Abstract
The “Bermuda triangle” of genetics, environment and autoimmunity is involved in the pathogenesis of rheumatoid arthritis (RA). Various aspects of genetic contribution to the etiology, pathogenesis and outcome of RA are discussed in this review. The heritability of RA has been estimated to be about 60 %, while the contribution of HLA to heritability has been estimated to be 11–37 %. Apart from known shared epitope (SE) alleles, such as HLA-DRB1*01 and DRB1*04, other HLA alleles, such as HLA-DRB1*13 and DRB1*15 have been linked to RA susceptibility. A novel SE classification divides SE alleles into S1, S2, S3P and S3D groups, where primarily S2 and S3P groups have been associated with predisposition to seropositive RA. The most relevant non-HLA gene single nucleotide polymorphisms (SNPs) associated with RA include PTPN22, IL23R, TRAF1, CTLA4, IRF5, STAT4, CCR6, PADI4. Large genome-wide association studies (GWAS) have identified more than 30 loci involved in RA pathogenesis. HLA and some non-HLA genes may differentiate between anti-citrullinated protein antibody (ACPA) seropositive and seronegative RA. Genetic susceptibility has also been associated with environmental factors, primarily smoking. Some GWAS studies carried out in rodent models of arthritis have confirmed the role of human genes. For example, in the collagen-induced (CIA) and proteoglycan-induced arthritis (PgIA) models, two important loci — Pgia26/Cia5 and Pgia2/Cia2/Cia3, corresponding the human PTPN22/CD2 and TRAF1/C5 loci, respectively — have been identified. Finally, pharmacogenomics identified SNPs or multiple genetic signatures that may be associated with responses to traditional disease-modifying drugs and biologics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is an autoimmune inflammatory rheumatic disease that affects approximately 0.5–1 % of the population and causes chronic synovial inflammation eventually leading to joint destruction and disability [1]. The “Bermuda triangle” of genetic, environmental factors and autoimmunity triggers the onset and perpetuation of synovitis underlying RA [2–10]. Early diagnosis and immediate, effective therapy are crucial in order to prevent joint deterioration, functional disability and unfavorable disease outcome. The optimal management of RA is needed within 3–6 months after the onset of disease; therefore, a very narrow "window of opportunity" is present to achieve remission [11].
The heritability of RA has been estimated to be about 60 % [12]. Genetic factors including class II major histocompatibility antigens/human leukocyte antigens (HLA-DR), as well as non-HLA genes have been implicated in the pathogenesis of RA [2, 3, 5, 6, 13]. Interestingly, HLA and some non-HLA genes have also been linked to autoimmunity to citrullinated proteins (ACPA [anti-citrullinated protein antibody]), as well as smoking. In brief, smoking and possibly other environmental and lifestyle-related factors may trigger ACPA production and the development of ACPA seropositive RA, at least in some cohorts [4, 5, 14–18]. Apart from susceptibility to RA, some genes may have relevance for disease outcome and prognosis [4, 5]. Certain gene profiles or signatures have also been associated with response or non-response to therapy (pharmacogenomics) [19–23].
For decades, the possible pathogenic effects of one or more disease-associated chromosomal regions (loci) have been investigated separately. These studies often led to controversies as some groups were able to confirm linkage of a certain single nucleotide polymorphism (SNP) to RA, while others could not. Recent genome-wide association studies (GWAS) and especially large-scale cohorts, such as the Wellcome Trust Case Control Consortium (WTCCC) database, have enabled the simultaneous assessment of thousands of genes, leading to more consequent results of genetic associations [24–26].
Animal studies may help to fill the gaps in human GWAS by allowing for gene mapping via congenic breeding and functional studies that will yield greater insights into the mechanisms of autoimmune responses in RA. Based upon the clinical, immunological and genetic components, the most appropriate animal models for RA seem to be those that use genetically controlled systemic autoimmune joint diseases, those in which both T and B cells are involved, and those that apply (auto)antigenic molecules of cartilage or joint tissues for provoking (“targeting”) synovial joint inflammation [27–31].
Here we will present a survey of recent data regarding the genetic contribution to RA. We will briefly review the heritability of RA followed by the most relevant and updated information on the pathogenic and prognostic role of some HLA and non-HLA genes. The “Bermuda triangle” of genes, ACPA, and environment (mentioned above) will also be discussed in more detail. Some aspects of GWAS studies carried out in “mice and men” will also be discussed. Finally, a brief outlook on pharmacogenomics will be presented. Due to space limitations, all these issues will only be discussed in brief, focusing on the most relevant information. Therefore, we apologize for any important contributions that have been omitted.
Heritability of RA
The overall heritability of ACPA positive and ACPA negative RA is similar, 60–70 % [3, 12]. However, there has been some controversy regarding the contribution of HLA to the genetic variability. Previously, the contribution of HLA has been estimated to be 37 % [32–34]. Recent studies have suggested that this figure may be overestimated. The contribution of SE HLA-DRB1 alleles to the genetic variance was found to be only 11 % [3, 34]. The difference between the two figures may be because RA susceptibility may also be associated with protective alleles, in addition to SE (see later). Moreover, while the general heritability is the same in ACPA positive and negative RA, the contribution of HLA alleles to the genetic variance is 40 % for seropositive, but only 2 % for ACPA negative disease [34].
The Pathogenic and Prognostic Roles of HLA Genes
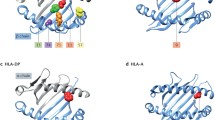
It was established decades ago that certain HLA-DRB1*01 (HLA-DR1) and HLA-DRB1*04 (HLA-DR4) alleles, containing the SE, are associated with susceptibility to RA. While the QKRAA, QQRAA and KKRAA amino acid motifs are known SEs conferring susceptibility, the DERAA motif is rather responsible for protective effects. HLA-DRB1*1001 is also a SE-containing allele, which can accommodate citrulline in its antigen anchoring pockets and thus stimulate citrullinated protein-specific T cell responses [3, 32, 33, 35].
