Abstract
Multiple sclerosis (MS) exhibits sex bias in disease clinical course as male MS patients develop severe, progressive clinical course with accumulating disability. So far, no factors have been found associating with this sex bias in MS severity. We set out to determine the genetic factor contributing to MS male-specific progressive disease. This is an MS cross-sectional study involving 213 Kuwaiti MS patients recruited at Dasman Diabetes Institute. Exome sequencing was performed on 18 females and 8 male MS patients’ genomic DNA. rs5945430 genotyping was performed using Taqman genotyping assay. Estradiol levels were determined by enzyme-linked immunosorbent assay. Exome analysis revealed a missense variant (rs5945430) in Plexin A3 (PLXNA3) gene (Xq28) associated with male-specific MS severity. Genotyping of 187 MS patients for rs5945430 confirmed the association of rs5945430G with increased disease severity in MS males (p = 0.013; OR 3.8; 95% CI 1.24–11.7) and disability (p = 0.024). Estradiol levels shown to effect PLXNA3 expression were lower in MS males compared to MS females, and they were lower than control rs5945430G males (p = 0.057), whereas MS females had similar estradiol levels to healthy females reducing the level of expressed PLXNA3 GG in MS females. PLXNA3 rs5945430G is associated with increased disease severity in MS male patients. Estradiol is a possible protective factor against the expression of rs5945430G in MS females.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) is a chronic complex demyelinating disorder resulting from central nervous system (CNS)-targeted autoimmunity manifesting primarily as lesions in the white matter and progressively in the gray matter of the CNS (Filippi et al. 2013; Perez-Miralles et al. 2013). MS is considered a multifactorial complex disorder, and many genetic and environmental factors are involved in MS pathogenesis. Among the strongest genetic susceptibility associations in MS are the major histocompatibility complex (MHC) genes. The most significant associations were seen in polymorphisms in HLA-DRB1 alleles albeit the association was variable among populations (Barcellos et al. 2006). Several genome-wide association studies have been performed in different MS populations; however, these studies produced different susceptibility markers and modifier genes highlighting ethnic, geo-epidemiological, and environmental factors associating with the disease (Kallaur et al. 2011).
The International Multiple Sclerosis Genetics Consortium (IMSGC) in a collaborative effort isolated 57 genetic variants of substantial evidence and assessed their association with MS risk in 13 European populations and US population (International Multiple Sclerosis Genetics et al. 2011). The majority of variants assessed were noncoding variants which the IMSGC have listed several nearby genes of functional relevance to MS that might be influenced by these variants; however, the 57 variants had variable associations with MS risk among the studied populations. To date, more than 100 genetic risk factors have been reported worldwide with individual MS population risk associations albeit the majority with negative or no result replication in other populations (Bashinskaya et al. 2015). While many genetic factors have been reported in association with MS disease risk, few were associated with MS clinical characteristics such as sex bias in disease progression and severity.
The general convention is that the incidence of MS among the sexes is at a ratio of 2:1, affecting females twice as many as males (Duquette et al. 1992). Male MS patients display a more progressive clinical course, with rapidly accumulating disability when compared to MS females (Ribbons et al. 2015; Al-Temaimi et al. 2015). The cause for gender bias in MS is unclear; however, it is suggested that an immunomodulatory sex-influenced genetic variant(s) have an attenuated expression in females, but a profound effect in males. Such genetic factors are still elusive for MS; however, several mechanisms that drive gender bias have been proposed as more gender-specific neuro- and immuno-physiological differences are discovered (Bordon 2014). The clinical value of such factor discovery is to anticipate rapid disease progression in predisposed male MS patients to offer better disease management options and aggressive treatment modalities in advance. Here we present a novel experimental design using next-generation technology (NGS) exome sequencing to compare male and female MS exomes. Our finding highlighted a variant of Plexin A3 (PLXNA3, Gene ID: 5558) as the variant associated with increased MS severity specifically in males. We investigated our finding in a larger cohort and found our results to be consistent in the replicate study.
