Abstract
Introduction
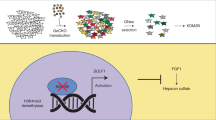
Human sulfatase 1 (SULF1) was recently identified and shown to desulfate cellular heparan sulfate proteoglycans (HSPGs). Since sulfated HSPGs serve as co-receptors for many growth factors and cytokines, SULF1 was predicted to modulate growth factor and cytokine signaling.
Discussion
The role of SULF1 in growth factor signaling and its effects on human tumorigenesis are under active investigation. Initial results show that SULF1 inhibits the co-receptor function of HSPGs in multiple receptor tyrosine kinase signaling pathways, particularly by the heparin binding growth factors—fibroblast growth factor 2, vascular endothelial growth factor, hepatocyte growth factor, PDGF, and heparin-binding epidermal growth factor (HB-EGF). SULF1 is downregulated in the majority of cancer cell lines examined, and forced expression of SULF1 decreases cell proliferation, migration, and invasion. SULF1 also promotes drug-induced apoptosis of cancer cells in vitro and inhibits tumorigenesis and angiogenesis in vivo.
Conclusion
Strategies targeting SULF1 or the interaction between SULF1 and the related sulfatase 2 will potentially be important in developing novel cancer therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sulfatases belong to a family of esterases that hydrolyze sulfate ester bonds of a wide range of substrates ranging from sulfated proteoglycans to conjugated steroids and sulfate esters of small aromatic compounds [1]. Cellular sulfatases work in concert with glycosidases in the degradation of sulfated molecules, including proteoglycans—the major class of sulfated macromolecules, sulfoglycolipids, and cytosolic steroid sulfates. Recently, full-length complementary DNAs (cDNAs) encoding two human proteins with sulfatase domains, designated sulfatase 1 (SULF1) and sulfatase 2 (SULF2), have been isolated and the proteins expressed and characterized [2]. SULF1 and SULF2 are extracellular heparan sulfate 6-O-endosulfatases that have unique structural features, enzymatic activities, and signaling functions [3]. In the first functional studies of this protein family, the quail homolog of SULF1 was shown to co-localize with Wnt pathway molecules at the cell surface of developing muscle cells during quail embryogenesis. Since Wnt ligands are known to bind to highly sulfated heparan sulfate proteoglycans (HSPGs) at the cell surface and within the extracellular matrix, Dhoot et al. proposed that QSulf1 functions as a heparin-degrading endosulfatase that releases Wnt ligands from cell surface and extracellular matrix stores and allows them to bind to their cognate-frizzled receptors [4]. HSPGs also act as co-receptors for heparin-binding receptor tyrosine kinase growth factors such as fibroblast growth factor (FGF), hepatocyte growth factor (HGF), and vascular endothelial growth factor (VEGF). The sulfation state of particular saccharide moieties of HSPGs regulates signaling by receptor tyrosine kinase growth factors [5–7]. Therefore, besides their potential role in activating Wnt signaling, which would predict an oncogenic effect on cells, enzymes that desulfate HSPGs could also have a tumor suppressor effect through abrogation of ligand–receptor binding and downstream signaling by receptor tyrosine kinases. In this review, we present recent studies on the roles of the heparin-degrading endosulfatase SULF1 in cancer, with particular reference to its in vitro and in vivo effects on cellular growth and survival signaling, tumor proliferation, migration, invasion, and angiogenesis.
Cloning and Normal Tissue-Specific Expression of Sulfatase 1
The expressed sequence tag KIAA1077 was initially identified as a coding sequence for a large protein from a size-fractionated human brain cDNA library during a search for genes encoding large proteins more than 50 kDa in size [8]. Protein motif searching predicted that KIAA1077 encoded an 818 amino acid sulfatase enzyme. Subsequently, the quail sulfatase QSulf1 was identified in a screen for hedgehog-induced genes and found to be expressed in muscle and neural progenitor cells in both embryonic and adult tissues [4]. Concurrently, KIAA1077 was shown to encode the human homolog of QSulf1, and additional studies led to the cloning of the rat, Drosophila, and mouse homologs [2, 5, 9, 10].
Expression and Regulation of SULF1 in Cancer
KIAA1077 transcripts were independently shown to be downregulated in libraries obtained by suppression subtraction hybridization of ovarian cancer specimens compared to normal ovarian epithelium [2]. In validation studies, although SULF1 messenger RNA (mRNA) was readily detected in all normal ovarian surface epithelial samples examined by semi-quantitative RT-PCR and Northern blotting, the message was undetectable in five of seven ovarian cancer cell lines. Further, SULF1 mRNA was found to be downregulated in 77% of ovarian cancers—completely undetectable in 40% and expressed at very low levels in an additional 37%. In particular, the clear cell histological subtype of ovarian cancer, which has a particularly poor prognosis, uniformly lacks detectable SULF1 expression [5]. To investigate the mechanisms of downregulation of SULF1 in ovarian cancer, allelic imbalance at the SULF1 locus and the effects of demethylating agents on SULF1 expression were examined. Ovarian cancer specimens showed increased loss of heterozygosity (LOH) in 44–53% of polymorphic markers spanning the SULF1 locus. Treatment with the demethylating agent 5-aza-deoxycytidine (5-aza-dC) led to reactivation of SULF1 expression in the SULF1-negative ovarian cancer cell lines SKOV3 and OV207. Further, treatment of OV207 cells with 5-aza-dC in combination with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) resulted in an additional increase in SULF1 expression compared to 5-aza-dC treatment alone. These results suggest that SULF1 is regulated epigenetically by DNA hypermethylation [11].
