Abstract
Background
Notch signaling is one of the main involved pathways in cell differentiation and organogenesis, and its deregulation may lead to tumorigenesis. In this pathway, targeted to the CSL (CBF1, Suppressor of Hairless or Lag-1) complex, notch intracellular domain (NICD) releases corepressors and recruits MAML1 as coactivator triggering the activation of notch signaling transcription complex. Hairy enhance of split-1 (HES1) is one of the notch signaling target genes which is a basic helix-loop-helix (bHLH) transcription factor acting as a proliferation stimulator through the suppression of cell cycle inhibitors such as p27 and p21.
Aims
In this study, we aimed to analyze the role of HES1 in the progression of esophageal squamous cell carcinoma (ESCC).
Methods
Messenger RNA (mRNA) expression of HES1 in fresh tumoral tissues and their margin normal samples were assessed in 50 ESCC patients by real-time polymerase chain reaction (RT-PCR).
Results
Thirteen out of 50 cases (26 %) had HES1 underexpression, while HES1 overexpression was observed only in 4 (8 %) samples. HES1 underexpression was significantly correlated with tumor depth of invasion (P = 0.035).
Conclusion
Although we have not observed any significant correlation between the HES1 expression and notch activation in ESCC, this study is the first report that elucidated the HES1 underexpression in ESCC and revealed its correlation with the invasiveness of ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the main reasons of cancer-related deaths in Iran and ranks as the fifth and sixth of the most common cancers in women and men, respectively [1, 2]. Developmental cell signaling pathways such as Wnt and notch play important roles in cell fate decision during development [3–5]. Notch family genes encode cell surface receptors. In a ligand binding-dependent manner, the receptors are activated through cleavages by some of the metalloproteases such as c-secretase and tumor necrosis factor-α-converting enzyme (TACE), releasing notch intracellular domain (NICD) into the cytoplasm. The NICD translocates into the nucleus where it functions as a transcription activator of notch signaling target genes. Targeted to the CSL (CBF1, Suppressor of Hairless or Lag-1) complex, NICD releases corepressors and recruits MAML1 coactivator, triggering the activation of notch signaling transcription complex [6, 7]. Hairy enhance of split-1 (HES1), cyclin D1, p21, p27, and c-Myc are some of the important notch target genes [8]. In the absence of NICD, the CSL complex binds to the regulatory region of target genes and recruits corepressors such as CBF1 and hairless/split ends (SPEN) [9, 10], which are associated with histone deacetylases to conserve the chromatin structure in a transcriptional inactive status.
Crosstalk between notch and other important cell signaling pathways such as Wnt and Sonic hedgehog (Shh) plays key roles in tumorigenesis [11–13]. The tumor growth and development is related to cancer stem-like cells (CSCs) where deregulated notch signaling leads to uncontrolled proliferation of CSCs to create a tumor mass [14]. The notch pathway keeps balance between cell proliferation and apoptosis. Therefore, it is clear that deregulation of the notch pathway may result in tumorigenesis [15]. The notch signaling pathway acts as a double-edged sword in tumorigenesis based on the cell context. In some malignancies such as lung, cervical, and colon cancers, it functions as an oncogene, while in some others such as hepatocellular carcinoma, it plays as a tumor suppressor [6, 7, 16]. Suppression of the notch signaling pathway will result in differentiation of cancer stem cells, and therefore, the notch pathway is suggested as an important target for cancer treatment [17].
HES proteins as basic helix-loop-helix (bHLH) transcription factors are one of the most important gene families which are targeted by the notch signaling pathway. This family consists of HES1–6 as members of the bHLH transcription factor repressors which play their roles through expressional regulation of cell cycle mediators such as p21 and cyclin D1. The HES1 has the most important role during the developmental processes [18, 19]. Although the expressional studies of HES1 in different malignancies have been reported, the impact of HES1 messenger RNA (mRNA) expression in ESCC progression and development is not elucidated yet. Therefore, in this study, we aimed to reveal the level of HES1 mRNA expression in ESCC and to evaluate its correlation with different clinicopathological features of the patients.