Methods
Case–Control Sample Collection
A total of 213 MS patients were randomly recruited for this study at Dasman Diabetes Institute while attending their routine monthly appointments. All study protocols were approved by Dasman diabetes institute ethical review committee which adheres to the Declaration of Helsinki’s Ethical Principles for Medical Research Involving Human Subjects. A hundred and five healthy control individuals were volunteers from the general population recruited by social networking and were recruited to determine Kuwaiti-specific data for variables included in this study. All information pertinent to study protocols was fully explained to all participants prior to procurement of their informed written consent. MS patients’ inclusion criteria were: a detailed clinical history (demographics, age of onset, disease duration, expanded disability status scale (EDSS) score, and treatment history), being a Kuwaiti born citizen, MS disease duration of ≥2 year, and the agreement to provide a blood sample. Exclusion criteria included having an EDSS score of 0, as MS severity score (MSSS) would be uninformative and MS type unclear. Healthy controls’ exclusion criteria included: being a non-Kuwaiti, having a family history of MS, and a diagnosis of an autoimmune or neurodegenerative disorder.
Exome Sequencing
Based on our Kuwaiti exome alignment of 250 Kuwaiti exomes, the majority of Kuwaitis align to European ancestry with a small percent (2.4%) aligning to admixed European/African genetic ancestry. Eight male MS patients and 18 female MS patients were selected for exome sequencing based on their EDSS score and having the disease for more than two years (Table 1). For every disease duration in years, one MS male of a given EDSS score was matched to two or three MS females of an approximate disease duration but a lower EDSS score. Whole blood was withdrawn in EDTA vacutainers and centrifuged at 2500 rpm at room temperature for fractionation. Plasma fractions were retained and stored at −80 °C, and buffy coat fractions were collected for DNA extraction. DNA extraction was performed using Qiagen DNA mini kit (Qiagen, Germany) according to manufacturer’s standard protocols. DNA concentration and quality were assessed using spectrophotometry and agarose gel electrophoresis. Exome sequencing was performed on an Illumina HiSeq2000 platform using SureSelectXT v5 library preparation with target coverage of 50× (Illumina, CA, USA). Sequences were checked for quality using FastQC software, and BWA software was used to map and align sequences to reference hg19 sequences. Picard was used to remove duplicate reads, and SAMtools was used to convert sequences from SAM format to sorted and indexed BAM files. GenomeAnalysisTK (GATK) was used to analyze the sequences for genotype calling, and SnpEff was used to annotate variants and predict their effects. Resultant exomes were uploaded for comparative analysis using Tute Genomics application (Tute Genomics, Inc., UT, USA). Genes with variants that have a Fisher exact test p values of highest significance were selected for further analysis in our MS cohort.
rs5945430 Genotyping
Remaining blood samples of MS patients and healthy controls were processed using the same protocol mentioned above for DNA extraction. A genetic variant (rs5945430) in PLXNA3 that had the most significant association with male severe phenotype as detected by exome sequencing was further investigated in MS patients and healthy controls. Taqman genotyping assay for single nucleotide polymorphism (SNP) rs5945430 was used to genotype the samples. In brief, 50 ng of DNA from every sample was used for genotyping using ABI7500 Fast Real-time PCR System (Applied Biosystems, CA, USA). Genotype zygosity was determined by SDS v1.4.1 software (Applied Biosystems, CA, USA). Males were scored hemizygous for this SNP, and females had biallelic genotypes.
ELISA
Human Estradiol ELISA (Invitrogen, CA, USA) kits were used to assay for estradiol levels in both cohorts. The assay is based on solid-phase competitive colorimetric immunoassay principle. A fixed amount of estradiol molecule was labeled with horseradish peroxidase (HRP) which competes with unlabeled estradiol present in standards or samples for a limited number of binding sites of a target-specific antibody. After 2-h incubation at room temperature, the microtiter plate will be washed to stop the competition reaction. The chromogen solution tetramethylbenzidine (TMB) will be added and incubated for 30 min. The reaction will be stopped with H2SO4, and the absorbance will be measured at the appropriate wavelength. The intensity of this colored product is inversely proportional to the concentration (pg/mL) of estradiol present in the original plasma fraction.