It is now clear that there is a complex hierarchy of risk factors conferred by different HLA-DRB1 genotypes. As discussed above, SE alleles (HLA-DRB1*01, DRB1*04, DRB1*10) exert the strongest association with disease susceptibility, however, recently, much attention has been drawn to two non-SE HLA-DRB1 alleles, DRB1*13 (HLA-DR13) and DRB*15 (HLA-DR15) [3, 15, 36, 37). DRB1*1301, *1302 and *1304 alleles have been linked to the DERAA motif described above [3, 36]. Although we reported an association between HLA-DRB1*13 and ACPA production in Hungarian RA patients [15], recent cohorts suggested that the HLA-DRB*1301 allele may protect against ACPA positive or ACPA negative RA [3, 5, 34, 37]. There has been some controversy regarding the role of HLA-DRB1*15; however, again, we found higher circulating ACPA levels associated with DRB1*15 positivity [15]. Recently, enhanced production of ACPA has been associated with HLA-DRB1*15 positivity in RA [35, 38]. We have also described a family with six members having RA in two generations. Interestingly, four out of the six RA patients carried the DRB1*15 genotype [39] (Table 1).
Regarding identical twins, in a cohort of 91 monozygotic twins pairs, increased concordance for RA was observed in SE positive pairs (RR 3.7). In addition, a 5-fold risk for RA concordance was seen in twins who were "homozygous" for the SE, compared with those negative for the SE [40].
Aiming for reconsideration of the SE hypothesis, a new classification system has been proposed and validated by French investigators [36]. In brief, the susceptibility risk represented by the RAA motif is modulated by amino acids at positions 70 and 71. Thus, at position 71, lysine (K) confers the highest risk, arginine (R) intermediate risk, while alanine (A) and glutamic acid (E) lower risk. At position 70, glutamine (Q) and R represent higher risk than D [36]. Based on the type of the amino acids at positions 70 and 71, the new classification system divides SE alleles into S1, S2, S3P and S3D groups and denotes all non-RAA motifs as X. A positive association with RA susceptibility was found for S2 and S3P allele carriers, while S1, S3D and X are low risk alleles pooled together as L alleles [36]. The new system has since successfully been validated on large international cohorts [36, 41]. The presence of S2 or S3P alleles has been correlated with ACPA production, whereas the presence of S3D and S1 alleles appeared to be protective. Our group has recently assessed the impact of S1, S2, S3P and S3D alleles on ACPA positivity. We found that not only S2 and S3P, but — to a lesser extent — S1 and S3D alleles also predisposed to ACPA (anti-CCP and anti-CV) production [41]. With regard to the above-mentioned non-SE alleles, HLA-DRB1*15, as well as the DRB1*1301, *1302 and *1304 alleles have also been classified in the S1 group that has strong association with susceptibility to RA [41].
With regard to prognosis and outcome, SE is a risk factor for more severe, destructive RA, as well as for the development of extraarticular manifestations. It is likely that the SE itself may not be directly responsible for poorer prognosis, but, as discussed later, may rather influence outcome indirectly, via ACPA production [3–5, 17, 34, 42].
In contrast to HLA-DRB1 alleles, HLA-DRB*03 (HLA-DR3) may rather be associated with ACPA negative RA and milder disease course [3]. In general, as discussed later, HLA-DRB1 associations are overwhelmingly observed in the ACPA positive RA subset [9, 17, 34].
Non-HLA Susceptibility Genes
In addition to HLA-DR alleles, several association studies have confirmed the role of other, non-HLA genes in susceptibility to RA. Among the 31 confirmed non-HLA loci contributing to RA risk, probably the strongest associations have been found with PTPN22 and IL23R genes [3, 9, 43–47]. We have also confirmed association of RA with these loci in Hungarian RA patients [44, 45]. The PADI4 gene encoding the peptidylarginine-deiminase 4 (PADI4) enzyme is involved in protein citrullination, a key event underlying the pathogenesis of RA. Association of the PADI4 haplotype with RA was suggested in Asian cohorts, but could not be confirmed in Caucasian populations including Hungarian cohorts [48–50]. As also discussed in relation to GWAS later, other important, confirmed loci include TRAF1, CTLA4, IRF5, STAT4, FCGR3A, IL6ST, IL2RA, IL2RB, CCL21, CCR6, CD40 and others [3, 4, 9, 34, 50–55] (Table 1).
Here, we give a short introduction to the most important loci in the list. The gene encoding the intracellular phosphatase protein tyrosine phosphatase non-receptor type 22 (PTPN22) shows the second strongest association with RA, right after HLA-DRB1. This allele has been linked to other autoimmune diseases such as type I diabetes, Graves’ disease, myasthenia gravis, systemic sclerosis, lupus and Addison’s disease, among others. The C1885T polymorphism of the PTPN22 gene leading to an amino acid change from Arg to Trp at amino acid position 620 confers risk for these diseases, mostly in Caucasian populations. This SNP has been associated by many groups with ACPA and rheumatoid factor (RF) positive RA and probably worse prognosis. The association of this locus with RA severity has been suggested, but it may be weak and could not be reproduced by some groups. The presence of PTPN22 C1885T polymorphism, in addition to SE and ACPA status strongly supports the early diagnosis of RA. In contrast to SE, PTPN22 may not be closely associated with smoking [18, 45, 47].
On the basis of recent GWAS data, TRAF1 in the TRAF1-C5 region may be the third most strongly RA-associated locus. This region has also been associated primarily with ACPA positive RA. The TNF receptor associated factor 1 (TRAF1) is an adaptor protein that links TNF family members, such as TNF-α, to downstream signaling networks. TRAF1 has been implicated in cell growth, proliferation, apoptosis, bone turnover, cytokine activation, and in the overall pathogenesis of RA. TRAF1 has been related to increased radiological progression; however, TRAF1/C5 may not be associated with RA mortality [9, 43, 53].
