Abstract
Objective To test if mitochondrial (mt)DNA deletions accumulate in brains of patients with multiple sclerosis (MS). Background Previous studies demonstrated an accumulation of oxidative damage to mtDNA and decreased activity of mitochondrial enzymes in lesions of MS, where activated immune cells produce increased amounts of reactive oxygen species and nitric oxide. The unknown link between oxidative damage and decreased activity of mitochondrial enzymes may be the accumulation of deletions in mtDNA molecules. mtDNA deletions in the brain have been associated with neurodegeneration and aging. Methods mtDNA deletions were quantified by using real-time PCR in laser-dissected, COX-positive and COX-negative single neuronal and glial cells from frozen postmortem brain tissue specimens including normal appearing gray (NAGM) and white matter (NAWM) regions and chronic active plaques of MS patients, and gray matter (GM) and white matter (WM) regions of age-matched controls. Three patients with advance Alzheimer’s and Parkinson’s diseases were included as positive controls. The proportion of deleted mtDNA molecules was correlated with pathology and age. Results We detected no pathology-related accumulation of mtDNA deletions when comparisons were made among NAGM, NAWM, and plaque of MS brains, or between NAGM–GM and NAWM–WM of patients and age-matched controls. However, an accumulation of mtDNA deletions was noted in non-neurological controls beyond 60 years of age and in patients with Alzheimer’s and Parkinson’s diseases. As expected, the rate of mtDNA deletions was higher in COX− than in COX+ cells. Conclusion While aging and neurodegeneration in PD and AD are associated with accumulation of COX− cells and mtDNA deletions, the pathology of MS is not.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammation and neurodegeneration underlie phenotypic presentations of multiple sclerosis (MS) (Lucchinetti et al. 2000; Bielekova et al. 2005). Inflammation can directly cause, but is not exclusively responsible for, the progressive neuroaxonal loss (Trapp et al. 1998; Naismith and Cross 2005; Richert et al. 2006). The ongoing CNS tissue loss, to some extent, is disconnected from inflammation and likely results from a complex process also related to demyelination, decreased trophic support, glutamate-induced cytotoxicity, and mitochondrial dysfunction (Thompson et al. 1991; Bjartmar and Trapp 2001; Bagnato et al. 2003; Richert et al. 2006). Our studies explore the role of mitochondria in MS.
We previously proposed that inflammatory macrophages and microglia produce increased amounts of reactive oxygen species (ROS) and nitric oxide (NO) which cause oxidative damage to macromolecules in myelin, oligodendrocytes, and neurons in MS (Cross et al. 1997; Vladimirova et al. 1999). Using high-pressure liquid chromatography, immunohistochemistry, and molecular techniques, we consistently demonstrated increased oxidative damage to mtDNA in association with decreased activity of Complex I in active plaques (Vladimirova et al. 1998; Lu et al. 2000). These data linking Complex I abnormalities to oxidative damage and inflammation strongly suggest that an acquired mitochondrial dysfunction contributes to neurodegeneration in MS (Kalman 2006; Kalman et al. 2007). Mitochondrial dysfunction, hypoxic deconditioning, and expression of HIF-1α/β were also demonstrated in histological type III plaques (Abdoul-Einen and Lassmann 2005). In addition, a decreased expression of several mitochondrial proteins and reduced activity of Complexes I and III were detected in the MS motor cortex (Dutta et al. 2006).
To further dissect the pathway of inflammation, mitochondrial dysfunction, and neurodegeneration, here we asked if there is an accelerated accumulation of deleted mtDNA molecules due to the increased oxidative damage in plaques. Accumulation of mtDNA deletions along with oxidative damage has been demonstrated in neurodegenerative disorders and aging (Bender et al. 2006; Kraytsberg et al. 2006). If mtDNA deletions are indeed induced by inflammation-related oxidative damage and underlie OXPHOS (oxidative phosphorylation) defects in MS (Lu et al. 2000), then more somatic mtDNA deletions should be present in severely affected than in less or non-affected regions of MS brains, and there should also be a higher proportion of mtDNA deletions in MS lesions than in corresponding brain regions of age-matched controls. Since both oxidative damage and the postulated deletions in mtDNA are expected to occur in a stochastic manner, we tested our hypothesis at a single-cell level.
