Abstract
Background
Aberrant expression of microRNAs (miRNAs) has been implicated in the etiopathogenesis and development of various cancers. Drosha and Dicer are the main components of the miRNA biosynthesis machine. Another enzyme, DGCR8, is the assistant of Drosha in the processing complex. Here, we tried to evaluate the mRNA transcript level of Drosha, Dicer, and DGCR8 genes in involved tissues from patients with gastric cancer.
Methods
Fifty tumoral and their marginal tissues, as the control group, were obtained from patients with gastric cancer. After RNA extraction from tissues and cDNA synthesis, quantification of mRNA expression of Drosha, Dicer, and DGCR8 was conducted using SYBR Green master mix and real-time PCR.
Results
It was observed that mRNA expression levels of Drosha, Dicer, and DGCR8 were significantly upregulated in tumoral tissues compared with marginal tissues. Upregulation of these genes was not correlated with clinical manifestations of the patients.
Conclusions
Upregulation of Drosha, Dicer, and DGCR8 plays a role in the development of cancer, probably through dysregulated the expression level of miRNAs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is one of the most common cancers throughout the world, which is ranked as the second cause of cancer-related death [1]. Like other cancers, gene expression patterns are changed in gastric cancer. In fact, there are mainly two types of genes, which are involved in cancer development, some with increased level of expression known as oncogenes while some with decreased expression level known as tumor suppressor genes [2].
MicroRNAs (miRNAs) are non-coding RNAs with 18–24 nucleotides, which are important in genes expression regulation and due to their roles in cell biology, they have a great role in cancer development [3]. About 1000 miRNAs are encoded in human genome and each of them regulates expression level of about 200 genes; hence, approximately 30% of all human genes are regulated by these small molecules [4]. With regard to their expression level and function, miRNAs may act like tumor suppressor genes or oncogenes [5]. All in all, cancer pathogenesis is related to mRNA-miRNA interaction and their expression levels [6]. Expression of miRNAs is regulated and processed by a complicated biological system. At the first step, they are transcribed by RNA polymerase 2 in nucleus and this immature molecule is known as primary miRNA (pri-miRNA) [7]. At the second step, an endonuclease 3 known as Drosha, which is linked with a nuclear protein known as DiGeorge syndrome critical region gene 8 (DGCR8 or Pasha), cutes and processes the pri-miRNA to pre-miRNA in cell nucleus which is about 75–90 nucleotide long. After processing by Drosha, the pre-miRNA transported to cytoplasm via exportin-5 where another endonuclease 3 known as Dicer cutes and converts pre-miRNA to mature and active microRNA [8,9,10].
As a result, Dicer, Drosha, and DGCR8 have an important role in the activation and expression profile of microRNAs. Previous studies have disclosed the dysregulation of microRNA in human cancers, such as gastric cancer [5, 11,12,13,14,15,16,17]. However, there is a gap in studying the expression level of these three genes, as most important part of microRNA maturing machinery, and their probable role in miRNA expression levels dysregulation. Herein, in this study, we evaluated the microRNA processing machinery system elements Drosha, Dicer, and DGCR8 in tumoral tissues from gastric cancer patients in order to fill this gap.
Material and Methods
Patients and Sampling
Fifty tumoral and their marginal tissues, as the normal control group, were collected from gastric cancer patients who had referred to Imam Reza Hospital of Tabriz University of Medical Sciences during 2012–2016. To get a pure sample population, all the patients were native to East Azerbaijan, North West of Iran. Through sample gathering, patients who had undergone chemotherapy and radiation therapy were excluded. All samples were collected during surgery and then transferred into RNAase inhibitor solation (Qiagen, Cat No. 76104) and stored in − 80 °C till RNA extraction. The clinical data of the patients were gathered and summarized in Table 1. The Human Research Ethics Committees from the Tabriz University of Medical Sciences approved the protocol of this study and written informed consent was taken by all patients.
RNA Extraction
Total RNA was extracted from tumoral and marginal tissues by Tripure isolation reagent (Roche, Cat No. 11667165001) according to the manufacturer’s protocol. Quantity of RNA samples was determined by Nanodrop and quality was examined by gel electrophoreses on 1% agarose. Afterwards, RNA samples were stored in − 80 °C till cDNA synthesis.