The association of RA with the STAT4 gene is relatively modest in comparison to associations discussed above. This locus also confers risk to SLE, scleroderma, type I diabetes, juvenile idiopathic arthritis, and possibly to inflammatory bowel disease (IBD). Signal transducer and activator of transcription 4 (STAT4) exerts a distinct role in the signaling of cytokines, primarily IL-12 through JAK2. Interestingly, different SNPs in the STAT4 gene may increase susceptibility to both ACPA positive and negative RA [43, 52].
The PADI4 enzyme mediates the citrullination of proteins (conversion of arginine residues to citrulline). Association between RA and the PADI4 gene was originally established in large Japanese and Korean cohorts. This genetic effect was much weaker in Caucasian populations. As discussed above, an association between RA and PADI4 could not be confirmed in Hungary either [9, 43, 49, 50].
Fcγ receptors are key players in antigen presentation and inflammation. In a cohort of 945 RA patients, Fcγ receptor IIIA (FCGR) 158 V/F polymorphism was assessed. The VV genotype was associated with ACPA seropositive RA in the Caucasian, but not the Asian, population [5, 43, 54].
The CD40 gene encodes a protein that as a member of the TNF-receptor superfamily is important in a broad range of immune responses, such as B-cell development and costimulation. CD40 is a known risk factor for RA. An SNP at the CD40 locus exerted association with RA severity [17, 43, 51].
Among the recently discovered new risk loci the CCR6 chemokine receptor gene may be of high interest [55, 56]. CCR6 is expressed by Th17 cells and is involved in IL-17-driven inflammation underlying RA and other chronic inflammatory diseases. In addition to CCR6 and its chemokine ligand CCL20, several chemokines and chemokine receptors have been implicated in RA [35, 55, 56].
In conclusion, several HLA and non-HLA genes have been implicated in susceptibility to or protection against RA. To date, more than 30 genes have been associated with the disease and these genetic factors account for about 50 % of the genetic variants linked to RA susceptibility [3–5, 9, 35, 43].
Interactions of Genes, Autoimmunity and the Environment in Disease Pathogenesis and Outcome
It is very likely that HLA-DR genes act indirectly, through the regulation of autoantibody production involved in the development and outcome of RA. Indeed, SE is probably the primary risk factor for increased ACPA production in RA [2, 4, 5, 8, 10]. SE was not only associated with positive ACPA titers, but also with absolute serum ACPA levels [15]. Among HLA-DRB1 alleles, ACPA production has been associated with HLA-DRB1*04, rather than HLA-DRB1*01. Amongst HLA-DRB1*04 subtypes, DRB1*0401, *0404, *0405 and *0408 SE exerted the closest association with positive ACPA [3, 35]. Of 2,221 SNPs within the MHC region, 299 showed significant associations with ACPA positive RA, while none of these SNPs could be associated with seronegative RA [57]. Among non-SE HLA-DRB1 alleles, HLA-DRB1*13 and DRB1*15 have also been implicated in ACPA production [15, 38]. However, recent studies suggested that HLA-DRB1*13, as well as HLA-DRB1*03 (already mentioned above) may rather be associated with ACPA negative RA [10, 34]. Moreover, HLA-DRB1*13 may predominantly account for protection against RA [37] (Table 2). Regarding various ACPAs, positive SE has been associated with anti-cyclic citrullinated peptide (CCP), anti-citrullinated vimentin (CV), anti-citrullinated fibrinogen (CF), anti-citrullinated α-enolase peptide (CEP) and anti-citrullinated myelin basic protein (MBP) production [10, 42, 48, 58, 59]. Interestingly, very recent data suggest that the triggering of autoimmunity and radiological outcome may not be associated with ACPA fine specificities but rather with ACPA development in general [58, 59].
Some non-MHC alleles may also differentiate between ACPA seropositive and seronegative RA. Among non-HLA susceptibility loci mentioned above, CTLA4 may be associated with ACPA positive, IRF5 with ACPA negative, while STAT4 with both seropositive and seronegative disease [5, 43, 52, 59] (Table 2).
Numerous studies have been performed with respect to the role of environmental and lifestyle-related factors, primarily smoking, in susceptibility to the development and progress of RA. At least in some geographical regions, smoking has been associated with the development of extraarticular manifestations, including nodulosis and cardiovascular complications, as well as with more progressive disease course. Smoking promotes the citrullination of synovial proteins and thus ACPA production. In Northern and Western European cohorts, a high risk for the development of ACPA positive RA was observed in smoking patients carrying one or two SE alleles. As already mentioned above, the interaction between tobacco exposure and HLA SE alleles may trigger autoimmunity not to specific citrullinated antigens, but rather to ACPA production in general [2, 10, 14, 16–18, 42, 60].
As most studies on the possible associations between genetics, autoimmunity and lifestyle-related factors underlying the pathogenesis of RA have been carried out in Western countries, we have recently carried out the very first Central-Eastern European cohort study in 91 Hungarian RA patients. We assessed interactions between HLA-DRB1, ACPA production and lifestyle-related factors including smoking and alcohol intake. With respect to SE, we used both the traditional and the new French SE classification system [36]. Detailed correlation analysis revealed positive correlations between SE and ACPA positivity or absolute serum levels of various ACPAs. This also applied for the new du Montcel SE classification [36]. The complex risk estimate of SE, ACPA and smoking revealed that SE, but not ACPA, positivity was independently associated with smoking. In contrast, absolute serum ACPA levels did correlate with smoking irrespective of SE [16].