Examination of SULF1 expression in other cancer types has shown complete loss or markedly diminished expression of SULF1 in cancer cell lines derived from breast, pancreas, kidney, and hepatocellular cancers, suggesting that downregulation of SULF1 is relatively widespread among epithelial cancers. In general, it appears that the frequency of loss of SULF1 expression is higher in cancer cell lines than in the corresponding primary tumors; for example, SULF1 mRNA was undetectable in nine of 11 (82%) hepatocellular carcinoma (HCC) cell lines examined, but the mRNA was downregulated in only 29% of surgically resected HCCs. This may be due to selection bias as resected cancers in general tend to be lower stage, lower grade cancers than those cancers that are capable of being established as in vitro cell lines. Lower SULF1 expression may therefore correlate with a more aggressive cancer phenotype [12]. SULF1 appears to be dysregulated both by genetic and epigenetic mechanisms in HCCs. In an analysis of 94 primary HCC tumors, LOH of markers surrounding the SULF1 gene ranged from 25% to 42%, with the peak of 42% occurring immediately centromeric to the SULF1 gene. Of the 31 HCCs in which SULF1 expression was assessed by real-time PCR, 14 showed LOH at the SULF1 locus. Seven of the 14 tumors with LOH (50%) also showed downregulation of SULF1 mRNA expression. Examination of SULF1 gene copy number using fluorescence in situ hybridization showed no loss or deletion of the SULF1 gene locus in the SULF1-negative HCC cell lines examined. Treatment with 5-aza-dC, either alone or in combination with TSA, led to reactivation of SULF1 in three SULF1-negative cell lines (Huh7, SK-Hep-1, and SNU449). This strongly suggests that either SULF1 or a SULF1-regulating gene is downregulated by epigenetic DNA hypermethylation [12]. In a study of squamous cell head and neck cancers, SULF1 was shown to function as a negative regulator of cell growth and loss of SULF1-potentiated growth factor signaling, enhanced cell motility and invasiveness, and inhibited stress-induced apoptosis with a resulting increase in tumorigenicity [6]. In a study of breast cancers, Narita et al. found that SULF1 mRNA expression was downregulated in 60% of primary invasive breast tumors [13]. The frequency of downregulation of SULF1 mRNA therefore appears to vary depending on the cancer cell type. In contrast to ovarian cancers and head and neck cancers, high SULF1 expression has been reported in the majority of pancreatic cancers [14]. SULF1 expression was found to be significantly increased (22.5-fold) when compared to normal controls, and SULF1 mRNA was localized in the cancer cells as well as in peritumoral fibroblasts. SULF1 expression was also found to reduce both anchorage-dependent and -independent cell growth and decreased FGF-2-mediated cell growth and invasion in pancreatic cancer cells [14, 15].
In addition to the variable SULF1 expression noted in epithelial malignancies, a number of benign tumors have also shown changes in SULF1 expression. For example, MCF10A cells, an epithelial cell line derived from normal human breast tissue, are generally regarded as relatively normal, but have a complete loss of SULF1 expression, possibly implicating loss of SULF1 as an early event in mammary carcinogenesis [16]. In contrast, and perhaps presaging the increase in SULF1 expression in pancreatic cancer, ten of 22 (45%) of chronic pancreatitis tissues showed increased SULF1 mRNA expression [14].
SULF1 Expression and DNA Hypermethylation
DNA hypermethylation regulates numerous tumor suppressor genes by silencing gene expression. Methylation of CpG islands prevents transcription factors from binding to their DNA-binding sites. Initial studies have explored the role of epigenetic DNA hypermethylation in regulating SULF1 gene expression. Sequence analysis of the SULF1 promoter region initially revealed a CpG-rich region in exon 1A which had previously been identified as a putative CpG island (Genbank Accession No. Z58846) [5, 17]. Further analysis of DNA methylation of 12 CpG dinucleotides in this CpG-rich region was performed on seven ovarian cell lines and 16 primary ovarian tumors by direct sequencing of bisulfite-modified DNA [11]. Two ovarian cell lines and four primary ovarian tumors showed 100% methylation levels; all of these hypermethylated samples for which mRNA expression was measured showed no SULF1 expression. This inverse correlation between DNA hypermethylation and SULF1 expression suggests that SULF1 is regulated by methylation in ovarian tumors. A recent study of breast and gastric cancers has shown that hypermethylation of the exon 1A CpG-rich region examined by Staub et al. in ovarian cancer was correlated with the downregulation of SULF1 mRNA in cell lines and tissues in both breast and gastric cancers [18]. Further evaluation suggested that of the 12 CpG dinucleotides, CpG sites 5–12 are the key nucleotides epigenetically regulating the transcription of SULF1 in ovarian cancer. Treatment of HCC cell lines that express SULF1 at low levels with the demethylating agent 5-Aza-dC showed expression of SULF1 in three of five cell lines, suggesting that SULF1 is regulated by epigenetic methylation in a proportion of HCCs. Additional studies of the 12 CpG dinucleotides of the exon 1A CpG island in 11 HCC cell lines (eight low SULF1 and three high SULF1-expressing cell lines) and ten primary HCCs (six low SULF1 and four high SULF1-expressing primary HCCs) using bisulfite genomic sequencing showed low CpG methylation rates in both HCC cell lines and primary HCCs. Further investigation of another CpG island in exon 5, near the translation start site, found a similar result with no inverse correlation of methylation rate and SULF1 mRNA expression in HCC. The absence of evidence of direct regulation of SULF1 gene transcription by DNA hypermethylation at both of these sites suggests that, at least in HCC, SULF1 may be regulated by a transcription factor encoded by an upstream gene which is itself subject to epigenetic regulation.