Statistical Analysis
Power analyses were conducted using GraphPad StatMate (GraphPad Software, CA, USA). According to 1000 genome data, rs5945430G frequency is 0.2, and an unequal cohort size at a ratio of 2 would have 95% power to detect an increase of 0.19 between MS cases and healthy control G allele frequencies. For estradiol levels power analysis, males and females were assessed separately. Estradiol levels in males (n = 65) have a 95% power to detect a difference between means of 1.78 (α = 0.05, two-tailed). For females (n = 122), we expected greater variability and a 90% power to detect a difference between means of 101.97 (α = 0.05, two-tailed) was accepted. For nonparametric data, Mann–Whitney U test was used. For exome and genotype analysis, Fisher’s exact test, Chi-square test, and linear regression analyses were used. A Bonferroni-adjusted p value <0.025 was considered significant for all tests. For statistical purposes, EDSS score was grouped into mild (0–2.5), intermediate (3–5.5) to severe (6–8), and MS severity score (MSSS) was assigned to each patient and an MSSS <3 was considered mild–moderate and >3 as severe (Roxburgh et al. 2005). For EDSS and MSSS association analyses PPMS cases (4 males), we excluded to prevent mismatching with MS females of similar scores but a different MS type. All statistical analyses were performed using GraphPad Prism v.6.05 (GraphPad Software, CA, USA).
Results
Exome Analysis
Aligned and annotated exomes were analyzed by comparing 8 MS males with 18 MS females. The selection was made to compare the genetic factors that predispose MS males to have a more severe MS clinical course than females when controlling for disease type, age of onset, sex, and age (Table 1). All 26 exomes were determined by sequence alignment to be of European genetic ancestry. The most significant genes with variants segregating our two cohorts ordered from highest to lowest significance were arylsulfatase D (ARSD, p = 1.2400 X 10−5), zinc finger protein 761 (ZNF761, p = 1.6900 × 10−5), and PLXNA3 (p = 3.63 × 10−5). ARSD (Xp22.33) was excluded after several failed genotyping attempts for this allele in male samples using primers specific to chromosome X, as ARSD has a pseudogene copy on the Y chromosome harboring numerous deleterious genetic mutations (Meroni et al. 1996). ZNF761 was excluded as the variant had a 1000 genome frequency of 1. PLXNA3 (Xq28) missense mutation rs5945430 was the best fit candidate for further investigation according to our criteria (, c.2589C > G; p.D863E), as the G allele was found in 8 females in heterozygosity and in 6 males in hemizygosity. We selected this variant for a replication study in the remaining 187 MS samples.
PLXNA3 rs5945430 is Associated with Severe MS Course in Males
PLXNA3 rs5945430 was genotyped in 65 male MS patients and 122 MS females. MS cohort demographics and clinical characteristics are shown in Table 2. rs5945430-specific frequency in the Kuwaiti population was estimated using genotypes of 105 healthy Kuwaiti controls. Healthy control cohort included 48 males and 65 females. rs5945430 allele frequencies in healthy controls and MS patients are shown in Table 3. ExAC database lists allele G as the minor allele for rs5945430 with a frequency of 0.22 across populations. In our sampled Kuwaiti population, the total frequency was higher (0.41). Allele frequency between the two cohorts did not differ significantly (p = 0.07), whereas genotype frequency differed (p = 0.014) with more CG and less CC/C representation in the MS cohort. Comparing rs5945430 genotype distribution in relation to MS clinical characteristics showed a significant association of allele G with higher MSSS in MS males (p = 0.0066). Allele G was found to increase the risk of an aggressive MS clinical course specifically in male MS patients (p = 0.013; OR 3.8; 95% CI 1.16–11.7). No significant association was found between rs5945430 genotype and MSSS in female MS patients (p = 0.065). Moreover, males hemizygous for allele G had higher EDSS scores (p = 0.024). Similarly, this association was male specific, and no association was found in females for EDSS (p = 0.76). We determined the expressed allele in heterozygous females by performing rs5945430 genotyping assays on cDNA synthesized from total RNA extracts using primers specific for our region of interest. Successful genotyping of heterozygous females showed a biallelic expression of this gene in the blood under the influence of random X-inactivation in blood cell lineages (data not shown).