Genomic Studies in Mice and Men
Genomic Studies in RA and Other Human Inflammatory Diseases
GWAS studies already mentioned above, such as the ones using the WTCCC database assessing eight common inflammatory diseases including RA, described three major loci in which copy number variants (CNVs) were associated with a disease. The three loci included HLA associated with RA [26]. In addition to the WTCCC database, 31 non-MHC RA risk alleles have been confirmed by numerous genomic studies and meta-analyses. For example, a large GWAS metaanalysis carried out on data derived from 14 centers identified seven new RA risk loci. These SNPs are near genes of known immune function, such as IL6ST, SPRED2, RBPJ, CCR6, IRF5 and PXK [46]. These human RA risk alleles were identified and confirmed using anywhere from a few to thousands of SNPs. The most relevant susceptibility loci confirmed by genomic studies include, among others, PADI4, PTPN22, IL23R, CTLA4, IRF5, STAT4, IL6ST, TRAF1, TNFAIP3, CD40, IL2RA, IL2RB, CCL21, CCR6, CD2 and FCGR2A [5, 6, 43, 46, 61]. Characteristics of the most common genes were described above (Table 1).
In order to characterize possible common “inflammatory” and disease specific genes, we assessed the expression of 96 genes known to be involved in inflammation in patients with RA, psoriasis and IBD. By using the TaqMan Low Density Array (TLDA) method, we identified five "inflammatory genes," ADM, AQP9, CXCL2, IL10 and NAMPT, encoding adrenomedullin, aquaporin 9, the CXCL2 (groβ) chemokine, IL-10 and visfatin, respectively, discriminating all patients with the three inflammatory diseases from healthy controls. In addition, 21 genes in RA, six genes in psoriasis and 11 genes in IBD separated the given disease from the other two diseases. "RA-specific" genes included the known PTPN22 and IL23R, as well as genes encoding ADAM proteases (ADAM12, ADAM19, ADAM33), the CXCL8 (IL-8), CCL4 (MIP-1β) and CCL5 (RANTES) chemokines, heat shock protein 90 (HSP90AA1) and the TLR4-ligand HMGB1. Furthermore, when comparing RA patients with erosive versus non-erosive disease, the gene encoding PADI4, an enzyme involved in protein citrullination, differentiated the two RA subsets [13].
There may be limitations of human RA genomic studies. SNPs define only certain regions where a number of genes or non-coding elements are located. These risk loci or alleles defined by various numbers and frequency of SNPs indicate only a chromosome region carrying potentially dozens or hundreds of genes, which is expected to have one or a few functionally defective genes involved in the pathomechanism of RA. Furthermore, some SNPs including those in the IL23R, PTPN22 or IRF5 genes have also been associated with autoimmune rheumatic diseases other than RA as well. Due to the extreme heterogeneity of the human population, the highly motivated and exciting early-stage studies led to the current “frustration” in the field, and only a few confirmatory or treatment-related meta-analysis studies have been published during the past few years [24, 27–31].
Murine GWAS Studies in Arthritis Models
Studies conducted in animal model of arthritis may help to fill the gaps in human GWAS by allowing high-resolution gene mapping and functional studies that will yield greater insights into the mechanisms of autoimmune T and B cell responses in RA. Numerous arthritis models have become available, yet, none of these models embodies the full spectrum of diseases collectively called RA. The most appropriate animal models for RA seem to be those that use genetically controlled systemic autoimmune joint diseases, those in which both T and B cells are involved, and those that apply (auto)antigenic molecules of cartilage or joint tissues for provoking synovial joint inflammation. Among the animal models of RA, especially from a genetic point of view, cartilage proteoglycan (PG) aggrecan-induced arthritis (PGIA) may be the closest genetic and clinical model of the human disease [27–31].
In contrast to human studies using heterogeneous populations, there is a chance to use genetic combinations of various arthritis susceptible and resistant inbred strains for GWAS to identify disease-associated quantitative trait loci (QTLs). There are over a hundred non-MHC genetic risk alleles identified in murine and rat arthritis models. Some, but rather a small number of these rodent QTLs correspond to, or are located in, close proximity of the human risk alleles in the corresponding human genomic regions. In contrast to human studies, many of these rodent QTLs have not been confirmed by independent laboratories. Another limitation of these animal studies is that a large number of QTLs represent over a dozen clinical phenotypes in combination with the presence or level of various autoantibody or cytokine biomarkers. In addition, the PCR-based single sequence length polymorphism (SSLP) method used for identification of QTLs in either mice or rats is conceptually different form SNP microarray-based screening of the human genome. Therefore there has been some confusion in the interpretation of animal GWAS, which makes it difficult to compare these valuable results with those obtained from human GWAS [27–31].
In recent years, we have carried out numerous GWAS studies in the murine models PGIA and collagen-induced arthritis (CIA). Over 5,000 wild-type parents, approximately 500 F1 hybrids negative for PGIA and 3,200 F2 hybrids of six different genetic intercrosses were screened using over 240 SSLP markers in order to identify genetic alterations including QTLs that are linked to murine arthritis susceptibility, disease severity and biomarkers. Not discussing the complex methodology in more detail, during the past 15 years, we have identified 29 Pgia and 14 new Cia loci in different genetic combinations of F2 hybrid mice [29, 31]. A total of 13 QTLs that correspond to at least one major RA risk allele in the human genome were identified on mouse chromosomes 1, 2, 3, 5, 6, 10, 13, 15 and 18. With regard to analogy between human RA and murine arthritis, we identified two matching risk alleles, the murine Pgia26/Cia5 on mouse chromosome 3 that corresponds to the PTPN22/CD2 allele on human chromosome 1, and Pgia2/Cia2/Cia3 on mouse chromosome 2 that corresponds to the TRAF1/C5 allele on human chromosome 9. These two major/dominant mouse QTLs were found to contain separate narrow sub-traits and were simultaneously tested for influence on arthritis susceptibility, disease onset and severity, also in combination with over 15 biomarkers that might have some potential relevance for RA [27, 29–31].