Methods
Tissue
We obtained frozen, postmortem specimens including two trio sets of chronic active plaques, nearby normal appearing gray matter (NAGM), and adjacent normal appearing white matter (NAWM) from frontal, parietal, and occipital lobes of 5 MS patients (Table 1). These specimens were selected based on Hematoxylin-Eosin and Luxol Fas Blue staining. Adjacent hemispheral normal gray matter (GM) and white matter (WM) specimens were also obtained from nine non-neurological controls who died either from systemic disorders (cancer, cardiovascular disease) or car accident. Four of the non-neurological controls were matched by age to MS patients, but five older controls were also included to encompass an age range of 34–80 years for the age—mtDNA deletion correlation study (Table 1). In addition, we obtained frozen brain tissue specimens from three other neurological disease controls (OND) that included cortex from two patients with advanced Alzheimer’s disease (AD) (90 and 94 years of age) and substantia nigra (SN) from one patient with advanced Parkinson’s disease (PD) (81 years of age). These specimens served as positive controls (Bender et al. 2006; Kraytsberg et al. 2006). All specimens were received from the Multiple Sclerosis Human Neurospecimen Bank, VAMC, Los Angeles, CA, and from the Rocky Mountain Multiple Sclerosis Tissue Bank, Denver, CO.
Preparation of Tissues
Frozen brain tissue specimens were placed in OCT and 16–20 μm sections were prepared using a Leica CM1900 Cryostate (Leica Microsystems, Bannockburn, IL). The sections were then mounted on slides with steel frames and PET membrane (Leica Microsystems, Bannockburn, IL) for staining and laser microdissection.
Cox/SDH Histochemistry
For Cox staining, sections were incubated in cytochrome c oxidase medium composed of 100 μM cytochrome c, 4 mM diaminobenzidine tetrahydrochloride, 20 μg/ml catalase in 0.2 M phosphate buffer, pH 7.0 at 37°C for 50 min. Subsequently, sections were washed in PBS (pH 7.4) three times for 10 min each. For SDH staining, sections were incubated in succinate dehydrogenase (SDH) medium composed of 130 mM sodium succinate, 200 μM phenazine methosulfate, 1 mM sodium azide, 1.5 mM nitroblue tetrazolium in 0.2 M phosphate buffer, pH 7.0 at 37°C for 40 min. Then, sections were washed again in PBS (pH 7.4) three times for 10 min each and let to dry at room temperature for 30 min (Sciacco et al. 1994; Taylor et al. 2003).
Cox+/− single neurons (NAGM, GM, SN) and glial cells (NAWM, WM, plaque) were collected according to the manufacturers’ protocol in 200-μl PCR tube caps by using a Leica Laser Microdissection Microscope (Leica Microsystems, Bannockburn, IL). Cresyl violet (Merck, Darmstadt, Germany) counter staining was applied for morphometric analyses of cells (Bender et al. 2006). In MS patients, we collected and analyzed at least 10 Cox positive and 10 Cox negative neurons and glial cells from each of the two sets of NAGM, NAWM, and plaque-containing specimens (altogether two sets of 3 × [10 + 10] = 120 single cells per individual) in the real-time PCR experiments. In normal controls, we twice collected and analyzed 10 Cox positive and 10 Cox negative neurons and glial cells from each set of GM and adjacent WM specimens (two sets of 2 × [10 + 10] = 80 single cells per individual) in the real-time PCR experiments.
Isolation of DNA from Single Cells
Each cell in the 200 μl PCR tube cap was covered with 1 μl of lysis buffer containing 1 μl of 10 mM EDTA, pH 8.0, 0.5% SDS, and 2 mg/ml proteinase K. After 30 min of incubation at 37°C, 10 μl of RNase/DNase free-water was added to the lysate and gently mixed by pipetting (Kraytsberg et al. 2006). To inactivate proteinase K, the lysate was incubated at 95°C for 2 min.
Real-time PCR
To quantify the proportion of deleted versus full-length mtDNA molecules per cell we used the ND4/ND1 assay (modified from Bender et al. 2006). This assay is based on the comparative amplification of two mtDNA regions: ND1 that is usually undeleted and ND4 that is commonly deleted in patients with large mtDNA rearrangements related to mitochondrial diseases, aging, and neurodegenerative diseases (Bender et al. 2006; Kraytsberg et al. 2006). Deletion rates were calculated as 1-ND4/ND1 copy number values (Bender et al. 2006).
