortical precursors correlates with differential expression of PACAP receptor (PAC1-R) isoforms. J Neurosci Res 53(6):651–662
Marquardt T, Gruss P (2002) Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci 25(1):32–38
Martins RA, Linden R, Dyer MA (2006) Glutamate regulates retinal progenitors cells proliferation during development. Eur J Neurosci 24(4):969–980
Martins RA, Pearson RA (2008) Control of cell proliferation by neurotransmitters in the developing vertebrate retina. Brain Res 1192:37–60
Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, Culler MD, Coy DH (1989) Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun 164(1):567–574
Miyata A, Jiang L, Dahl RD, Kitada C, Kubo K, Fujino M, Minamino N, Arimura A (1990) Isolation of a neuropeptide corresponding to the N-terminal 27 residues of the pituitary adenylate cyclase activating polypeptide with 38 residues (PACAP38). Biochem Biophys Res Commun 170(2):643–648
Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL (2009) KLF family members regulate intrinsic axon regeneration ability. Science 326(5950):298–301
Moore DL, Goldberg JL (2011) Multiple transcription factor families regulate axon growth and regeneration. Dev Neurobiol 71(12):1186–1211
Nicot A, Lelievre V, Tam J, Waschek JA, DiCicco-Bloom E (2002) Pituitary adenylate cyclase-activating polypeptide and sonic hedgehog interact to control cerebellar granule precursor cell proliferation. J Neurosci 22(21):9244–9254
Njaine B, Martins RA, Santiago MF, Linden R, Silveira MS (2010) Pituitary adenylyl cyclase-activating polypeptide controls the proliferation of retinal progenitor cells through downregulation of cyclin D1. Eur J Neurosci 32(3):311–321
Qin S, Liu M, Niu W, Zhang CL (2011) Dysregulation of Kruppel-like factor 4 during brain development leads to hydrocephalus in mice. Proc Natl Acad Sci U S A 108(52):21117–21121
Qin S, Zhang CL (2012) Role of Kruppel-like factor 4 in neurogenesis and radial neuronal migration in the developing cerebral cortex. Mol Cell Biol 32(21):4297–4305
Reglodi D, Kiss P, Szabadfi K, Atlasz T, Gabriel R, Horvath G, Szakaly P, Sandor B, Lubics A, Laszlo E, Farkas J, Matkovits A, Brubel R, Hashimoto H, Ferencz A, Vincze A, Helyes Z, Welke L, Lakatos A, Tamas A (2012) PACAP is an endogenous protective factor-insights from PACAP-deficient mice. J Mol Neurosci 48(3):482–492
Rehen SK, Varella MH, Freitas FG, Moraes MO, Linden R (1996) Contrasting effects of protein synthesis inhibition and of cyclic AMP on apoptosis in the developing retina. Development 122(5):1439–1448
Rocha-Martins M, Njaine B, Silveira MS (2012) Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and western blot throughout rat retinal development. PLoS One 7(8):e43028
Rowland BD, Bernards R, Peeper DS (2005) The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 7(11):1074–1082
Shimizu Y, Takeuchi T, Mita S, Notsu T, Mizuguchi K, Kyo S (2010) Kruppel-like factor 4 mediates anti-proliferative effects of progesterone with G(0)/G(1) arrest in human endometrial epithelial cells. J Endocrinol Invest 33(10):745–750
Silveira MS, Costa MR, Bozza M, Linden R (2002) Pituitary adenylyl cyclase-activating polypeptide prevents induced cell death in retinal tissue through activation of cyclic AMP-dependent protein kinase. J Biol Chem 277(18):16075–16080
Suh J, Lu N, Nicot A, Tatsuno I, DiCicco-Bloom E (2001) PACAP is an anti-mitogenic signal in developing cerebral cortex. Nat Neurosci 4(2):123–124
Szabadfi K, Szabo A, Kiss P, Reglodi D, Setalo G Jr, Kovacs K, Tamas A, Toth G, Gabriel R (2014) PACAP promotes neuron survival in early experimental diabetic retinopathy. Neurochem Int 64:84–91
Sze KL, Lee WM, Lui WY (2008) Expression of CLMP, a novel tight junction protein, is mediated via the interaction of GATA with the Kruppel family proteins, KLF4 and Sp1, in mouse TM4 Sertoli cells. J Cell Physiol 214(2):334–344
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–872
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676
Varella MH, Correa DF, Campos CB, Chiarini LB, Linden R (1997) Protein kinases selectively modulate apoptosis in the developing retina in vitro. Neurochem Int 31(2):217–227
Varella MH, de Mello FG, Linden R (1999) Evidence for an antiapoptotic role of dopamine in developing retinal tissue. J Neurochem 73(2):485–492
Vaudry D, Falluel-Morel A, Bourgault S, Basille M, Burel D, Wurtz O, Fournier A, Chow BK, Hashimoto H, Galas L, Vaudry H (2009) Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol Rev 61(3):283–357
Wada Y, Nakamachi T, Endo K, Seki T, Ohtaki H, Tsuchikawa D, Hori M, Tsuchida M, Yoshikawa A, Matkovits A, Kagami N, Imai N, Fujisaka S, Usui I, Tobe K, Koide R, Takahashi H, Shioda S (2013) PACAP attenuates NMDA-induced retinal damage in association with modulation of the microglia/macrophage status into an acquired deactivation subtype. J Mol Neurosci 51(2):493–502
Wei D, Kanai M, Jia Z, Le X, Xie K (2008) Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res 68(12):4631–4639
Yoon HS, Chen X, Yang VW (2003) Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem 278(4):2101–2105
Young TL, Cepko CL (2004) A role for ligand-gated ion channels in rod photoreceptor development. Neuron 41(6):867–879
Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A 102(12):4459–4464
Acknowledgments
We thank José Nilson dos Santos, José Francisco Tibúrcio, Damaris Sincorá, Gildo Brito de Souza, and Talita Martins for technical assistance and the Bioinformatics Core Facility at University of Vermont, USA. Support for the University of Vermont Neuroscience Core Facility from NIH NCRR P30RR032135/NIGMS P30GM103498 is also gratefully acknowledged. This investigation was supported by grants from CAEN/International Society for Neurochemistry, CNPq, and FAPERJ.
Conflict of Interest
There is no conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Brian Njaine and Maurício Rocha-Martins contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(GIF 32 kb)
Rights and permissions
About this article
Cite this article
Njaine, B., Rocha-Martins, M., Vieira-Vieira, C.H. et al. Pleiotropic Functions of Pituitary Adenylyl Cyclase-Activating Polypeptide on Retinal Ontogenesis: Involvement of KLF4 in the Control of Progenitor Cell Proliferation. J Mol Neurosci 54, 430–442 (2014). https://doi.org/10.1007/s12031-014-0299-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-014-0299-2