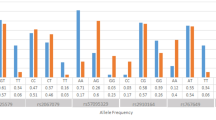
To further investigate the sex influence on PLXNA3 association with MS severity, we assessed estradiol levels in our cohort. It has been shown that PLXNA3 levels are reduced by increasing estradiol levels in vitro (GEO profile ID GDS3600) (Sobrino et al. 2009). Our hypothesis was that PLXNA3 GG effects in MS females are masked due to increased levels of estradiol, therefore mitigating its effect. Overall estradiol levels were lower in MS males compared to MS females. Male MS patients’ estradiol levels differed significantly from healthy male controls (p < 0.001). Moreover, MS allele G males had lower estradiol levels than their healthy male G counterparts albeit the mean difference (7.347 pg/mL) did not reach significance (p = 0.057) due to low sample number in the MS male G cohort (Fig. 1). This suggests that the effect of PLXNA3 G allele is possibly augmented by reduced estradiol levels specifically in MS males. No significant difference in estradiol levels was found between healthy females and MS females (p = 0.19). However, due to the cross-sectional nature of our study we could not determine the effect of menopause on rs5945430GG MS females’ EDSS and MSSS scores. In addition, our cohort only has 12.3% of females at or above the age of 45 years, and only one of those patients was genotype GG.
Discussion
Recent reports have placed plexins at the forefront of neuroinflammation research (Belyk et al. 2015; Ito et al. 2014; Okuno et al. 2010; Kremer et al. 2015). In the present study, a unique approach to analyze sex bias in MS severity was conducted utilizing exome sequencing. MS exome analysis revealed a novel association of PLXNA3 missense variant rs5945430G with MS male increased disease severity. The increased disease severity was assessed by two MS severity quantifiable variables EDSS and MSSS and was found to be consistently significant in MS males. PLXNA3 is located on chromosome Xq28 and is a class 3 semaphorin receptor and is the most ubiquitously expressed plexin in the developing mammalian CNS. PLXNA3 mediates its functions by binding semaphorin 3A and 3F (SEMA3A, SEMA3F) simultaneously with neuropilin-2 (NPN-2) (Cheng et al. 2001; Yaron et al. 2005). Evidence-based studies have shown PLXNA3 to be involved in axon pathfinding, axonal branches and mossy fiber pruning, oligodendrocyte cell migration, and regulation of bone innervation (Bagri et al. 2003; Yaron et al. 2005; Gomez et al. 2005; Waimey et al. 2008; Xiang et al. 2012). Other functions include induction of apoptosis in macrophages and regulation of the immune response (Ji et al. 2009; Fiedler et al. 2010). Our findings suggest that rs5945430 is a disease-modifying variant contributing to the bias in MS severity among the sexes. It is likely that the change from aspartic acid to glutamic acid alters the protein folding efficiency under specific temperatures (Lee et al. 2004). The missense residue is located in the first IPT (Ig-like plexins repeat) domain which has an immunoglobulin-like fold. This domain is part of PLXNA3 ectodomain that binds semaphorins, and is found in cell surface receptors and intracellular transcription factors (Janssen et al. 2010).