ND1 forward nt3485–3504 primer: 5′ CCCTAAAACCCGCCACATCT 3′
ND1 reverse nt3553–3532 primer: 5′ GAGCGATGGTGAGAGCTAAGGT 3′
ND4 forward nt12087–12109 primer: 5′ CCATTCTCCTCCTATCCCTCAAC 3′
ND4 reverse nt12170–12140 primer: 5′ CACAATCTGATGTTTTGGTTAAACTATAATT3 3′
PCR reactions were carried out in 25-μl volume using the QuantiFast Sybr Green PCR kit (Qiagen, Hilden, Germany) for each reaction having 2× QuantiFast Sybr Green PCR Master Mix, 20–20 pmol of the forward and reverse primers, 5–5 μl of DNA from the 11 μl total DNA lysate of each Cox+ and Cox− cell, and RNase/Dnase-free water was added as needed. An internal standard including the human beta-globin gene was amplified yielding a 110-bp PCR product (Roche Applied Science, Indianapolis, IN). Logarithmic dilutions of the beta-globin gene were prepared to result in five standards in the range of 15 ng and 1.5 pg included in each PCR run. The beta-globin-specific primers were provided by the manufacturer (Roche Applied Science, Indianapolis, IN).
DNA amplification was carried out in an Applied Biosystems 7000 Real Time PCR machine using the following cycling conditions: 95°C for 5 min followed by 42 cycles of 95°C for 10 s, 60°C for 30 s, and then 1 cycle at 60°C for 2 min, and finally hold at 4°C. In each reaction, a negative (no DNA template) and a non-deleted mtDNA control (mtDNA obtained from peripheral blood of a healthy control aged < 40 years) were included.
Validation of the ND4/ND1 test was performed using a mtDNA template with a 7.44-kb deletion between nt [8,637–8,648] and [16,073–16,084] flanked by a 12-bp direct repeat at both ends. Southern blot analyses determined 35%, while our ND4/ND1 test defined 37% deletion rate in this heteroplasmic mtDNA specimen that was generously provided by Dr. Shulin Zhang, Dr. Lee-Jun Wong and Sandra Peacock at the Medical Genetics Laboratories of the Baylor College of Medicine.
A quality control experiment was performed to establish the inherent technical variations in the ND4/ND1 test. We measured mtDNA deletions in 20 Cox positive single neurons from the cortex of a young normal control that was not expected to have deletions. This study revealed a mean ratio of ND4/ND1 = 1.0 ± 0.06 (mean ± SE), translating into 0 ± 0.06 deletion.
Statistics
For statistical analyses the non-parametric Wilcoxon signed-rank test was used for intra-individual comparisons of mtDNA deletion values defined by real-time PCR. For the inter-group comparisons of mtDNA deletion values, t-test and ANOVA analyses were performed using the SPSS v 14 package.
Results
Proportion of Deleted and Undeleted mtDNA in MS and Control Brains
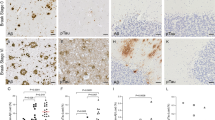
For quantifying deleted and full-length mtDNA molecules on COX/SDH-stained neurons and glial cells, we used the method of real-time PCR. While all patients and controls up to 60 years of age appeared to have a very low proportion of Cox− relative to Cox+ cells (ranging between 0.008 and 0.07) irrespective of brain segment (NAWM, NAGM, plaque; or WM, GM), the proportion of Cox− cells gradually increased after 60 years of age reaching 0.20 in the oldest NC (80 years old). The ratio of Cox−/Cox+ neurons was also high (0.2–0.35) in the two patients with AD (91 and 94 years of age) and in the patient with PD (81 years of age) (Table 1, Fig. 1a and b).