Pharmacogenomics in RA: A Step Towards Personalized Medicine
As described by Cronstein [20] as an analogy to fashion design, ready-to-wear or “prêt-a-porter” clothing is designed for different people with different body shapes, while made-to-order or “haute couture” clothing is individually designed for a specific person. Personalized medicine uses haute couture drugs fitted to the needs of an individual RA patient, while prêt-a-porter compounds would work well for most patients, but would be ineffective for some of them [20]. There is indeed a need for biomarkers associated with better or worse response to a given treatment modality. These biomarkers may include cellular responses, autoantibodies, genes and other molecular structures. For example, higher versus lower synovial B-cell numbers or ACPA positivity versus negativity have been associated with differential responses to biologics, such as rituximab [19, 20, 62].
Pharmacogenetics or pharmacogenomics focus on the associations of expression of a single gene or multiple genetic signatures with responses to drugs (Tables 3 and 4). There has been major advance in this field, yet, we are still very far from finding the optimal genetic biomarkers. Regarding traditional DMARDs, several SNPs may be related to altered efficacy or toxicity of methotrexate (MTX) such as those in genes encoding proteins involved in the folate pathway, drug transport, nucleotide synthesis, and cytokine production. Without going into details, among folate pathway-associated variants. the most widely studied SNPs are C677T and A1298C mutations in the methylene tretrahydrofolate reductase (MTHFR) gene, the G80A amino acid change in the reduced folate career 1 (RFC-1) gene (SLC19A1) and the C1420T mutation in the serine hydroxymethyltransferase (SHMT) gene [19, 63–65]. The C3435T SNP in the gene encoding the MDR1 efflux pump (ABCB1) is involved in drug transport and increases MTX efflux [66]. Nucleotide synthesis-related genes implicated in MTX activity include ATIC and TYMS. Mutations in these genes lead to altered mRNA stability and enzyme activity and thus improved efficacy, but increased toxicity [19]. Finally, an SNP in the IL1RN gene was also associated with decreased efficacy of MTX [19, 67]. Increased toxicity of azathioprine has been associated with variations in the genes encoding thiopurine S-methyltransferase (TMPT) or ITPase [19]. Mutations in the IL10 (A1082G, C819T, C592A) or the TNF gene (A308G) results in increased efficacy of hydroxychloroquine, while increased efficacy of leflunomide has been linked to variations in the dihydroorotate dehydrogenase (DHODH, C19A), estrogen receptor (ESR1) or the cytochrome P450 (CYP1A2) gene [19] (Table 3).
Recently, therapeutic responses to biologic agents have drawn more attention. According to the recent treat-to-target and EULAR/ACR treatment recommendations [11], treatment strategies should aim at remission or at least low disease activity (LDA). While the administration of anti-tumor necrosis factor α (TNF-α) show sustained efficacy in many patients, remission or LDA cannot be reached in as much as 60 % of RA patients [68]. Reasons for switching biologics include primary lack and secondary loss of response, partial response or side effects [68]. There is an immense need to find biomarkers including pharmacogenomic ones that are able to predict inefficacy or side effects early during the course of treatment. The first studies on single SNPs were followed by more detailed pharmacogenomic investigations on multiple gene signatures. Most studies have been conducted on the G308A SNP in the gene encoding TNF-α. This SNP has been associated with clinical responses to infliximab, etanercept and adalimumab, and most studies reported increased efficacy. Other polymorphisms associated with clinical outcome during anti-TNF treatment include other changes in the TNF locus (e.g., A238G), T196G change in the TNFRSF1B gene, Val158Phe change in the FCGR gene, an SNP in the receptor-type tyrosine-protein phosphatase C gene (PTPRC) [19, 21]. SNPs in the MAPK14 gene lead to increased efficacy of anti-TNF antibodies, but not to etanercept [19, 21]. Responses to the B-cell-depleting antibody rituximab have been linked to SNPs in the FCGR gene encoding FcγRIIIA (Val158Phe amino acid change), the IL6 gene and the gene encoding the B lymphocyte stimulator (BLyS) [19–21, 54] (Table 4).
Expression of multiple genes and genetic signatures have been studied in association with responses to biologics using microarray technique. To date, most studies have been conducted on responses to infliximab and etanercept. Unfortunately, these studies yielded to very inconclusive results as different groups reported different signatures. A few examples are presented below.
Genes that have been associated with responses to etanercept include primarily cytokine genes (TNF, lymphotoxin α [LTA], IL10, TGFB1, IL1RN), cytokine receptor genes (TNFRSF1A, TNFRSF1B) and Fc receptor alleles (FCGR2A, FCGR3A, FCGR3B) [21]. In a study conducted in 19 patients treated with etanercept, good clinical response was associated with SNPs in the CCL4, IL8, IL1B, TNFAIP3 and some other genes. Four SNPs (TNF, IL10, TGFB1, IL1RN) have been studied in relation to etanercept responses in 123 RA patients. Some TNF and IL10 SNPs could be associated with better responses, while the combination of IL1RN and TGFB1 SNPs indicated unfavorable responses. In 457 early-stage RA patients, HLA-DRB1, TNF and LTA SNPs were related to clinical response to etanercept (reviewed in [21]).
With respect to infliximab, in a study conducted on 44 RA patients, clinical responses were associated with eight genes including HLADRB3. In another cohort, 41 genes including HLADPB1 and PTPN12 could be linked to infliximab responses. The function of most genes related to clinical responses has not yet been identified in RA. Polymorphisms in absolutely different genes including chemokine and chemokine receptor genes (CCL4, CX3CR1) and interferon-induced genes were associated with responses to infliximab in yet another study. Pharmacogenomic studies in infliximab-treated patients have been conducted in several other cohorts (reviewed in [19, 20]).
To date the largest published pharmacogenomic GWAS included 566 RA patients. Changes in altogether 460,000 SNPs were assessed in patients treated with etanercept, infliximab or adalimumab. Prediction analysis revealed seven loci that may be related to responses to these biologics. Two loci, PDZD2 encoding PDZ domain-containing protein 2 and EYA4 encoding the protein “eyes absent 4” showed the strongest correlation with biologic responses. Yet, the role of these two genes in RA is unknown [19].