Complementary DNA Syntheses and Real-time PCR Quantification
In this study, 2 step real time PCR was used for quantitative measurement of target genes expression level. We applied TAKARA complementary DNA (cDNA) syntheses kit (TAKARA, Cat No. 6130) to synthesize cDNA. After that, quantitative real-time PCR was conducted by SYBR green PCR master mix (TAKARA, Cat No. RR820W) and specific primer set for each gene (Table 2). Primers were adopted from the studies by Sand et al. to evaluate Drosha, Dicer, DGCR8, and RNA induced silencing complex (RISC) components in epithelial skin cancer [18, 19]. For normalizing, the expression level of target genes GAPDH (housekeeping gene) expression level was used. At the end, the average of duplicated Ct values was measured and the relative expression level of target genes was determined by comparative Ct method [20].
Statistical Analysis
Statistical analysis was performed using the Graph Pad Prism 6 (Graph Pad Software Inc. San Diego, CA, USA). Kolmogorov-Smirnov’s normality test was applied for evaluating normality of data. Independent sample t test was conducted to compare target gene expression level between cancerous tissues and their paired marginal tissues. Pearson’s correlation test was performed for evaluating the correlation between expression of target genes and patient’s clinical parameters. All results were expressed as mean ± standard deviation (SD). Statistical significance level for all P value was less than 0.05.
Results
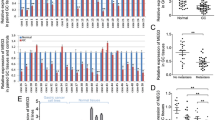
Analysis of mRNA expression level of Dicer, Drosha, and DGCR8 in tumoral tissues from gastric cancer patients and comparison of those with marginal tissues indicated that mRNA expression levels of Drosha (fold change = 2.8, P = 0.0023), Dicer (fold change = 2.1, P = 0112), and DGCR8 (fold change = 3.7, P = 0.0014) were significantly upregulated in tumoral tissues in relation to marginal tissues (Fig. 1).
The correlation analysis between the mRNA expression level of Dicer, Drosha, and DGCR8 and clinicopathological characters was conducted. It was observed that mRNA expression levels of Dicer, Drosha, and DGCR8 were not significantly correlated with clinicopathological and demographic parameters with regard to age, sex, smoking, alcohol consumption, nodal status, distant metastasis, and tumor grade in the patients (Table 3).
Discussion
Epidemiological evidence reports that gastric cancer is the third leading cause of cancer-related death in all over the world. In spite of a decreased incidence in some geographical regions, there is a challenge in clinical aspects of gastric cancer due to the disease diagnosis in an advanced stage, poor prognosis, and inefficient treatment choices. Helicobacter pylori infection has been considered as the most common cause of the gastric cancer, and then Epstein-Barr virus infection as well as familial genetic background is important. Among the most critical predisposing factors for gastric cancer are smoking, high salt intake and other dietary contributing factors [21]. The prevalence of the gastric cancer in East-Azerbaijan, Iran, where the patients in this study were taken from, is 8.3% (the most prevalent after skin, esophageal, and breast cancers) in female and 15.6% in males [22]. Therefore, sophisticated hygiene programing appears to be crucial in reducing the prevalence as well as treating the patients. This could be possible through identification of novel diagnostic and prognostic approaches, such as miRNAs, which seems to be promising within this context [16, 23].
Previous studies have documented the crucial role of miRNAs machinery system components in some types of cancers [24]. According to these studies, Drosha, Dicer, and DGCR8 demonstrated different expression pattern in each tumor type. Furthermore, aberrant expression profile of these genes has been associated with dysregulation of miRNAs, which in turn was related to the onset and development of malignancies [19, 25]. However, there is no obvious evidence indicating how expression pattern of miRNAs is controlled directly by these genes [26, 27].
In the ongoing study, we evaluated the mRNA expression level of Dicer, Drosha, and DGCR8 in tumoral tissues of gastric cancer patients in comparison to marginal non tumor tissues. It was found that there were significant upmodulations in expression levels of these three genes in tumoral samples in comparison to marginal tissues. Alteration in Dicer, Drosha, and DGCR8 expression levels has previously been demonstrated in gastric and hepatocellular cancer cell lines [28]. Moreover, the altered expression of Dicer and Drosha has been associated with deregulated expression pattern of some miRNAs in gastric cancer [16, 27].
Our experiments also demonstrated that there was no any relation between expression levels of these genes and clinical symptoms of the gastric cancer patients, which was also previously shown [5].