Cox/SDH histochemistry in brain specimens of MS and AD patients. COX/SDH histochemistry was performed for the selection of Cox+ and Cox− cells in MS patients and controls. COX+ cells (brown staining) are seen without the occurrence of COX− (purple) cells in corresponding NAGM, NAWM, and a perivascular plaque of a MS patient (a), this representative cortical specimen from a 94-year-old patient with AD depicts the significant accumulation of Cox− cells (purple) relative to the Cox+ cells (brown) (b)
Because of the sparse occurrence of Cox− cells in MS patients (all below 60 years) and controls matched to patients by age, only cells with strong and weak Cox staining could have been collected in sufficient numbers for the real-time PCR studies in these cohorts. In contrast, from both the normal controls beyond 60 years of age and OND controls with AD or PD, Cox− cells were available in sufficient numbers for the real-time PCR studies.
Neurons and glial cells sorted by COX staining were isolated by laser dissection for DNA extraction. The proportion of deleted mtDNA molecules was initially quantified in all specimens of the MS “a” and “b” tissue series separately, along with separately isolated cells from matched control tissues (Table 1). Since there was no difference in the corresponding data of “a” and “b” groups, we discuss here the results from the combined (a + b) analyses.
Figure 2a and b summarizes the results of the real-time PCR quantitation of deleted mtDNA molecules in NAGM, NAWM, and plaques of MS patients. Figure 2c shows the results of real-time PCR in GM and WM of all controls. No significant differences are observed when data from either the high or from the low Cox positive trios of NAGM, NAWM, and plaques are compared with each other, or when similar data from WM and GM of age-matched normal controls are compared with each other using the Wilcoxon signed-rank test (intra-individual comparison among NAWM, NAGM, and plaque; or between WM and GM). Similarly, no differences are found in the proportion of deleted mtDNA molecules when values in the NAGM and NAWM of MS patients are compared to GM and WM of age matched controls, respectively, using the t-test or ANOVA analyses (inter-group comparisons). Nevertheless, the proportion of deleted mtDNA molecules is higher in the Cox low cells as compared to Cox high cells in all tissue regions of patients (NAGM, NAWM, plaques) and controls (GM, WM), with significant P-values (P ≤ 0.008, Wilcoxon signed-rank test) in the MS NAWM, and control GM and WM (Fig. 2a–c). These observations suggest that the proportion of deleted mtDNA molecules is similar in various regions of MS brains (NAGM–NAWM–plaque), and in MS brains and control brains matched by age (NAWM–WM, NAGM–GM). Cox low positive cells tend to have higher proportion of deleted mtDNA molecules than Cox high positive cells derived from the same tissue specimen.
Figure 2 shows the mean ± SE of deleted mtDNA molecules (defined as 1-ND4/ND1) in strongly COX+ cells of MS NAGM, NAWM, and plaques (a); in weakly Cox+ cells of MS NAGM, NAWM. and plaques (b); in Cox+ and Cox− neurons and glial cells of non-neurological controls (c); and in Cox+ and Cox− neurons of patients with AD and PD (d). From each tissue category, at least 10 Cox+ and 10 Cox− cells were collected for real-time PCR analyses
In contrast, as the proportion of Cox− neurons and glial cells increases with age, the proportion of deleted mtDNA molecules also increases beyond 60 years of age in the control cohort (Fig. 2c). Statistical significance is observed when the proportion of deleted mtDNA molecules in Cox− GM (P = 0.041, ANOVA) and Cox− WM cells (P = 0.000, ANOVA) are compared in the groups below versus above 60 years of age. No differences are observed in the proportion of mtDNA deletions in Cox+ cells from controls below versus above 60 years of age. Similarly, Fig. 2d shows that Cox− neurons of the three oldest patients with neurodegenerative diseases (AD2985, AD2943, and PD4078) display marked deletions in mtDNA molecules as compared to Cox+ neurons (P = 0.0003 [1-tail], P = 0.0007 [2-tail] Wilcoxon signed-rank test).
Discussion
The complex process of neurodegeneration seems to be both directly and indirectly linked to inflammation in MS, but the underlying mechanisms are not fully defined (Kalman 2006; Kalman et al. 2007). A potential subcellular mechanism that can lead to apoptosis of oligodendrocytes and neurons in acute inflammatory lesions as well as be responsible for a delayed neuroaxonal degeneration long after the decline of inflammation may involve mitochondrial abnormalities (Bjartmar et al. 2001; Barnett and Prineas 2004; Kalman 2006; Kalman et al. 2007). Previously, we observed that inflammatory ROS and NO cause oxidative damage to mtDNA, which is associated with OXPHOS defects and likely contributes to neurodegeneration in MS (Lu et al. 2000). Here we tested if oxidative damage to mtDNA is associated with a decreased activity of OXPHOS enzymes because of an induced deletion of mtDNA-encoded genes of Complexes I, III, IV, and V (Lu et al. 2000; Dutta et al. 2006).