SULF1 Desulfates HSPGs and Downregulates Heparin-Binding Growth Factor Pathways
Effects of SULF1 on HSPG Sulfation
Most of the previously identified cellular sulfatases are localized in the lysosomal compartment and participate in the degradation of sulfated steroids and macromolecules. Initial studies of the quail and Drosophila homologs of SULF1 suggested that they were localized at the cell surface. Biochemical studies have established that quail sulfatase 1 (QSulf1) is a heparan sulfate (HS) 6-O-endosulfatase with preference, in particular, toward trisulfated IdoA2S-GlcNS6S disaccharide units within HS chains. In cells, QSulf1 can function autonomously to remodel the sulfation of cell surface HS when localized either on the cell surface or in the Golgi apparatus [19]. Dhoot et al. also proposed that QSulf1 might be involved in desulfation of cell surface HSPGs, the major sulfated macromolecule at the cell surface, and consequently in the regulation of HSPG-mediated growth factor signaling [4]. A number of direct and indirect sources of evidence have provided substantial support for this hypothesis. To determine whether SULF1 expression causes desulfation of cell surface HSPGs, Lai et al. performed immunocytochemistry using the 10E4 anti-HSPG monoclonal antibody that recognizes native heparan sulfate containing the N-sulfated glucosamine moiety. The HCC cell line SNU182, which expresses a high level of endogenous SULF1, was compared with parental SNU449 cells or vector-transfected SNU449-Vector cells, which do not express SULF1, and to three SNU449 stable clones expressing full-length SULF1. The parental SNU449 and SNU449-Vector cells showed cell surface staining for N-sulfated glucosamine-containing HSGAGs; in contrast, cell surface staining was diminished or absent in the SNU182 cell line and all three SNU449–SULF1 clones [12]. As a control to confirm the presence of HSPGs on the surface of all the cell lines, Lai et al. used the 3G10 “anti-stub” antibody after heparitinase I treatment, which confirmed the presence of HSPG stubs on both sulfatase-expressing and sulfatase-negative cell lines. Transient expression of a construct expressing antisense SULF1 mRNA restored the cell surface 10E4 anti-HSPG immunoreactivity of the sulfatase-positive cell lines. To investigate whether QSulfs dynamically modify the 6-O-sulfation patterns on the cell surface of living cells, Ai et al. used radiolabeled GAGs. They prepared extracellular matrix (ECM)-coated plates coated with 35S-radiolabeled GAG substrates by metabolically labeling the 293T cells with 35SO4 and then lysing the labeled cells, leaving ECM bound to the surface of the plate. 293T cells that stably express either QSulf1, QSulf2, or enzymatically inactive QSulf1 were then plated on the 35S-radiolabeled ECM, cultured overnight, and assayed for 35S release into the culture medium. Cells expressing QSulf1 or QSulf2 actively released 35S radioactivity, whereas control cells expressing enzymatically inactive QSulf1 released only background levels of 35S radioactivity. These and similar results from other groups provide strong direct evidence that SULF1 desulfates HSGAGs at the cell surface [12, 14, 15, 19].
Effects of SULF1 on Fibroblast Growth Factor Receptor Tyrosine Kinase Signaling
It is now evident that HSPGs serve as key regulators of heparin-binding growth factor signaling. The best studied example of this regulation is the role of HSPGs in FGF-2 signaling. HSPGs serve as co-receptors for binding of FGF-2 to its cognate FGF receptors, leading to the formation of a ternary complex that is essential for cell proliferation and angiogenesis [20, 21]. Sulfation of specific sites on the HSPGs is critical for this interaction. The interaction of FGF-2 with FGFR and HSPGs leads to receptor dimerization, activation, and autophosphorylation, followed by activation of downstream signaling pathways, including the mitogen-activated protein kinase (MAPK) pathway.