While the exact molecular mechanism for this variant is unclear, it is plausible that this mutation results in PLXNA3 protein of altered stability that possibly increases/decreases its interaction with semaphorins and possibly neuropilin. Both SEMA3A and SEMA3F can regulate synaptic transmission in the brain and promote regulatory cytoskeletal remodeling in the adult CNS, and dysfunctional regulation of their effects will ultimately impact physiological state of the CNS (Sahay et al. 2005; Kolk et al. 2009; Tran et al. 2009). Moreover, plexins and their substrate semaphorins play a pivotal role in oligodendrocyte remyelination of MS demyelinated plaques and migratory behavior, as SEMA3A interaction with PLXNA3 impairs oligodendroglial migration to demyelination sites contrary to SEMA3F (Piaton et al. 2011). Our findings elude to a molecular mechanism that hinders appropriate remyelination/axonal growth possibly due to sustained interaction with SEMA3A or preferential interaction with SEMA3A than SEMA3F resulting in excessive or premature axonal pruning of demyelinated axons instead of repair resulting in accumulation of neuro-functional disability (Coleman and Perry 2002). However, this hypothesis remains to be investigated although recent evidence suggests that semaphorins, specifically SEMA3A, and their receptors have altered expression in MS lesions which strengthens our finding’s credibility and warrants further investigation (Williams et al. 2007; Costa et al. 2015).
Furthermore, the sex-specific association of this variant suggests the influence of a discriminatory factor. We sought to investigate the most obvious candidate, namely sex hormones. Estradiol and testosterone aside from their sexual regulatory functions also have immunomodulatory and CNS functions (Gubbels Bupp 2015; Spence and Voskuhl 2012). Estradiol, the female sex hormone, was investigated in the animal model of MS and in MS patients for its potential effects on MS course (Kipp et al. 2016). Those studies show that clinical severity of MS was reduced by estradiol treatment, and a significant reduction in MS relapse and symptoms in pregnant MS patients during which the female sex hormone is high (Bebo et al. 2001; Confavreux et al. 1998; Runmarker and Andersen 1995). Postmenopausal MS females also report worsening of MS symptoms and rapid progression in disability in conjunction with reduced estradiol levels (Christianson et al. 2015; Bove et al. 2015). Moreover, studies have conclusively shown that estradiol levels are lower in MS males compared to controls (Caruso et al. 2014). Furthermore, in an in vitro study using microarray expression profiling of polymorphonuclear cells from MS patients treated with estradiol reduced the expression of PLXNA3 (Sobrino et al. 2009). It is possible that low estradiol levels in MS males allow for PLXNA3 expression in higher magnitude than MS females. A dysfunctional PLXNA3 would further complicate the overexpression in an already damaged CNS that is undergoing neurodegeneration and inflammation. While our hypothesis is not efficiently proven due mostly to low sample size, it is a plausible explanation for the lack of association of PLXNA3 variant with disease severity in MS females. Our future plan is to fully investigate the role of sex hormones on this variant’s expression and fully characterize this variant in vitro.
In conclusion, we presented a novel experimental approach utilizing exome sequencing and sub-cohort classification to better analyze gender clinical course disparities in MS. A major finding of this study is the association of a missense mutation in PLXNA3 with increased disability in MS males. The effect of PLXNA3 rs5945430G allele is only seen in low-estradiol MS patients and specifically males. Further investigation of this variant as a potential prognostic marker for MS disease, and treatment modalities to counteract its effect using estradiol offer valuable disease management protocols for MS.
References
Al-Temaimi, R., Alroughani, R., Jacob, S., & Al-Mulla, F. (2015). Gender influence in EBV antibody response in multiple sclerosis patients from Kuwait. Journal of Neuroimmunology, 285, 57–61. doi:10.1016/j.jneuroim.2015.05.021.
Bagri, A., Cheng, H. J., Yaron, A., Pleasure, S. J., & Tessier-Lavigne, M. (2003). Stereotyped pruning of long hippocampal axon branches triggered by retraction inducers of the semaphorin family. Cell, 113, 285–299.
Barcellos, L. F., Sawcer, S., Ramsay, P. P., Baranzini, S. E., Thomson, G., Briggs, F., et al. (2006). Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Human Molecular Genetics, 15, 2813–2824. doi:10.1093/hmg/ddl223.
Bashinskaya, V. V., Kulakova, O. G., Boyko, A. N., Favorov, A. V., & Favorova, O. O. (2015). A review of genome-wide association studies for multiple sclerosis: Classical and hypothesis-driven approaches. Human Genetics, 134, 1143–1162. doi:10.1007/s00439-015-1601-2.