With the exception of Complex II that has exclusively nDNA-encoded subunits, the other enzyme complexes of OXPHOS (Complexes I, III, IV, and V) have both nDNA and mtDNA-encoded subunits (Kalman 2006). If mtDNA deletions develop secondary to oxidative damage with the involvement of genes in the latter group of enzyme complexes, it may cause a reduction of OXPHOS activity observed in MS lesions (Lu et al. 2000).
Complete sequencing of mtDNA in six patients with MS and Devic’s disease (four specimens derived from plaques and two specimens from blood), and high-resolution restriction endonuclease scans of mtDNA derived from peripheral blood of cohorts of MS patients and matched controls identified no inherited pathogenic mutations only disease-associated single nucleotide polymorphisms (SNPs), similar to that reported by others (Kalman et al. 1996, 1997, 1999; Kalman and Alder 1998, Bosley et al. 2007). We also detected weak associations between MS and SNPs in 3 nDNA-encoded genes of Complex I in families (Vyshkina et al. 2005). However, the pathological significance of these inherited variants remains speculative and cannot explain the regional variations in Complex I activity in MS brains (in plaques < NAWM) (Lu et al. 2000). The regional decreases in the activity of mitochondrial enzymes must be related to acquired abnormalities either in their genes, mRNA expression, or proteins. Here we tested if inflammation and oxidative damage are associated with somatic mtDNA deletions in MS lesions similar to that described in aging and Parkinson’s disease (Bender et al. 2006; Kraytsberg et al. 2006).
These analyses revealed that the accumulation of mtDNA deletions does not show pathology-related regional variations in MS brains (NAGM, NAWM, plaques), and the proportion of mtDNA deletion is similar in MS and control brains matched by age. This finding is in agreement with a recent report on the lack of mtDNA deletions in MS muscle where decreased activity of Complex I was noted (Kumleh et al. 2006).
However, our analyses showed that while the proportion of deleted mtDNA molecules is negligible in younger individuals, the proportion of mutated molecules dramatically increases after 60 years of age along with the growing numbers of Cox− cells. This observation is in consensus with that reported by Bender et al. (2006) and Kraytsberg et al. (2006). As expected, the proportion of deleted mtDNA molecules is higher in Cox− than in Cox+ cells in both MS patients and controls. This difference becomes more pronounced in individuals above 60 years of age. The number of Cox− cells and the proportion of deleted mtDNA molecules are the highest in the OND patients with AD or PD, who are also the oldest subjects in this study.
The argument may be raised that we missed mtDNA deletions in MS brains because we selected cells based on COX staining instead of oxidative damage (e.g. defined by anti-8-OH-deoxi-guanosine immunohistochemistry) (Lu et al. 2000). We believe that COX/SDH staining provided a more stringent selection of cell with potential mitochondrial impairment than immunohistochemistry for oxidative damage could have accomplished. The frequency of COX negative cells was much lower than the frequency of cells with signs of oxidative damage in plaques, suggesting that not every cell with oxidative damage to mtDNA develops COX deficiency and mitochondrial functional defect (Lu et al. 2000 and unpublished observations). Therefore, in the light of the above negative observations, it is not likely that studying mtDNA in cells with signs of oxidative damage (as opposed to cells with COX low or negative staining) would reveal mtDNA deletions in MS lesions.
As we here excluded the possibility of an inflammation-induced accelerated accumulation of somatic mtDNA deletions in MS brains, the question remains as to how oxidative damage affects the activity of enzymes involved in the mitochondrial electron transfer. Possible pathways include an altered mRNA expression of OXPHOS protein subunits due to either oxidative damage to DNA/mtDNA or to regulatory effects of inflammatory products in the CNS (Dutta et al. 2006). In addition, post-translational modifications (oxidation, nitration) in OXPHOS proteins can be the consequences of the exposure to inflammatory ROS and NO (Qi et al. 2006, 2007). Since neurodegeneration is the major pathological correlate of disability in MS, elucidation of the molecular cascade underlying neurodegeneration merits further studies.