Materials and Methods
Tissue Samples
Fifty ESCC patients from the Emam Reza and Omid Hospitals of Mashhad University of Medical Sciences (MUMS) were enrolled in this study. RNA extraction from the tumoral and corresponding normal tissues was performed using the RNeasy Mini Kit (Qiagen, Hilden, Germany). All the cases had not received any chemo- and radiotherapy before the surgery. The ethic committee of MUMS confirmed the study, and all the patients gave their written consent to be enrolled. The normal and tumoral microdissected samples were conserved in RNAlater solution (Qiagen, Hilden, Germany) and histologically examined by expert pathologists based on the last TNM classification guideline [20].
RNA Extraction, cDNA Synthesis, and Quantitative RT-PCR
cDNA synthesis was performed after RNA extraction from the normal and tumoral tissues as described before [21]. Assessment of the level of HES1 mRNA expression was done using the primer sequences presented in Table 1 through a duplicate comparative real-time PCR (Stratagene Mx3000P, La Jolla, CA) SYBR Green (GENETBIO, Korea) method. The data were normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) which has the lowest variability between the normal and tumor tissues of ESCC in comparison with the other housekeeping genes [22, 23]. The thermal profile involved an initial denaturation step of 95 °C for 10 min followed by 40 cycles of 95 °C (15 s), 52 °C (30 s), and 72 °C (30 s). Fold changes in HES1 mRNA expression were calculated using the ΔΔCt method [24], and overexpression and underexpression of the gene were defined for the tumors with more than +2-fold and less than −2-fold in the level of HES1 mRNA expression, respectively. The range in between was considered as normal expression.
Statistical Analysis
Statistical analysis was done using SPSS 16.0 (SPSS, Chicago, IL). Due to normal distribution of the samples, assessment of correlations between the level of HES1 mRNA expression and clinicopathological features was performed by the Pearson test, independent sample t test, and ANOVA. Statistical tests were considered significant with a p value <0.05.
Results
Study Population
Fifty ESCC patients including 26 males (52 %) and 24 females (48 %) were enrolled in this study. The patients’ age was scattered between 37 and 83 years (mean ± SD 62.80 ± 10.89). The tumor size ranged from 1 to 12 cm (mean ± SD 4.12 ± 1.95). The tumors were almost equally distributed in the lower 23 (46 %) and middle 27 (54 %) parts of the esophagus. Most of the tumors (49/50, 98 %) were classified into tumor stages of ΙΙ/ΙΙΙ, and 40 out of 50 tumor samples (80 %) were invaded to adventitia (T3). The majority of cases were moderately differentiated (32/50, 64 %). The number of tumors without any metastasis to the regional lymph nodes was more than the tumors with lymph node metastasis (28 vs. 22 cases, respectively).
Significant Downregulation of HES1 in ESCC
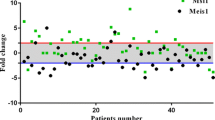
Assessment of HES1 expression was performed using real-time PCR (SYBR Green method), in which the level of HES1 expression in tumor samples was compared to the corresponding normal (tumor-free) esophageal tissues. The fold changes of HES1 mRNA expression in patients are shown as a scatterplot in Fig. 1. Thirteen out of 50 cases (26 %) had significant HES1 underexpression (P = 0.001), while HES1 overexpression was observed only in 4 (8 %) samples, and the other tumor tissues showed normal expression of HES1 mRNA (mean ± SD 0.30 ± 0.20). Figure 2 compares the different levels of HES1 mRNA expression in three groups (underexpressed, normal, and overexpressed samples), schematically.