Bebo, B. F., Jr., Fyfe-Johnson, A., Adlard, K., Beam, A. G., Vandenbark, A. A., & Offner, H. (2001). Low-dose estrogen therapy ameliorates experimental autoimmune encephalomyelitis in two different inbred mouse strains. J Immunol, 166, 2080–2089.
Belyk, M., Kraft, S. J., Brown, S., Pediatric Imaging, N., & Genetics, S. (2015). PlexinA polymorphisms mediate the developmental trajectory of human corpus callosum microstructure. Journal of Human Genetics, 60, 147–150. doi:10.1038/jhg.2014.107.
Bordon, Y. (2014). Autoimmunity: A breakthrough to explain sex bias? Nature Reviews Immunology, 14, 355. doi:10.1038/nri3696.
Bove, R., Healy, B. C., Musallam, A., Glanz, B. I., De Jager, P. L., & Chitnis, T. (2015). Exploration of changes in disability after menopause in a longitudinal multiple sclerosis cohort. Mult Scler. doi:10.1177/1352458515606211.
Caruso, D., Melis, M., Fenu, G., Giatti, S., Romano, S., Grimoldi, M., et al. (2014). Neuroactive steroid levels in plasma and cerebrospinal fluid of male multiple sclerosis patients. Journal of Neurochemistry, 130, 591–597. doi:10.1111/jnc.12745.
Cheng, H. J., Bagri, A., Yaron, A., Stein, E., Pleasure, S. J., & Tessier-Lavigne, M. (2001). Plexin-A3 mediates semaphorin signaling and regulates the development of hippocampal axonal projections. Neuron, 32, 249–263.
Christianson, M. S., Mensah, V. A., & Shen, W. (2015). Multiple sclerosis at menopause: Potential neuroprotective effects of estrogen. Maturitas, 80, 133–139. doi:10.1016/j.maturitas.2014.11.013.
Coleman, M. P., & Perry, V. H. (2002). Axon pathology in neurological disease: A neglected therapeutic target. Trends in Neurosciences, 25, 532–537.
Confavreux, C., Hutchinson, M., Hours, M. M., Cortinovis-Tourniaire, P., & Moreau, T. (1998). Rate of pregnancy-related relapse in multiple sclerosis: Pregnancy in multiple sclerosis group. New England Journal of Medicine, 339, 285–291. doi:10.1056/NEJM199807303390501.
Costa, C., Martinez-Saez, E., Gutierrez-Franco, A., Eixarch, H., Castro, Z., Ortega-Aznar, A., et al. (2015). Expression of semaphorin 3A, semaphorin 7A and their receptors in multiple sclerosis lesions. Multiple Sclerosis Journal, 21, 1632–1643. doi:10.1177/1352458515599848.
Duquette, P., Pleines, J., Girard, M., Charest, L., Senecal-Quevillon, M., & Masse, C. (1992). The increased susceptibility of women to multiple sclerosis. Canadian Journal of Neurological Sciences, 19, 466–471.
Fiedler, S. E., Schillace, R. V., Daniels, C. J., Andrews, S. F., & Carr, D. W. (2010). Myeloid translocation gene 16b is a dual A-kinase anchoring protein that interacts selectively with plexins in a phospho-regulated manner. FEBS Letters, 584, 873–877. doi:10.1016/j.febslet.2010.02.007.
Filippi, M., Preziosa, P., Copetti, M., Riccitelli, G., Horsfield, M. A., Martinelli, V., et al. (2013). Gray matter damage predicts the accumulation of disability 13 years later in MS. Neurology, 81, 1759–1767. doi:10.1212/01.wnl.0000435551.90824.d0.
Gomez, C., Burt-Pichat, B., Mallein-Gerin, F., Merle, B., Delmas, P. D., Skerry, T. M., et al. (2005). Expression of Semaphorin-3A and its receptors in endochondral ossification: Potential role in skeletal development and innervation. Developmental Dynamics, 234, 393–403. doi:10.1002/dvdy.20512.
Gubbels Bupp, M. R. (2015). Sex, the aging immune system, and chronic disease. Cellular Immunology, 294, 102–110. doi:10.1016/j.cellimm.2015.02.002.