Very recently, we have conducted the first pharmacogenomic study in 13 tocilizumab-treated RA patients. The expression levels of altogether 59 genes showed significant changes between baseline and after 4 weeks of treatment. Four genes (DHFR, CCDC32, EPHA, TRAV8) determined responders after correction for multiple testing. Again, very little is known about the function of these genes in RA. Dihydrofolate reductase (DHFR) was identified as putative predictor for MTX response, T cell receptor alpha variable 8–3 (TRAV8-3) is involved in CD8+ T-cell responses, the ephrin receptor A4 (EPHA) plays a role in the nervous system, while the function of CCDC32 is unknown [22].
Thus, pharmacogenomics may bring us closer to personalized medicine; however, recent studies yielded to inconclusive results reporting substantially different gene patterns in various cohorts. In addition, although numerous genes have been picked up that may predict therapeutic responses, the role of many of these genes in the pathogenesis of RA is unknown. Therefore, the breakthrough is yet to come. Further studies, possibly using pre-selected and well-characterized gene sets, are needed to determine the value of pharmacogenomics in RA [19, 22].
Conclusions
The “Bermuda triangle” of genetic, environmental factors and autoimmunity has been associated with susceptibility to and outcome of RA. The heritability of RA has been estimated to be about 60 %. In addition to HLA-DRB1*01 and HLA-DRB1*04 SE alleles, recently, much attention has been drawn to two non-SE HLA-DRB1 alleles, DRB1*13 and DRB*15. The recent du Montcel SE classification system divides SE alleles into S1, S2, S3P and S3D groups. It seems that S2 and S3P, but, to a lesser extent, S1 and S3D alleles may also predispose to ACPA positive RA. SE is a risk factor for more severe, destructive, as well as “systemic” RA. GWAS also confirmed the association of RA with at least 30 alleles, most importantly PTPN22, TRAF1, CTLA4, IRF5, STAT4, IL23R, CCR6, CD40 and PADI4. There is also significant interaction between genes, ACPA status and smoking. In CIA and PgIA models of arthritis, susceptibility loci that correspond to the human PTPN22/CD2 and TRAF1/C5 alleles were identified. Finally, pharmacogenomics focuses on the associations of SNPs or multiple genetic signatures with responses to drugs. Yet, more large-scale confirmatory studies need to be conducted in the field of genetics and genomics, in association with inflammatory rheumatic diseases.
References
Alamanos Y, Drosos AA (2005) Epidemiology of adult rheumatoid arthritis. Autoimmun Rev 4(3):130–136
Klareskog L, Padyukov L, Alfredsson L (2007) Smoking as a trigger for inflammatory rheumatic diseases. Curr Opin Rheumatol 19(1):49–54
van der Helm-van Mil AH, Wesoly JZ, Huizinga TW (2005) Understanding the genetic contribution to rheumatoid arthritis. Curr Opin Rheumatol 17(3):299–304
van der Woude D, Alemayehu WG, Verduijn W, de Vries RR, Houwing-Duistermaat JJ, Huizinga TW et al (2010) Gene–environment interaction influences the reactivity of autoantibodies to citrullinated antigens in rheumatoid arthritis. Nat Genet 42(10):814–816, author reply 816
Szodoray P, Szabo Z, Kapitany A, Gyetvai A, Lakos G, Szanto S et al (2010) Anti-citrullinated protein/peptide autoantibodies in association with genetic and environmental factors as indicators of disease outcome in rheumatoid arthritis. Autoimmun Rev 9(3):140–143
de Vries R (2011) Genetics of rheumatoid arthritis: time for a change! Curr Opin Rheumatol 23(3):227–232
Cooles FA, Isaacs JD. Pathophysiology of rheumatoid arthritis. Curr Opin Rheumatol;23(3):233–40
Szekanecz Z, Soos L, Szabo Z, Fekete A, Kapitany A, Vegvari A et al (2008) Anti-citrullinated protein antibodies in rheumatoid arthritis: as good as it gets? Clin Rev Allergy Immunol 34(1):26–31
Klareskog L, Padyukov L, Lorentzen J, Alfredsson L (2006) Mechanisms of disease: genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract Rheumatol 2(8):425–433
Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L (2004) A gene–environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum 50(10):3085–3092
Smolen JS, Landewe R, Breedveld FC, Dougados M, Emery P, Gaujoux-Viala C et al (2010) EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs. Ann Rheum Dis 69(6):964–975
MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K et al (2000) Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 43(1):30–37
Mesko B, Poliska S, Szegedi A, Szekanecz Z, Palatka K, Papp M et al (2010) Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med Genomics 3:15
Lee HS, Irigoyen P, Kern M, Lee A, Batliwalla F, Khalili H et al (2007) Interaction between smoking, the shared epitope, and anti-cyclic citrullinated peptide: a mixed picture in three large North American rheumatoid arthritis cohorts. Arthritis Rheum 56(6):1745–1753
Kapitany A, Szabo Z, Lakos G, Aleksza M, Vegvari A, Soos L et al (2008) Associations between serum anti-CCP antibody, rheumatoid factor levels and HLA-DR4 expression in Hungarian patients with rheumatoid arthritis. Isr Med Assoc J 10(1):32–36
Besenyei T, Gyetvai A, Szabo Z, Fekete A, Kapitany A, Szodoray P et al (2011) Associations of HLA-shared epitope, anti-citrullinated peptide antibodies and lifestyle-related factors in Hungarian patients with rheumatoid arthritis: data from the first Central-Eastern European cohort. Joint Bone Spine 78(6):652–653
Scott IC, Steer S, Lewis CM, Cope AP (2011) Precipitating and perpetuating factors of rheumatoid arthritis immunopathology: linking the triad of genetic predisposition, environmental risk factors and autoimmunity to disease pathogenesis. Best Pract Res Clin Rheumatol 25(4):447–468
Vittecoq O, Lequerre T, Goeb V, Le Loet X, Abdesselam TA, Klemmer N (2008) Smoking and inflammatory diseases. Best Pract Res Clin Rheumatol 22(5):923–935
Davila L, Ranganathan P (2011) Pharmacogenetics: implications for therapy in rheumatic diseases. Nat Rev Rheumatol 7(9):537–550
Cronstein BN (2006) Pharmacogenetics in the rheumatic diseases, from pret-a-porter to haute couture. Nat Clin Pract Rheumatol 2(1):2–3
Danila MI, Hughes LB, Bridges SL (2008) Pharmacogenetics of etanercept in rheumatoid arthritis. Pharmacogenomics 9(8):1011–1015