To explore the role of desulfation of cell surface HSGAGs by SULF1 in cellular growth control, several groups have investigated the effect of SULF1 expression on FGF-2 signaling in cancer cell lines [12, 14]. In all cases, formation of the FGF-2–HSGAG–FGFR ternary complex was abrogated by SULF1 expression, with subsequent inhibition of receptor dimerization and inactivation of the intracellular FGFR tyrosine kinase. The FGFR tyrosine kinase phosphorylates tyrosines in the cytoplasmic domain of the receptor as well as in the adaptor SNT/FRS, a docking protein for initiators of intracellular signaling pathways, including the MAPK pathway. Decreased phosphorylation of p44/42 (ERK1/2) MAP kinases consequently leads to decreased cell proliferation and sensitizes cells to drug-induced apoptosis [5, 6, 12, 14]. Additional data suggest that during embryonic development, SULF1 inhibits FGF-2- and FGF-4-induced mesoderm formation in the Xenopus embryo and FGF-dependent angiogenesis in the chicken embryo through 6-O-desulfation of cell surface HSGAGs and consequent inhibition of HSPG-mediated FGFR1 activation through interference with FGF–HS–FGFR1 ternary complex formation [22]. In addition, soluble heparin that is enzymatically modified by SULF1 acts as a potent inhibitor of FGF-2-induced angiogenesis in the chicken embryo [22].
SULF1 Downregulates Hepatocyte Growth Factor Signaling
Like FGF-2, HGF is also postulated to play an integral role in the unconstrained proliferation of solid tumors. HGF (also known as scatter factor) is a stromal-derived heparin-binding pleiotropic polypeptide, and its receptor c-Met is overexpressed in a variety of human cancers [23]. Binding of HGF to c-Met activates the catalytic tyrosine kinase domain of c-Met and subsequently activates the MEK and PI3K signal pathways, contributing to the expression of the pro-angiogenic factors IL-8 and VEGF [23]. Tumors that overexpress c-Met exhibit angiogenic factor expression and enhanced scattering in response to HGF in vitro, and tumorigenesis and metastasis in response to HGF in the tumor microenvironment in vivo [24]. While the overexpression of HGF and c-Met is associated with tumor metastasis and angiogenesis, the mechanisms underlying these effects remain unclear. HGF has the capacity to mediate “invasive growth”, a hallmark of metastasis, due to coordinated induction of cell motility and degradation of the extracellular matrix [25, 26]. While HGF-mediated activation of extracellular signal-regulated kinase (ERK) is postulated to be important for tumorigenesis, there is a paucity of information on the activation of ERK mediated through HGF signaling in human cancers.
HGF-mediated signaling appears to play an important role in the pathogenesis of head and neck cancer (SCCHN), and Lai et al. examined the effect of SULF1 on HGF signaling in SCCHN. SULF1 expression resulted in the abrogation of HGF-mediated activation of both the ERK and PI3K/Akt pathways, with a resultant decrease in proliferation and mitogenicity in vitro. Concomitantly, HGF-mediated cell motility and invasiveness were attenuated in SULF1-expressing head and neck cancer cell lines. Further, SULF1 expression increased the sensitivity of SCCHN cells lines to staurosporine- and cisplatin-induced apoptotic cell death. Therefore, loss of SULF1 in SCCHN leads to enhanced HGF signaling and activation of cell survival pathways [12].
SULF1 Downregulates Signaling by Heparin-Binding Epidermal Growth Factor
To determine whether SULF1 modulates HB-EGF signaling, Lai et al. examined the action of HB-EGF in parental and SULF1-transfected ovarian cancer cells. HB-EGF was chosen because of its dependence on heparin binding for its action and because of its postulated role in ovarian carcinogenesis. Overexpression of HER2 and HER4, which mediate the effects of heparin-independent EGF and HB-EGF, respectively, is well documented in ovarian cancer cells [12]. HB-EGF treatment of serum-starved parental and vector-transfected cells resulted in sustained phosphorylation of the epidermal growth factor receptor (EGFR) on both Tyr992 and Tyr1068. Both the Tyr992 and Tyr1068 amino acid residues have been implicated in EGFR-induced ERK pathway activation. Consistent with the enhanced phosphorylation of the receptor, increased ERK phosphorylation was demonstrated in these cells after HB-EGF treatment. In contrast, SULF1-expressing clones demonstrated diminished EGFR and ERK phosphorylation after HB-EGF treatment, but SULF1 did not affect the response to treatment with regular EGF [5].
SULF1 Downregulates Vascular Endothelial Growth Factor Signaling
VEGF is also a heparin-binding growth factor. Because VEGF produced by epithelial tumor cells acts on endothelial cells to stimulate angiogenesis, Narita et al. investigated the effect of SULF1 on VEGF-mediated signaling in human vascular endothelial cells (HUVECs) in culture. HUVECs constitutively express SULF1, therefore Narita and colleagues knocked down SULF1 expression using retroviral vectors expressing short hairpin RNA (shRNA) targeting two different regions of the SULF1 mRNA (pSR-SULF1) [27]. Knockdown of SULF1 significantly increased VEGF165-mediated cell proliferation as measured by [3H]thymidine incorporation in HUVEC cells. In contrast, only a minor difference in proliferation was observed between cells treated with pSR-SULF1 and pSR vector alone when proliferation was stimulated by VEGF121, an isoform lacking the heparin-binding domain. When HUVECs were treated with pSR-SULF1 together with sodium chlorate, which inhibits sulfation of HSPGs, the level of VEGF165-stimulated [3H]thymidine incorporation was comparable to that of vector-treated cells. Therefore, the increased HUVEC cell proliferation produced by knockdown of SULF1 is mediated by the heparan sulfate-binding isoform of VEGF [27].