International Multiple Sclerosis Genetics Consortium, and Wellcome Trust Case Control Consortium, Sawcer, S., Hellenthal, G., Pirinen, M., Spencer, C. C., et al. (2011). Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature, 476, 214–219. doi:10.1038/nature10251.
Ito, T., Morita, T., Yoshida, K., Negishi, T., & Yukawa, K. (2014). Semaphorin 3A-Plexin-A1 signaling through ERK activation is crucial for Toll-like receptor-induced NO production in BV-2 microglial cells. International Journal of Molecular Medicine, 33, 1635–1642. doi:10.3892/ijmm.2014.1727.
Janssen, B. J., Robinson, R. A., Perez-Branguli, F., Bell, C. H., Mitchell, K. J., Siebold, C., et al. (2010). Structural basis of semaphorin-plexin signalling. Nature, 467, 1118–1122. doi:10.1038/nature09468.
Ji, J. D., Park-Min, K. H., & Ivashkiv, L. B. (2009). Expression and function of semaphorin 3A and its receptors in human monocyte-derived macrophages. Human Immunology, 70, 211–217. doi:10.1016/j.humimm.2009.01.026.
Kallaur, A. P., Kaimen-Maciel, D. R., Morimoto, H. K., Watanabe, M. A., Georgeto, S. M., & Reiche, E. M. (2011). Genetic polymorphisms associated with the development and clinical course of multiple sclerosis (review). International Journal of Molecular Medicine, 28, 467–479. doi:10.3892/ijmm.2011.731.
Kipp, M., Hochstrasser, T., Schmitz, C., & Beyer, C. (2016). Female sex steroids and glia cells: Impact on multiple sclerosis lesion formation and fine tuning of the local neurodegenerative cellular network. Neuroscience and Biobehavioral Reviews, 67, 125–136. doi:10.1016/j.neubiorev.2015.11.016.
Kolk, S. M., Gunput, R. A., Tran, T. S., van den Heuvel, D. M., Prasad, A. A., Hellemons, A. J., et al. (2009). Semaphorin 3F is a bifunctional guidance cue for dopaminergic axons and controls their fasciculation, channeling, rostral growth, and intracortical targeting. Journal of Neuroscience, 29, 12542–12557. doi:10.1523/JNEUROSCI.2521-09.2009.
Kremer, D., Hartung, H. P., & Kury, P. (2015). Targeting semaphorins in MS as a treatment strategy to promote remyelination: A tale of mice, rats and men. Multiple Sclerosis, 21, 1616–1617. doi:10.1177/1352458515608693.
Lee, D. Y., Kim, K. A., Yu, Y. G., & Kim, K. S. (2004). Substitution of aspartic acid with glutamic acid increases the unfolding transition temperature of a protein. Biochemical and Biophysical Research Communications, 320, 900–906. doi:10.1016/j.bbrc.2004.06.031.
Meroni, G., Franco, B., Archidiacono, N., Messali, S., Andolfi, G., Rocchi, M., et al. (1996). Characterization of a cluster of sulfatase genes on Xp22.3 suggests gene duplications in an ancestral pseudoautosomal region. Human Molecular Genetics, 5, 423–431.
Okuno, T., Nakatsuji, Y., Moriya, M., Takamatsu, H., Nojima, S., Takegahara, N., et al. (2010). Roles of Sema4D-plexin-B1 interactions in the central nervous system for pathogenesis of experimental autoimmune encephalomyelitis. The Journal of Immunology, 184, 1499–1506. doi:10.4049/jimmunol.0903302.
Perez-Miralles, F., Sastre-Garriga, J., Tintore, M., Arrambide, G., Nos, C., Perkal, H., et al. (2013). Clinical impact of early brain atrophy in clinically isolated syndromes. Multiple Sclerosis, 19, 1878–1886. doi:10.1177/1352458513488231.
Piaton, G., Aigrot, M. S., Williams, A., Moyon, S., Tepavcevic, V., Moutkine, I., et al. (2011). Class 3 semaphorins influence oligodendrocyte precursor recruitment and remyelination in adult central nervous system. Brain, 134, 1156–1167. doi:10.1093/brain/awr022.