References
Aboul-Enein, F., & Lassmann, H. (2005). Mitochondrial damage and histotoxic hypoxia: A pathway of tissue injury in inflammatory brain disease? Acta Neuropathologica (Berl)., 109, 49–55.
Bagnato, F., Jeffries, N., Richert, N. D., Stone, R. D., Ohayon, J. M., McFarland, H. F., & Frank, J. A. (2003). Evolution of T1 black holes in patients with multiple sclerosis imaged monthly for 4 years. Brain, 126, 1782–1789.
Barnett, M. H., & Prineas, J. W. (2004). Relapsing and remitting multiple sclerosis: Pathology of the newly forming lesion. Annals of Neurology, 55, 458–468.
Bender, A., Krishnan, K. J., Morris, C. M., Taylor, G. A., Reeve, A. K., Perry, R. H., Jaros, E., Hersheson, J. S., Betts, J., Klopstock, T., Taylor, R. W., & Turnbull, D.M. (2006). High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nature Genetics, 38, 507–508.
Bjartmar, C., & Trapp, B. D. (2001). Axonal and neuronal degeneration in multiple sclerosis: Mechanisms and functional consequences. Current Opinion in Neurology, 14, 271–278.
Bielekova, B., Kadom, N., Fisher, E., Jeffries, N., Ohayon, J., Richert, N., Howard, T., Bash, C. N., Frank, J. A., Stone, L., Martin, R., Cutter, G., & McFarland, H. F. (2005). MRI as a marker for disease heterogeneity in multiple sclerosis. Neurology, 65, 1071–1076.
Bosley, T. M., Constantinescu, C. S., Tench, C. R., & Abu-Amero, K. K. (2007). Mitochondrial changes in leukocytes of patients with optic neuritis. Molecular Vision, 13, 1516–1528.
Cross, A. H., Manning, P. T., Stern, M. K., & Misko, T. P. (1997). Evidence for the production of peroxynitrite in inflammatory CNS demyelination. Journal of Neuroimmunology, 80, 121–130.
Dutta, R., McDonough, J., Yin, X., Peterson, J., Chang, A., Torres, T., Gudz, T., Macklin, W. B., Lewis, D. A., Fox, R. J., Rudick, R., Mirnics, K., & Trapp, B. D. (2006). Mitochondrial dysfunction as a cause of axonal degeneration in multiple sclerosis patients. Annals of Neurology, 59, 478–489.
Kalman, B. (2006). Role of mitochondria in MS. Current neurology and neuroscience reports. Section: Demyelinating Disorders, 6, 244–252.
Kalman, B., & Alder, H. (1998). Is the mitochondrial DNA involved in determining susceptibility to multiple sclerosis? Acta Neurologica Scandinavica, 98, 232–237.
Kalman, B., Laitinen, K., & Komoly, S. (2007). The involvement of mitochondria in the pathogenesis of multiple sclerosis. Journal of Neuroimmunology, 188, 1–12.
Kalman, B., Lublin, F. D., & Alder, H. (1996). Characterization of the mitochondrial DNA in patients with multiple sclerosis. Journal of Neurological Science, 140, 75–84.
Kalman, B., Li, S., Chatterjee, D., O’Connor, J., Voehl, M. R., Brown, M. D., & Alder, H. (1999). Large scale screening of the mitochondrial DNA reveals no pathogenic mutations but a haplotype associated with multiple sclerosis in Caucasians. Acta Neurologica Scandinavica, 99, 16–25.
Kalman, B., Rodriguez-Valdez, J. L., Bosch, U., & Lublin, F. D. (1997). Screening for Leber hereditary optic neuropathy associated mitochondrial DNA mutations in patients with prominent optic neuritis. Multiple Sclerosis, 2, 279-282.
Kraytsberg, Y., Kudryavtseva, E., McKee, A. C., Geula, C., Kowall, N. W., & Khrapko, K. (2006). Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nature Genetics, 38, 507–508.