Mesko B, Poliska S, Szamosi S, Szekanecz Z, Podani J, Varadi C et al (2012) Peripheral blood gene expression and IgG glycosylation profiles as markers of tocilizumab treatment in rheumatoid arthritis. J Rheumatol 39(5):916–928
Plant D, Bowes J, Potter C, Hyrich KL, Morgan AW, Wilson AG et al (2011) Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum 63(3):645–653
Centola M, Szekanecz Z, Kiss E, Zeher M, Szegedi G, Nakken B et al (2007) Gene expression profiles of systemic lupus erythematosus and rheumatoid arthritis. Expert Rev Clin Immunol 3(5):797–806
Feng T, Zhu X (2010) Genome-wide searching of rare genetic variants in WTCCC data. Hum Genet 128(3):269–280
Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S et al (2010) Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464(7289):713–720
Adarichev VA, Vermes C, Hanyecz A, Mikecz K, Bremer EG, Glant TT (2005) Gene expression profiling in murine autoimmune arthritis during the initiation and progression of joint inflammation. Arthritis Res Ther 7(2):R196–R207
Ahlqvist E, Hultqvist M, Holmdahl R (2009) The value of animal models in predicting genetic susceptibility to complex diseases such as rheumatoid arthritis. Arthritis Res Ther 11(3):226
Glant TT, Finnegan A, Mikecz K (2003) Proteoglycan-induced arthritis: immune regulation, cellular mechanisms, and genetics. Crit Rev Immunol 23(3):199–250
Glant TT, Mikecz K, Arzoumanian A, Poole AR (1987) Proteoglycan-induced arthritis in BALB/c mice. Clinical features and histopathology. Arthritis Rheum 30(2):201–212
Glant TT, Adarichev VA, Nesterovitch AB, Szanto S, Oswald JP, Jacobs JJ et al (2004) Disease-associated qualitative and quantitative trait loci in proteoglycan-induced arthritis and collagen-induced arthritis. Am J Med Sci 327(4):188–195
Deighton CM, Walker DJ, Griffiths ID, Roberts DF (1989) The contribution of HLA to rheumatoid arthritis. Clin Genet 36(3):178–182
Gregersen PK, Silver J, Winchester RJ (1987) The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30(11):1205–1213
van der Woude D, Houwing-Duistermaat JJ, Toes RE, Huizinga TW, Thomson W, Worthington J et al (2009) Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum 60(4):916–923
van der Helm-van Mil AH, Verpoort KN, le Cessie S, Huizinga TW, de Vries RR, Toes RE (2007) The HLA-DRB1 shared epitope alleles differ in the interaction with smoking and predisposition to antibodies to cyclic citrullinated peptide. Arthritis Rheum 56(2):425–432
du Montcel ST, Michou L, Petit-Teixeira E, Osorio J, Lemaire I, Lasbleiz S et al (2005) New classification of HLA-DRB1 alleles supports the shared epitope hypothesis of rheumatoid arthritis susceptibility. Arthritis Rheum 52(4):1063–1068
van der Woude D, Lie BA, Lundstrom E, Balsa A, Feitsma AL, Houwing-Duistermaat JJ et al (2010) Protection against anti-citrullinated protein antibody-positive rheumatoid arthritis is predominantly associated with HLA-DRB1*1301: a meta-analysis of HLA-DRB1 associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in four European populations. Arthritis Rheum 62(5):1236–1245
Laki J, Lundstrom E, Snir O, Ronnelid J, Ganji I, Catrina AI et al (2012) Very high levels of anti-citrullinated protein antibodies are associated with HLA-DRB1*15 non-shared epitope allele in patients with rheumatoid arthritis. Arthritis Rheum 64(7):2078–2084
Zsilak S, Gal J, Hodinka L, Rajczy K, Balog A, Sipka S et al (2005) HLA-DR genotypes in familial rheumatoid arthritis: increased frequency of protective and neutral alleles in a multicase family. J Rheumatol 32(12):2299–2302
Jawaheer D, Thomson W, MacGregor AJ, Carthy D, Davidson J, Dyer PA et al (1994) "Homozygosity" for the HLA-DR shared epitope contributes the highest risk for rheumatoid arthritis concordance in identical twins. Arthritis Rheum 37(5):681–686
Gyetvai A, Szekanecz Z, Soos L, Szabo Z, Fekete A, Kapitany A, et al. New classification of the shared epitope in rheumatoid arthritis: impact on the production of various anti-citrullinated protein antibodies. Rheumatology (Oxford) 2009
Huizinga TW, Amos CI, van der Helm-van Mil AH, Chen W, van Gaalen FA, Jawaheer D et al (2005) Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum 52(11):3433–3438
Bax M, van Heemst J, Huizinga TW, Toes RE (2011) Genetics of rheumatoid arthritis: what have we learned? Immunogenetics 63(8):459–466
Farago B, Magyari L, Safrany E, Csongei V, Jaromi L, Horvatovich K et al (2008) Functional variants of interleukin-23 receptor gene confer risk for rheumatoid arthritis but not for systemic sclerosis. Ann Rheum Dis 67(2):248–250
Farago B, Talian GC, Komlosi K, Nagy G, Berki T, Gyetvai A et al (2009) Protein tyrosine phosphatase gene C1858T allele confers risk for rheumatoid arthritis in Hungarian subjects. Rheumatol Int 29(7):793–796
Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP et al (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42(6):508–514
Goeb V, Dieude P, Daveau R, Thomas-L'otellier M, Jouen F, Hau F et al (2008) Contribution of PTPN22 1858T, TNFRII 196R and HLA-shared epitope alleles with rheumatoid factor and anti-citrullinated protein antibodies to very early rheumatoid arthritis diagnosis. Rheumatology (Oxford) 47(8):1208–1212
Cha S, Choi CB, Han TU, Kang CP, Kang C, Bae SC (2007) Association of anti-cyclic citrullinated peptide antibody levels with PADI4 haplotypes in early rheumatoid arthritis and with shared epitope alleles in very late rheumatoid arthritis. Arthritis Rheum 56(5):1454–1463
Poor G, Nagy ZB, Schmidt Z, Brozik M, Meretey K, Gergely P Jr (2007) Genetic background of anticyclic citrullinated peptide autoantibody production in Hungarian patients with rheumatoid arthritis. Ann N Y Acad Sci 1110:23–32
Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M et al (2003) Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet 34(4):395–402
van der Linden MP, Feitsma AL, le Cessie S, Kern M, Olsson LM, Raychaudhuri S et al (2009) Association of a single-nucleotide polymorphism in CD40 with the rate of joint destruction in rheumatoid arthritis. Arthritis Rheum 60(8):2242–2247
Liang YL, Wu H, Shen X, Li PQ, Yang XQ, Liang L, et al. Association of STAT4 rs7574865 polymorphism with autoimmune diseases: a meta-analysis. Mol Biol Rep 2012.
Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B et al (2007) TRAF1-C5 as a risk locus for rheumatoid arthritis—a genomewide study. N Engl J Med 357(12):1199–1209
Lee YH, Ji JD, Song GG (2008) Associations between FCGR3A polymorphisms and susceptibility to rheumatoid arthritis: a metaanalysis. J Rheumatol 35(11):2129–2135
Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, Takahashi A et al (2010) A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet 42(6):515–519
Szekanecz Z, Koch AE, Tak PP (2011) Chemokine and chemokine receptor blockade in arthritis, a prototype of immune-mediated inflammatory diseases. Neth J Med 69(9):356–366
Ding B, Padyukov L, Lundstrom E, Seielstad M, Plenge RM, Oksenberg JR et al (2009) Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum 60(1):30–38
Verpoort KN, Cheung K, Ioan-Facsinay A, van der Helm-van Mil AH, de Vries-Bouwstra JK, Allaart CF et al (2007) Fine specificity of the anti-citrullinated protein antibody response is influenced by the shared epitope alleles. Arthritis Rheum 56(12):3949–3952
Daha NA, Toes RE (2011) Rheumatoid arthritis: Are ACPA-positive and ACPA-negative RA the same disease? Nat Rev Rheumatol 7(4):202–203
Pedersen M, Jacobsen S, Klarlund M, Pedersen BV, Wiik A, Wohlfahrt J et al (2006) Environmental risk factors differ between rheumatoid arthritis with and without auto-antibodies against cyclic citrullinated peptides. Arthritis Res Ther 8(4):R133
Perricone C, Ceccarelli F, Valesini G. An overview on the genetic of rheumatoid arthritis: a never-ending story. Autoimmun Rev, 10(10):599–608
Ruyssen-Witrand A, Rouanet S, Combe B, Dougados M, Le Loet X, Sibilia J et al (2012) Fcgamma receptor type IIIA polymorphism influences treatment outcomes in patients with rheumatoid arthritis treated with rituximab. Ann Rheum Dis 71(6):875–877
van Ede AE, Laan RF, Blom HJ, Huizinga TW, Haagsma CJ, Giesendorf BA et al (2001) The C677T mutation in the methylenetetrahydrofolate reductase gene: a genetic risk factor for methotrexate-related elevation of liver enzymes in rheumatoid arthritis patients. Arthritis Rheum 44(11):2525–2530
Berkun Y, Levartovsky D, Rubinow A, Orbach H, Aamar S, Grenader T et al (2004) Methotrexate related adverse effects in patients with rheumatoid arthritis are associated with the A1298C polymorphism of the MTHFR gene. Ann Rheum Dis 63(10):1227–1231
Dervieux T, Kremer J, Lein DO, Capps R, Barham R, Meyer G et al (2004) Contribution of common polymorphisms in reduced folate carrier and gamma-glutamylhydrolase to methotrexate polyglutamate levels in patients with rheumatoid arthritis. Pharmacogenetics 14(11):733–739
Pawlik A, Wrzesniewska J, Fiedorowicz-Fabrycy I, Gawronska-Szklarz B (2004) The MDR1 3435 polymorphism in patients with rheumatoid arthritis. Int J Clin Pharmacol Ther 42(9):496–503
Tolusso B, Pietrapertosa D, Morelli A, De Santis M, Gremese E, Farina G et al (2006) IL-1B and IL-1RN gene polymorphisms in rheumatoid arthritis: relationship with protein plasma levels and response to therapy. Pharmacogenomics 7(5):683–695
van Vollenhoven RF (2007) Switching between anti-tumour necrosis factors: trying to get a handle on a complex issue. Ann Rheum Dis 66(7):849–851
Acknowledgments
This work was supported by research grants ETT 315/2009 from the Medical Research Council of Hungary (Z.S.); by the TÁMOP 4.2.1/B-09/1/KONV-2010-0007 and TÁMOP-4.2.2.A-11/1/KONV-2012-0031 projects co-financed by the European Union and the European Social Fund (Z.S.), the Bridging Fund provided by the University of Debrecen, Medical and Health Sciences Center (Z.S.), and a grant (R01 AR059356) awarded by the National Institutes of Health, USA (T.T.G.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kurkó, J., Besenyei, T., Laki, J. et al. Genetics of Rheumatoid Arthritis — A Comprehensive Review. Clinic Rev Allerg Immunol 45, 170–179 (2013). https://doi.org/10.1007/s12016-012-8346-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12016-012-8346-7