Effects of SULF1 on Tumor Cell Growth, Migration, and Apoptosis In Vitro and In Vivo
Hepatocellular Carcinoma
SULF1 expression significantly inhibits the growth of HCC cell lines in vitro and HCC xenografts in nude mice in vivo (Fig. 1). Because HSPGs are expressed not only on epithelial cells, but also in the extracellular matrix, on endothelial cells, stellate cells, and other stromal cells, it is possible that SULF1 expression on the HCC cell surface has a paracrine effect and results in decreased sensitivity of endothelial and stromal cells to pro-angiogenic heparin-binding growth factors such as FGF-2 and VEGF. While investigating the epigenetic regulation of SULF1, Lai et al. discovered that histone H4 acetylation is upregulated by SULF1 in HCC cells. Consequently, SULF1 potentiates the inhibition of HCC tumorigenesis by HDAC inhibitors both in vitro and in vivo (Figs. 2 and 3). The potential exists for future translation of these effects into therapeutic strategies for HCC [28, 29].
Forced expression of SULF1 significantly delays growth of HCC xenografts in nude mice. a Two million Huh7 cells stably transfected with empty plasmid vector (injected into the left side of the mouse) or with a plasmid expressing the full-length SULF1 (injected into the right side of the mouse) were inoculated subcutaneously into the flanks of nude mice and the resulting xenografts photographed on the 30th day after inoculation. b The graph shows the results from xenografts implanted into ten nude mice; there was profound inhibition of growth of Huh7 xenografts expressing SULF1 (P < 0.001). c, d Immunohistochemistry was performed to confirm human SULF1 protein expression in the xenografts from cell lines stably transfected with empty vector (c) and SULF1 (d) using a rabbit polyclonal antibody against SULF1
Apicidin inhibits growth and induces apoptosis in Huh7 xenografts in nude mice. a, b Five million Huh7 cells were inoculated subcutaneously into the right flanks of nude mice. When tumor sizes reached 400–800 mm3, the mice were randomly grouped and treated by intraperitoneal injection of either 1% DMSO or apicidin at 2.5 mg/kg daily for the first week and every other day for the second week. Nude mice bearing xenografts were then killed. Tissue from the xenografts was fixed with formalin and paraffin-embedded. After H&E staining, the percent apoptosis in 500 nuclei from six randomly selected areas of each slide was counted. The relative rate of apoptosis in DMSO vs. apicidin treated mice was graphed. c Apicidin significantly induced apoptosis in Huh7 xenografts (P = 0.0007)
SULF1 enhances the efficacy of apicidin in growth inhibition of HCC xenografts. Huh7 xenografts with or without SULF1 expression were generated. When the volume of xenografts reached 400 to 800 mm3, the nude mice were grouped randomly and treated with 100 μl 10% DMSO for controls or 100 μl of apicidin dissolved in 10% DMSO at a total dose of 2.5 mg/kg body weight. The mice were treated IP daily for 1 week, every other day for 1 week and every 3 days for 2 weeks. Tumor size was measured with calipers every 3 days and the mice were killed when their tumor volume reached 4,000 mm3. The combination of SULF1 and apicidin inhibits tumor growth in mice bearing Huh7 xenografts
Pancreatic Cancer
Pancreatic cancers expressed significantly (22-fold) increased SULF1 mRNA levels compared to normal control tissues, and four out of eight pancreatic cancer cell lines examined constitutively expressed SULF1. Stable transfection of the SULF1-negative Panc-1 pancreatic cancer cell line with a full-length SULF1 expression vector resulted in increased sulfatase activity and decreased sulfation of cell surface HSPGs. Expression of SULF1 reduced both anchorage-dependent and -independent cell growth and decreased FGF-2-mediated cell growth and invasion in this cell line [14]. In contrast, transfection of Panc-1 pancreatic cancer cells with a full-length SULF1 expression vector resulted in increased invasiveness and adhesiveness. An in vivo nude mouse xenograft tumor model revealed a markedly reduced growth potential of SULF1-expressing Panc-1 cells, which correlated with the significantly lower proliferation rate in vitro. Interestingly, SULF1-positive nude mouse tumors displayed better development of interstitial matrix structures, with increased blood vessel density within these tumors. Further, in an orthotopic model, SULF1-positive tumors exhibited enhanced local invasiveness. In human primary pancreatic cancers, there was strong staining for sulfated heparan sulfate proteoglycans, which was markedly reduced in metastatic tissue samples, suggesting that increased expression of SULF1 may promote tumor metastasis. These results suggest that there are tissue-specific differences in the actions of SULF1 in cancer and also that SULF1 likely has effects on multiple cell signaling pathways that are modulated by HSPG sulfation [30].