Ribbons, K. A., McElduff, P., Boz, C., Trojano, M., Izquierdo, G., Duquette, P., et al. (2015). Male sex is independently associated with faster disability accumulation in relapse-onset ms but not in primary progressive MS. PLoS ONE, 10, e0122686. doi:10.1371/journal.pone.0122686.
Roxburgh, R. H., Seaman, S. R., Masterman, T., Hensiek, A. E., Sawcer, S. J., Vukusic, S., et al. (2005). Multiple sclerosis severity score: Using disability and disease duration to rate disease severity. Neurology, 64, 1144–1151. doi:10.1212/01.WNL.0000156155.19270.F8.
Runmarker, B., & Andersen, O. (1995). Pregnancy is associated with a lower risk of onset and a better prognosis in multiple sclerosis. Brain, 118(Pt 1), 253–261.
Sahay, A., Kim, C. H., Sepkuty, J. P., Cho, E., Huganir, R. L., Ginty, D. D., et al. (2005). Secreted semaphorins modulate synaptic transmission in the adult hippocampus. Journal of Neuroscience, 25, 3613–3620. doi:10.1523/JNEUROSCI.5255-04.2005.
Sobrino, A., Mata, M., Laguna-Fernandez, A., Novella, S., Oviedo, P. J., Garcia-Perez, M. A., et al. (2009). Estradiol stimulates vasodilatory and metabolic pathways in cultured human endothelial cells. PLoS ONE, 4, e8242. doi:10.1371/journal.pone.0008242.
Spence, R. D., & Voskuhl, R. R. (2012). Neuroprotective effects of estrogens and androgens in CNS inflammation and neurodegeneration. Frontiers in Neuroendocrinology, 33, 105–115. doi:10.1016/j.yfrne.2011.12.001.
Tran, T. S., Rubio, M. E., Clem, R. L., Johnson, D., Case, L., Tessier-Lavigne, M., et al. (2009). Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature, 462, 1065–1069. doi:10.1038/nature08628.
Waimey, K. E., Huang, P. H., Chen, M., & Cheng, H. J. (2008). Plexin-A3 and plexin-A4 restrict the migration of sympathetic neurons but not their neural crest precursors. Development Biology, 315, 448–458. doi:10.1016/j.ydbio.2008.01.002.
Williams, A., Piaton, G., Aigrot, M. S., Belhadi, A., Theaudin, M., Petermann, F., et al. (2007). Semaphorin 3A and 3F: Key players in myelin repair in multiple sclerosis? Brain, 130, 2554–2565. doi:10.1093/brain/awm202.
Xiang, X., Zhang, X., & Huang, Q. L. (2012). Plexin A3 is involved in semaphorin 3F-mediated oligodendrocyte precursor cell migration. Neuroscience Letters, 530, 127–132. doi:10.1016/j.neulet.2012.09.058.
Yaron, A., Huang, P. H., Cheng, H. J., & Tessier-Lavigne, M. (2005). Differential requirement for Plexin-A3 and -A4 in mediating responses of sensory and sympathetic neurons to distinct class 3 Semaphorins. Neuron, 45, 513–523. doi:10.1016/j.neuron.2005.01.013.
Acknowledgements
This research was funded by Kuwait Foundation for the Advancement of Science (KFAS) Grant Number 2012-1302-02.
Author’s Contributions
MQ and MH performed DNA extraction and genotyping experiments. SPJ performed the ELISA experiment. RA facilitated patient sample collection and provided clinical data of MS patients. RAA conceived the project’s design, implementation, exome sequencing, data analysis and wrote the manuscript. All authors have reviewed and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Qureshi, M., Hatem, M., Alroughani, R. et al. PLXNA3 Variant rs5945430 is Associated with Severe Clinical Course in Male Multiple Sclerosis Patients. Neuromol Med 19, 286–292 (2017). https://doi.org/10.1007/s12017-017-8443-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-017-8443-0