Studies have shown that knockdown of miRNA machinery system components can culminate in total decreased expression level of miRNAs in various cell types [29, 30]. Furthermore, it has been observed that functional mutations may occur in genes encoding miRNA machinery components and may be contributing to cancer development [31]. It seems logical that increased levels of Drosha, Dicer, and DGCR8 result in increased production of miRNAs in general. However, even though the miRNA biogenesis enzymes are upregulated, there might be decreased levels of several miRNAs. This is because of regulatory mechanisms, including that occur after the biogenesis of miRNAs, such as untemplated nucleotidyl addition to the 3′ end of miRNA, miRNA editing through the conversion of adenosine to inosine, miRNA methylation, regulation through miRNA stability that contains are cleavage at the terminal loop, etc. [32].
In consideration of all, Dicer, Drosha, and DGCR8 expression patterns, as the most important enzymes in miRNA processing system, were upregulated in tumoral tissues compared with normal marginal tissues from patients with gastric cancer. But there were no significant communications between expression profile of these genes and clinical and pathological symptoms of the patients. However, this conclusion is premature and could not be determined based on our data only and further investigations are still mandatory.
References
Alberts S, Cervantes A, Van de Velde C. Gastric cancer: epidemiology, pathology and treatment. Ann Oncol. 2003;14(90002):31–6.
Kobayashi T, Kikuchi S, Lin Y, Yagyu K, Obata Y, Ogihara A, et al. Trends in the incidence of gastric cancer in Japan and their associations with Helicobacter pylori infection and gastric mucosal atrophy. Gastric Cancer. 2004;7(4):233–9.
Cowland JB, Hother C, Grønbaek K. MicroRNAs and cancer. Apmis. 2007;115(10):1090–106.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24(4):652–7.
Landi D, Gemignani F, Barale R, Landi S. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2008;27(1):35–43.
Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–70.
Gregory RI, K-p Y, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40.
Han J, Lee Y, Yeom K-H, Kim Y-K, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27.
Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14(23):2162–7.
Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, et al. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem. 2008;104(3):805–17.
Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34(2):537.
Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–43.
Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129(11):2600–10.
Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4(6):e659.
Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11(2):136–46.
Asadi M, Shanehbandi D, Mohammadpour H, Hashemzadeh S, Sepehri B. Expression level of miR-34a in tumor tissue from patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2018. https://doi.org/10.1007/s12029-018-0060-0.
Sand M, Skrygan M, Georgas D, Arenz C, Gambichler T, Sand D, et al. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51(11):916–22.
Sand M, Gambichler T, Skrygan M, Sand D, Scola N, Altmeyer P, et al. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Investig. 2010;28(6):649–53.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):e45.
Rugge M, Fassan M, Graham D. Epidemiology of gastric cancer. In: Strong V, editor. Gastric Cancer. Cham: Springer; 2015.
Somi MH, Farhang S, Mirinezhad SK, Naghashi S, Seif-Farshad M, Golzari M. Cancer in East Azerbaijan, Iran: results of a population-based cancer registry. Asian Pac J Cancer Prev. 2008;9(2):327–30.
DanM F. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2009;2008:175497.
Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14(23):7956–62.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–9.
Volinia S, Calin GA, Liu C-G, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61.
Jafari N, Dogaheh HP, Bohlooli S, Oyong GG, Shirzad Z, Alibeiki F, et al. Expression levels of microRNA machinery components Drosha, Dicer and DGCR8 in human (AGS, HepG2, and KEYSE-30) cancer cell lines. Int J Clin Exp Med. 2013;6(4):269–74.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9.
Grelier G, Voirin N, Ay A, Cox D, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101(4):673–83.
Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67(5):2345–50.
Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24.
Acknowledgements
We are deeply thankful of our patients for their contribution.
Funding
This study was supported by a grant from Tabriz University of Medical Sciences (Grant No. 94-06-21-4112).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The Human Research Ethics Committees from the Tabriz University of Medical Sciences approved the protocol of this study and written informed consent was taken by all patients.
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Asadi, M., Shanehbandi, D., Zafari, V. et al. Transcript Level of MicroRNA Processing Elements in Gastric Cancer. J Gastrointest Canc 50, 855–859 (2019). https://doi.org/10.1007/s12029-018-0154-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-018-0154-8