Other Malignancies
Multiple Myeloma
Multiple myeloma is a clonal plasma cell malignancy that accounts for slightly more than 10% of all hematologic malignancies. To assess the effect of SULF expression on tumor growth in vivo, Dai et al. implanted human myeloma cells transfected with the cDNA for either SULF1 or SULF2 into severe combined immunodeficient mice. Tumor growth was dramatically reduced to the order of 5- to 10-fold in cells with enhanced expression of either sulfatase as compared with controls. In addition to inhibition of tumor growth, SULF1 also promoted a marked increase in extracellular matrix deposition within tumors that may, along with attenuated growth factor signaling, contribute to the reduction in tumor growth. These findings demonstrate that dynamic regulation of heparan sulfate structure by SULF1 present within the tumor microenvironment can have a dramatic impact on the growth and progression of malignant cells in vivo [31].
Breast Cancer
Lai et al. reported that SULF1 is downregulated in breast cancer cell lines. They also showed that MCF10A cells derived from fibrocystic breast tissue, which is generally regarded as relatively normal, also had a complete loss of SULF1 expression, possibly implicating SULF1 loss as an early event in mammary carcinogenesis [5]. A subsequent study by Narita et al. showed that SULF1 inhibits autocrine activation of the EGFR–ERK pathway induced by serum withdrawal in MDA-MB-468 breast cancer cells. Additionally, SULF1-mediated inhibition of autocrine signaling was associated with reduced cyclin D1 levels, decreased S phase and increased G2-M fractions on flow cytometry, and increased cell death. SULF1 expression was downregulated in the majority (60%) of primary invasive breast tumors, with a predominant association with lobular histology. These findings were consistent with downregulation of SULF1 in mammary carcinogenesis [13]. A similar tumor suppressor role of SULF1 was also found in head and neck squamous cell cancer [12] and ovarian cancer [5].
SULF1 Potentiates the Efficacy of Histone Deacetylase Inhibitors
Histones undergo extensive posttranslational modifications that regulate gene expression. Acetylation is a key histone modification that is primarily controlled by two enzyme families, histone deacetylases (HDACs) and histone acetyltransferases (HATs). The activity of HDACs causes transcriptional silencing of DNA. Eleven distinct zinc-dependent histone deacetylase isoforms have been identified in humans [32]. HDACs are responsible for the regulation of many genes involved in cancer cell proliferation; subsequently, HDAC inhibitors have been shown to be effective anti-cancer agents. The HDAC inhibitor, apicidin, induces acetylation of histone H4 in HCC cells. Lai et al. reported that forced expression of SULF1 enhanced apicidin-induced acetylation of histone H4 in both Huh7 and Hep3B cells. Expression of SULF1 also significantly increased apicidin-induced apoptosis in the Huh7, Hep3B, and SNU449 HCC cell lines. SULF1 and its quail homolog QSULF1 have been shown to downregulate MAPK kinase through decreased phosphorylation of ERK44/42 [5, 12, 22]. Lai et al. showed that knockdown of SULF1 by SULF1-targeting shRNA constructs upregulated phosphorylation of MEK, ERK 44/42, and AKT and attenuated apicidin-induced downregulation of MEK, ERK 44/42, and AKT phosphorylation in Hep3B and Huh7 cells. Knockdown of SULF1 significantly diminished apicidin-induced apoptosis in both Huh7 and Hep3b cells. Further, SULF1 enhanced apicidin-induced inhibition of HCC cell migration and the anti-tumor effects of apicidin [28] (Fig. 4). It has also recently been shown that combination of the anti-cancer agent doxorubicin with apicidin significantly increased the anti-tumor effect in SULF1-expressing Huh7 and Hep3B cells, as compared to treatment with either apicidin or doxorubicin alone, both in vitro and in vivo (Fig. 5) [29]. Clearly, the potential exists for future translation of the combined effects of SULF1, HDAC inhibitors, and other anti-cancer agents into therapeutic strategies for cancer.
Working model for the effect of SULF1 and HDAC inhibitors on histone acetylation and downstream pathways. Forced expression of SULF1 disrupts the balance between HDACs and HATs by inhibiting HDACs and/or by upregulating HAT activity. This leads to an increase in the acetylation of histone H4 and to subsequent downregulation of AKT and MAPK kinase pathways. Therefore, SULF1 potentiates the effects of HDAC inhibitors both in vitro and in vivo
Additive effect of apicidin and doxorubicin (DOX) in SULF1-expressing HCC cells in vivo. Huh7 xenografts with or without SULF1 expression were generated. When the volume of xenografts reached 400 to 800 mm3, the nude mice were grouped randomly and treated with 100 μl 10% DMSO for controls, 100 μl of apicidin dissolved in 10% DMSO at a dose of 2.5 mg/kg body weight, 100 μl of DOX at a dose of 3 mg/kg body weight, or the combination of apicidin with DOX. The mice were treated IP with DMSO or apicidin daily for 1 week, every other day for 1 week, and every 3 days for 2 weeks, and IV with DOX via tail vein every 6 days for four times. Tumor size was measured with calipers every 3 days and the mice were killed when their tumor volume reached 4,000 mm3. The experiments were repeated and the mice were killed at the end of the 4-week treatment. Compared to the DMSO control, DOX inhibited tumor growth of Huh7 vector xenografts (P < 0.05), but these effects were not statistically further enhanced by combination of the DOX with apicidin (a, c) [28]. Compared to the Huh7 vector xenografts, Huh7 SULF1-expressing xenografts showed more profound inhibition of tumor growth after treatment with DOX (P < 0.05). Further, in SULF1-expressing Huh7 xenografts, the combination of apicidin with DOX showed significantly enhanced anti-tumor effects as compared with the effect of either apicidin or DOX alone (P < 0.05; b, d) [28]
Evidence for Actions on SULF1 from Knockout Mouse Models
To investigate the role of SULF1 and SULF2 in vivo, Lamanna et al. generated mSulf1 and mSulf2 knockout mice by the classical approach of insertion of a neomycin-resistance cassette into exon 2 of the murine Sulf1 gene and exon 1 of the murine Sulf2 gene. They found that FGF-2 induced a 3-fold increase in proliferation of mouse embryonic fibroblasts from mSULF1−/− knockout animals compared with cells from wild-type animals [33, 34]. The focus of future studies will be analysis of the knockout mouse phenotypes, particularly to characterize the ability of the mSulfs to regulate tumorigenesis in vivo.