Correlations Between HES1 mRNA Expression and Clinicopathological Features
Correlations between the level of HES1 mRNA expression and clinicopathological features including the depth of tumor invasion, lymph node metastasis, tumor stage, grade, and location are shown in Table 2. There was a significant correlation between the depth of tumor invasion (T) and HES1 mRNA expression in which underexpressed tumors in T3 had lower levels of HES1 mRNA expression in comparison with the underexpressed tumors in T2 (−4.23 ± 0.763 vs. −2.55 ± 0.304, fold change) (p = 0.035). Most of underexpressed cases (9 out of 13, 69.2 %) were invaded to the adventitia classified as T3, histologically. There was not any significant correlation between the HES1 underexpression and stage of tumor. Although there was not any significant correlation between the tumor grade and HES1 mRNA expression (p = 0.45), most of HES1 underexpressed tumors were moderately differentiated (9 out of 13, 69.2 %). The underexpressed tumors were observed in the lower and middle esophagus equally. There was not any significant correlation between the level of HES1 mRNA expression and sex, tumor size, location, and metastasis status. HES1 overexpressed tumors had higher mean size in comparison with the underexpressed tumors (mean ± SD 5.63 ± 2.13 vs. 3.97 ± 0.50 cm), and the biggest tumor sample in size (12 cm) belonged to the HES1 overexpressed tissue.
Discussion
The notch signaling pathway is one of the most important developmental signaling cascades involved in cell fate decision and differentiation in both embryonic and adult stem cells [25, 26]. It functions via the basic helix-loop-helix (bHLH) transcription factors such as HES gene family [27]. Among the members of HES family, HES1 has a key role in embryogenesis and regulation of differentiation in blood and a variety of tissues such as muscles [28–30]. Therefore, it can be expected that deregulation of HES1 expression may lead to disorganization of normal cell growth program [31]. It has been shown that HES1 functions as a tumor suppressor in some malignancies such as lung cancer and hepatocarcinoma [32]; however, different reports have showed the role of notch signaling in the progression and development of solid tumors including colon, prostate, and cervix cancers [33–35].
In this study, it was observed that the level of HES1 mRNA expression in ESCC was significantly lower compared to the corresponding margin normal esophageal tissues, suggesting that HES1 underexpression is probably related to ESCC progression. Such finding may ignore the function of the notch pathway in tumor progression through the HES1 gene expression [36]. Recently, we showed significant overexpression of MAML1 in ESCC and its correlation with aggressiveness and metastasis of the tumor cells [37]. Since MAML1 is the main coactivator of notch signaling transcription machinery, its overexpression leads us to hypothesize that notch signaling is active in ESCC. Therefore, underexpression of HES1, as one of the main notch signaling target genes, is apparently in contrast with the activity of this pathway in ESCC, suggesting that HES1 is not an ideal indicator of notch signaling activation in the disease. In this study, there was not only a significant correlation between the level of HES1 expression and depth of tumor invasion but also the number of HES1 underexpressed samples was more than the overexpressed cases. Since our previous report regarding overexpression of MAML1 [37] was done on the same study population, we compared the results and interestingly found that most of the HES1 underexpressed cases also showed normal or underexpression of MAML1. Having combined these data, we may extrapolate that HES1, as a suppressor transcription factor, normally suppresses some involved genes in ESCC progression and development, and when HES1 is underexpressed through deregulation of notch signaling pathway, the HES1 target genes experience uncontrolled expression leading to ESCC progression through advanced stages. Another interesting issue that can complicate judgment about HES1 gene expression status is the controlling role of other cell signaling pathways in HES1 expression. It has been shown that the HES1 expression is also regulated by the hedgehog signaling pathway, suggesting that the notch activation via the MAML1 is not lonely a justified guaranty for the HES1 overexpression and activation [38]. In hedgehog cell signaling pathway, HES1 gene expression is positively regulated by Gli1/2 in a notch-independent manner, relying to the complicated crosstalk between the signaling pathways [39, 40].