Kumleh, H. H., Riazi, G. H., Houshmand, M., Sanati, M. H., Gharagozli, K., & Shafa, M. (2006). Complex I deficiency in Persian multiple sclerosis patients. Journal of Neurological Sciences, 243, 65–69.
Lu, F., Selak, M., O’Connor, J., Croul, S., Butunoi, C., & Kalman, B. (2000). Oxidative damage to mitochondrial DNA and activity of mitochondrial enzymes in lesions of multiple sclerosis. Journal of Neurological Sciences, 177, 95-103.
Lucchinetti, C., Bruck, W., Parisi, J., Scheithauer, B., Rodriguez, M., & Lassmann, H. (2000). Heterogeneity of multiple sclerosis lesions: Implication for the pathogenesis of demyelination. Annals of Neurology, 47, 707–717.
Naismith, R. T., & Cross, A. H. (2005). Multiple sclerosis and black holes. Connecting pixels. Archives of Neurology, 62, 1666–1668.
Qi, X., Lewin, A. S., Sun, L., Hauswirth, W. W., & Guy, J. (2006). Mitochondrial protein nitration primes neurodegeneration in experimental autoimmune encephalomyelitis. Journal of Biological Chemistry, 281, 31950–31962.
Qi, X., Lewin, A. S., Sun, L., Hauswirth, W. W., & Guy, J. (2007). Suppression of mitochondrial oxidative stress provides long-term neuroprotection in experimental optic neuritis. Investigative Ophthalmology Visual Science, 48, 681–691.
Richert, N. D., Howard, T., Frank, J. A., Stone, R., Ostuni, J., Ohayon, J., Bash, C., & McFarland, H. F. (2006). Relationship between inflammatory lesions and cerebral atrophy in multiple sclerosis. Neurology, 66, 551–556.
Sciacco, M., Bonilla, E., Schon, E. A., DiMauro, S., & Moraes, C. T. (1994). Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Human Molecular Genetics, 3, 13–19.
Taylor, R. W., Barron, M. J., Borthwick, G. M., Gospel, A., Chinnery, P. F., Samuels, D. C., Taylor, G. A., Plusa, S. M., Needham, S. J., Greaves, L. C., Kirkwood, T. B., & Turnbull, D. M. (2003). Mitochondrial DNA mutations in human colonic crypt stem cells. Journal of Clinical Investigation, 112, 1351–1360.
Thompson, A. J., Kermode, A. G., Wicks, D., MacManus, D. G., Kendall, B. E., Kingsley, D. P., & McDonald, W. I. (1991). Major differences in the dynamics of primary and secondary progressive multiple sclerosis. Annals of Neurology, 29, 53–62.
Trapp, B. D., Peterson, J., Ransohoff, R. M., Rudick, R., Mork, S., & Bo L. (1998). Axonal transection in the lesions of multiple sclerosis. New England Journal of Medicine, 338, 278–285.
Vladimirova, O., Lu, F. M., Shawver, L., & Kalman, B. (1999). The activation of protein kinase C induces higher production of reactive oxygen species by mononuclear cells in patients with multiple sclerosis than in controls. Inflammation Research, 48, 412–416.
Vladimirova, O., O’Connor, J., Cahill, A., Alder, H., & Kalman, B. (1998). Oxidative damage to DNA in plaques of MS brains. Multiple Sclerosis, 4, 413–418.
Vyshkina, T., Banisor, I., Yao Shugart, Y., Leist, T. P., & Kalman, B. (2005). Genetic variants of Complex I in multiple sclerosis. Journal Neurological Science, 228, 55–64.
Acknowledgments
This study was supported by grants from the National Multiple Sclerosis Society (PP1335), Serono, and by the Syracuse VAMC. The studied brain tissue specimens were obtained from the invaluable collections of the Multiple Sclerosis Human Neurospecimen Bank, VAMC, Los Angeles, CA, and of the Rocky Mountain Multiple Sclerosis Tissue Bank, Denver, CO.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Blokhin, A., Vyshkina, T., Komoly, S. et al. Lack of Mitochondrial DNA Deletions in Lesions of Multiple Sclerosis. Neuromol Med 10, 187–194 (2008). https://doi.org/10.1007/s12017-008-8025-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-008-8025-2