Unanswered Questions and Future Directions for Research
SULF1 appears to act predominantly through 6-O-desulfatation of heparan sulfate proteoglycans at the cell surface or in the extracellular matrix. Because of the complex domain structure of heparan sulfate, it is as yet unknown which specific subdomains of heparan sulfate act as substrates for SULF1, and also what the specific mechanisms and determinants are for recognition of heparan sulfate by SULF1. Clearly, there is much that is yet to be learned about this enzyme; however, the profound effects demonstrated thus far suggest that it is critically important to understand the molecular functions of SULF1, as well as the roles of the related heparan-degrading endosulfatase SULF2, in development and carcinogenesis.
Potential Therapeutic Implications of Recent Advances in Sulfatase Biology
Evidence from both embryology and cancer biology suggests that there are profound and complex effects of sulfatases on avian and mammalian development and carcinogenesis. The tumor suppressor effect of SULF1 is increasingly being recognized. Target pathways implicated in SULF1 effects include the hedgehog, Wnt and multiple heparan sulfate-dependent receptor tyrosine kinase pathways. Recently, DNA methylation inhibitors such as zebularine have shown promising activity against a variety of cancer cells both in vitro and in vivo. DNA methylation and HDAC inhibitors in combination with agents derived from a better molecular understanding of the endosulfatase genes SULF1 and SULF2 may prove to be important modalities for prevention and treatment of cancer.
References
Diez-Roux G, Ballabio A. Sulfatases and human disease. Annu Rev Genomics Hum Genet. 2005;6:355–79. doi:10.1146/annurev.genom.6.080604.162334.
Shridhar V, Sen A, Chien J, Staub J, Avula R, Kovats S, et al. Identification of underexpressed genes in early- and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002;62 1:262–70.
Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP Jr. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 2006;281 8:4969–76. doi:10.1074/jbc.M511902200.
Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP Jr. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293 5535:1663–6. doi:10.1126/science.293.5535.1663.
Lai J, Chien J, Staub J, Avula R, Greene EL, Matthews TA, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278 25:23107–17. doi:10.1074/jbc.M302203200.
Lai JP, Chien JR, Moser DR, Staub JK, Aderca I, Montoya DP, et al. hSulf1 sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126 1:231–48. doi:10.1053/j.gastro.2003.09.043.
Forsten-Williams K, Chu CL, Fannon M, Buczek-Thomas JA, Nugent MA. Control of growth factor networks by heparan sulfate proteoglycans. Ann Biomed Eng. 2008;36 12:2134–48. doi:10.1007/s10439-008-9575-z.
Kikuno R, Nagase T, Ishikawa K, Hirosawa M, Miyajima N, Tanaka A, et al. Prediction of the coding sequences of unidentified human genes. XIV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6 3:197–205. doi:10.1093/dnares/6.3.197.
Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277 51:49175–85. doi:10.1074/jbc.M205131200.
Ohto T, Uchida H, Yamazaki H, Keino-Masu K, Matsui A, Masu M. Identification of a novel nonlysosomal sulphatase expressed in the floor plate, choroid plexus and cartilage. Genes Cells. 2002;7 2:173–85. doi:10.1046/j.1356-9597.2001.00502.x.
Staub J, Chien J, Pan Y, Qian X, Narita K, Aletti G, et al. Epigenetic silencing of HSulf-1 in ovarian cancer: implications in chemoresistance. Oncogene. 2007;26 34:4969–78. doi:10.1038/sj.onc.1210300.
Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, et al. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23 7:1439–47. doi:10.1038/sj.onc.1207258.
Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, et al. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282 19:14413–20. doi:10.1074/jbc.M611395200.
Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, Felix K, et al. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer. 2005;4 1:14. doi:10.1186/1476-4598-4-14.
Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS ONE. 2007;2 4:e392. doi:10.1371/journal.pone.0000392.
Soule HD, Maloney TM, Wolman SR, Peterson WD Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50 18:6075–86.
Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6 3:236–44. doi:10.1038/ng0394-236.