Our results showed that there was an inverse correlation between the HES1 gene expression and depth of tumor invasion especially in advanced stages of the disease. Therefore, such underexpression of HES1 in aggressive ESCC may be reflected from malfunction of the Shh pathway in these patients. Moreover, expression of HES1 is strictly dependent on the microRNA (miRNA) signature. It has been shown that several miRNAs such as miR-124 and miR-23b can regulate HES1 gene expression [41, 42]. Such regulation may be associated to the level of HES1 mRNA expression in ESCC.
In conclusion, we revealed the significant underexpression of HES1 in ESCC samples and showed its significant association with the depth of tumor invasion. To the best of our knowledge, this is the first report elucidating the expressional status of HES1 and its correlation with indices of poor prognosis in ESCC patients. Although these results cannot confirm the role of notch signaling in ESCC progression, definitely it cannot be ignored due to existent crosstalk between the notch and hedgehog cell signaling pathways to control HES1 gene expression as well as miRNA signature. Despite the lack of a consistent relation between the HES1 expression and notch pathway in ESCC patients, our study introduces HES1 underexpression as an independent prognostic marker for the disease.
References
Forghanifard MM, Gholamin M, Farshchian M, Moaven O, Memar B, Forghani MN, et al. Cancer-testis gene expression profiling in esophageal squamous cell carcinoma: identification of specific tumor marker and potential targets for immunotherapy. Cancer Biol Ther. 2011;12(3):191–7.
Mousavi SM, Gouya MM, Ramazani R, Davanlou M, Hajsadeghi N, Seddighi Z. Cancer incidence and mortality in Iran. Ann Oncol. 2009;20(3):556–63.
Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–87.
Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–7.
Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22(42):6598–608.
Miele L. Notch signaling. Clin Cancer Res. 2006;12(4):1074–9.
Miele L, Miao H, Nickoloff BJ. NOTCH signaling as a novel cancer therapeutic target. Curr Cancer Drug Targets. 2006;6(4):313–23.
Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27(38):5124–31.
Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16(15):1964–76.
Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002;21(20):5417–26.
Nakamura T, Tsuchiya K, Watanabe M. Crosstalk between Wnt and Notch signaling in intestinal epithelial cell fate decision. J Gastroenterol. 2007;42(9):705–10.
Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88(1):11–7.
Moghbeli M, Moghbeli F, Forghanifard MM, Garayali A, Abbaszadegan MR. Cancer stem cell markers in esophageal cancer. Am J Cancer Sci. 2013;2(1):37–50.
Wang Z, Li Y, Kong D, Ahmad A, Banerjee S, Sarkar FH. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. 2010;292(2):141–8.
Wang Z, Li Y, Banerjee S, Sarkar FH. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009;279(1):8–12.
South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012;23(4):458–64.
Ma J, Xia J, Miele L, Sarkar FH, Wang Z. Notch signaling pathway in pancreatic cancer progression. Pancreat Disord Ther. 2013;3(114).
Roma J, Almazan-Moga A, Sanchez de Toledo J, Gallego S. Notch, Wnt, and hedgehog pathways in rhabdomyosarcoma: from single pathways to an integrated network. Sarcoma. 2012;2012:695603.
Wang X, Fu Y, Chen X, Ye J, Lu B, Ye F, et al. The expressions of bHLH gene HES1 and HES5 in advanced ovarian serous adenocarcinomas and their prognostic significance: a retrospective clinical study. J Cancer Res Clin Oncol. 2010;136(7):989–96.
Wittekind C. 2010 TNM system: on the 7th edition of TNM classification of malignant tumors. Pathologe. 2010;31(5):331–2.
Moghbeli M, Abbaszadegan MR, Farshchian M, Montazer M, Raeisossadati R, Abdollahi A, et al. Association of PYGO2 and EGFR in esophageal squamous cell carcinoma. Med Oncol. 2013;30(2):516.
Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, et al. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes. 2005;19(2):101–9.
Moghbeli M, Forghanifard MM, Aarabi A, Mansourian A, Abbaszadegan MR. Clinicopathological sex-related relevance of Musashi1 mRNA expression in esophageal squamous cell carcinoma patients. Pathol Oncol Res. 2014;20(2):427–33.
Edmunds RC, McIntyre JK, Luckenbach JA, Baldwin DH, Incardona JP. Toward enhanced MIQE compliance: reference residual normalization of qPCR gene expression data. J Biomol Tech. 2014;25(2):54–60.
Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14(5):341–7.
Lee JB, Werbowetski-Ogilvie TE, Lee JH, McIntyre BA, Schnerch A, Hong SH, et al. Notch-HES1 signaling axis controls hemato-endothelial fate decisions of human embryonic and induced pluripotent stem cells. Blood. 2013;122(7):1162–73.
Axelson H. The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 2004;204(2):171–8.
Miyazaki M, Kawamoto H, Kato Y, Itoi M, Miyazaki K, Masuda K, et al. Polycomb group gene mel-18 regulates early T progenitor expansion by maintaining the expression of Hes-1, a target of the Notch pathway. J Immunol. 2005;174(5):2507–16.
Ross DA, Hannenhalli S, Tobias JW, Cooch N, Shiekhattar R, Kadesch T. Functional analysis of Hes-1 in preadipocytes. Mol Endocrinol. 2006;20(3):698–705.
Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65(1):43–53.
Liu J, Ye F, Chen H, Lu W, Zhou C, Xie X. Expression of differentiation associated protein Hes1 and Hes5 in cervical squamous carcinoma and its precursors. Int J Gynecol Cancer. 2007;17(6):1293–9.
Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14(1):48–54.
Axelson H. Notch signaling and cancer: emerging complexity. Semin Cancer Biol. 2004;14(5):317–9.
Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107(6):2223–33.
Nam Y, Aster JC, Blacklow SC. Notch signaling as a therapeutic target. Curr Opin Chem Biol. 2002;6(4):501–9.
Beatus P, Lundkvist J, Oberg C, Lendahl U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126(17):3925–35.
Forghanifard MM, Moaven O, Farshchian M, Montazer M, Raeisossadati R, Abdollahi A, et al. Expression analysis elucidates the roles of MAML1 and Twist1 in esophageal squamous cell carcinoma aggressiveness and metastasis. Ann Surg Oncol. 2012;19(3):743–9.
Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med. 2010;16(1):17–26.
Katoh Y, Katoh M. Integrative genomic analyses on GLI1: positive regulation of GLI1 by Hedgehog-GLI, TGFbeta-Smads, and RTK-PI3K-AKT signals, and negative regulation of GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol. 2009;35(1):187–92.
Wall DS, Mears AJ, McNeill B, Mazerolle C, Thurig S, Wang Y, et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184(1):101–12.
Kimura H, Kawasaki H, Taira K. Mouse microRNA-23b regulates expression of Hes1 gene in P19 cells. Nucleic Acids Symp Ser (Oxf). 2004;48:213–4.
Wang C, Yao N, Lu CL, Li D, Ma X. Mouse microRNA-124 regulates the expression of Hes1 in P19 cells. Front Biosci (Elite Ed). 2010;2:127–32.
Acknowledgments
The authors gratefully acknowledge the colleagues at the Division of Human Genetics and Department of Surgery and Pathology of Emam Reza and Omid Hospitals (MUMS) for their assistances in gathering and preparing the ESCC samples.
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Taleb, S., Abbaszadegan, M.R., Moghbeli, M. et al. HES1 as an Independent Prognostic Marker in Esophageal Squamous Cell Carcinoma. J Gastrointest Canc 45, 466–471 (2014). https://doi.org/10.1007/s12029-014-9648-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-014-9648-1