Chen Z, Fan JQ, Li J, Li QS, Yan Z, Jia XK, et al. Promoter hypermethylation correlates with the Hsulf-1 silencing in human breast and gastric cancer. Int J Cancer. 2009;124 3:739–44. doi:10.1002/ijc.23960.
Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP Jr. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162 2:341–51. doi:10.1083/jcb.200212083.
Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, et al. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem. 2000;275 32:24653–60. doi:10.1074/jbc.M908930199.
Delehedde M, Lyon M, Gallagher JT, Rudland PS, Fernig DG. Fibroblast growth factor-2 binds to small heparin-derived oligosaccharides and stimulates a sustained phosphorylation of p42/44 mitogen-activated protein kinase and proliferation of rat mammary fibroblasts. Biochem J. 2002;366 Pt 1:235–44.
Wang S, Ai X, Freeman SD, Pownall ME, Lu Q, Kessler DS, et al. QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc Natl Acad Sci USA. 2004;101 14:4833–8. doi:10.1073/pnas.0401028101.
Dong G, Chen Z, Li ZY, Yeh NT, Bancroft CC, Van Waes C. Hepatocyte growth factor/scatter factor-induced activation of MEK and PI3K signal pathways contributes to expression of proangiogenic cytokines interleukin-8 and vascular endothelial growth factor in head and neck squamous cell carcinoma. Cancer Res. 2001;61 15:5911–8.
Dong G, Lee TL, Yeh NT, Geoghegan J, Van Waes C, Chen Z. Metastatic squamous cell carcinoma cells that overexpress c-Met exhibit enhanced angiogenesis factor expression, scattering and metastasis in response to hepatocyte growth factor. Oncogene. 2004;23 37:6199–208. doi:10.1038/sj.onc.1207851.
Matsumoto K, Matsumoto K, Nakamura T, Kramer RH. Hepatocyte growth factor/scatter factor induces tyrosine phosphorylation of focal adhesion kinase (p125FAK) and promotes migration and invasion by oral squamous cell carcinoma cells. J Biol Chem. 1994;269 50:31807–13.
Galeazzi E, Olivero M, Gervasio FC, De Stefani A, Valente G, Comoglio PM, et al. Detection of MET oncogene/hepatocyte growth factor receptor in lymph node metastases from head and neck squamous cell carcinomas. Eur Arch Otorhinolaryngol. 1997;254 Suppl 1:S138–43. doi:10.1007/BF02439745.
Narita K, Staub J, Chien J, Meyer K, Bauer M, Friedl A, et al. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66 12:6025–32. doi:10.1158/0008-5472.CAN-05-3582.
Lai JP, Yu C, Moser CD, Aderca I, Han T, Garvey TD, et al. SULF1 inhibits tumor growth and potentiates the effects of histone deacetylase inhibitors in hepatocellular carcinoma. Gastroenterology. 2006;130 7:2130–44. doi:10.1053/j.gastro.2006.02.056.
Lai J-P, Sandhu D, Moser C, Cazanave S, Oseini A, Shire A, Shridhar V, Sanderson S, Roberts L. Additive effect of apicidin and doxorubicin in sulfatase 1 (SULF1)-expressing hepatocellular carcinoma in vitro and in vivo. J Hepatol. 2009; in press.
Abiatari I, Kleeff J, Li J, Felix K, Buchler MW, Friess H. Hsulf-1 regulates growth and invasion of pancreatic cancer cells. J Clin Pathol. 2006;59 10:1052–8. doi:10.1136/jcp.2005.031716.
Dai Q, Qian SB, Li HH, McDonough H, Borchers C, Huang D, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280 46:38673–81. doi:10.1074/jbc.M507986200.
Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des. 2008;14 6:505–28. doi:10.2174/138161208783885353.
Lamanna WC, Baldwin RJ, Padva M, Kalus I, Ten Dam G, van Kuppevelt TH, et al. Heparan sulfate 6-O-endosulfatases: discrete in vivo activities and functional co-operativity. Biochem J. 2006;400 1:63–73. doi:10.1042/BJ20060848.
Lamanna WC, Frese MA, Balleininger M, Dierks T. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J Biol Chem. 2008;283 41:27724–35. doi:10.1074/jbc.M802130200.
Acknowledgments
This study was supported by the Mayo Clinic and Mayo Clinic Cancer Center, NIH Grants CA82862 and CA100882, an Industry Research Scholar Award from the Foundation for Digestive Health and Nutrition, a Harold Amos Medical Faculty Development Award from The Robert Wood Johnson Foundation, a generous gift from The Richard M. Schulze Family Foundation, and the Miles and Shirley Fiterman Center for Digestive Diseases at the Mayo Clinic, Rochester, MN, USA. Dr. Roberts and Mayo Clinic have a potential financial interest associated with technology used in research referenced in this review. The authors thank Vicki Campion for secretarial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lai, JP., Sandhu, D.S., Shire, A.M. et al. The Tumor Suppressor Function of Human Sulfatase 1 (SULF1) in Carcinogenesis. J Gastrointest Canc 39, 149–158 (2008). https://doi.org/10.1007/s12029-009-9058-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-009-9058-y