Abstract
Dorsal root ganglia (DRG) sensory neurons can transmit information about noxious stimulus to cerebral cortex via spinal cord, and play an important role in the pain pathway. Alterations of the pain pathway lead to CIPA (congenital insensitivity to pain with anhidrosis) or chronic pain. Accumulating evidence demonstrates that nerve damage leads to the regeneration of neurons in DRG, which may contribute to pain modulation in feedback. Therefore, exploring the regeneration process of DRG neurons would provide a new understanding to the persistent pathological stimulation and contribute to reshape the somatosensory function. It has been reported that a subpopulation of satellite glial cells (SGCs) express Nestin and p75, and could differentiate into glial cells and neurons, suggesting that SGCs may have differentiation plasticity. Our results in the present study show that DRG-derived SGCs (DRG-SGCs) highly express neural crest cell markers Nestin, Sox2, Sox10, and p75, and differentiate into nociceptive sensory neurons in the presence of histone deacetylase inhibitor VPA, Wnt pathway activator CHIR99021, Notch pathway inhibitor RO4929097, and FGF pathway inhibitor SU5402. The nociceptive sensory neurons express multiple functionally-related genes (SCN9A, SCN10A, SP, Trpv1, and TrpA1) and are able to generate action potentials and voltage-gated Na+ currents. Moreover, we found that these cells exhibited rapid calcium transients in response to capsaicin through binding to the Trpv1 vanilloid receptor, confirming that the DRG-SGC-derived cells are nociceptive sensory neurons. Further, we show that Wnt signaling promotes the differentiation of DRG-SGCs into nociceptive sensory neurons by regulating the expression of specific transcription factor Runx1, while Notch and FGF signaling pathways are involved in the expression of SCN9A. These results demonstrate that DRG-SGCs have stem cell characteristics and can efficiently differentiate into functional nociceptive sensory neurons, shedding light on the clinical treatment of sensory neuron-related diseases.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability to detect noxious stimuli is critical to wellbeing and survival of organisms, which is illustrated by examination of individuals who suffer from congenital insensitivity to pain with anhidrosis (CIPA). These people cannot engage appropriate protective behaviors against noxious stimuli, such as retraction from open flames or sharp objects, due to incapability of detecting painful stimuli [1, 2]. Scientists defined the neural apparatus responsible for detecting noxious stimulus as nociceptors [3]. The nociceptors’ cell bodies are located in the dorsal root ganglia (DRG) or trigeminal ganglion (TG), and have a central and peripheral axonal branch, which respectively innervate the spinal cord and its target organs. Thus, in the pain pathway, DRG or TG sensory neurons can transmit the information about the location, type, and intensity of noxious stimulus to the cerebral cortex via spinal cord [1, 2].
Alterations of the pain pathway can lead to CIPA or chronic pain. If CIPA is considered to be a loss of pain sensation, then at some level, chronic pain can be thought to be the hyperactivity of tissue or nerve damage to promote guarding of the injured area [1]. Evidence shows that in low- and middle-income countries, the prevalence of chronic pain was 33% of the general adult population [4]. However, no matter in the development of CIPA or chronic pain, there is no denying that DRG or TG sensory neurons play an important role, which is confirmed by numerous studies about DRG or TG neurons [5,6,7,8]. Accumulating evidence demonstrates that nerve damage leads to the regeneration of neurons in DRG, which may contribute to pain modulation in feedback [9,10,11,12]. Therefore, exploring the regeneration process of DRG neurons would provide a new understanding to the persistent pathological stimulation, and contribute to reshape the somatosensory function.
Satellite glial cells (SGCs) are a kind of cells around sensory neurons in DRG and originate from neural crest cells (NCCs) [13, 14]. Studies have found that SGCs can regulate chronic pain by communicating with sensory neurons through gap junction or releasing neural modulating factors, such as NO, ATP, TNF-α [15,16,17,18]. After nerve injury, SGCs express high levels of BDNF, p75 and fibroblast growth factor 2 (FGF2), indicating that SGCs have the functions of supporting, nourishing and protecting neurons [19, 20]. In addition, there is evidence showing that SGCs have differentiation potential. In the DRG explants culture, a subpopulation of SGCs expressing Nestin and p75 migrated from explants, and could differentiate into glial cells and neurons in the presence of serum [21]. After peripheral nerve injury, SGCs around sensory neurons in vivo express Nestin and Sox2 [11, 22, 23], suggesting that DRG may contain a group of incompletely differentiated cells.
In this study, we cultured the neonatal rat DRG and found that the migrated DRG-SGCs highly express the NCC markers Nestin, p75, Sox10, Sox2. Furthermore, by activating the Wnt signaling pathway and inhibiting the FGF and Notch signaling pathways, DRG-SGCs can be differentiated into functional nociceptive sensory neurons. Our results demonstrate that DRG-SGCs have the characteristics of stem cells and can differentiate into sensory neurons. This provides us with a new understanding of sensory neuron-related diseases and is of great significance for the effective use of DRG-SGCs in the treatment of CIPA or chronic pain.
Materials and Methods
DRG-SGCs Culture
DRGs were isolated from Sprague Dawley (SD) rats (post-natal 1–2 d). All animal experimental protocols were approved by the guidelines of the Institutional Medical Experimental Animal Use Committee of Soochow University. The nerve roots and epineurium were carefully removed. For explants method, DRG were plated at a density of approximately 30 explants per 60-mm dish and fresh DRG-SGCs medium was then added. For trypsinization approach, the DRGs were digested with collagenase (Sigma C5138-100 mg) at 37oC for 30 min, and then transferred to 0.25% trypsin (Gibco) for 20 min. At the end of the incubation period, the solution was spun at 1,000 rpm for 5 min. The cells were resuspended in 3 ml of DRG-SGCs medium and inoculated into a 60 mm dish. The DRG-SGCs medium comprised DMEM/F12 (Corning, 10-092-CVR), 2% B27 (Invitrogen, 17504-044), L-Glutamine (Gibco), penicillin/streptomycin (Gibco), 20 ng/ml NRG1 (neuregulin1, 11,609-H01H2, Sino Biological Inc., China), 10 ng/ml T3 (Sigma, T2877), 10 ng/ml T4 (MCE, HY-18,341), 0.3 mg/ml BSA (Sigma, A1933), 10− 6 M Insulin (Sigma, I1882), 38 ng/ml Dexamethasone (Sigma, D4902).
When a large number of cells had migrated from the DRG explants, the cells were passaged. After removing the DRG explants, SGCs were digested with 0.25% trypsin (Gibco) at 37oC for 2 min. Digestion was terminated by the addition of 10% fetal bovine serum (FBS, BI, 04-001-1ACS) in DMEM/F12 medium. The cell suspension was centrifuged at 1,000 rpm for 5 min, and then cells were seeded into the poly-L-lysine (PLL)-coated plates (Sigma, P7280-5 mg) at a density of 100,000 cells per well of a 35 mm dish. The culture medium was changed every 2 d.
Neural Induction
After cell passage for 24 h, the culture medium was replaced with neuronal differentiation medium (NDM). 3 d later, 10 µM forskolin (Selleck, S2449) was added to the medium, and the treatment was continued for 21 d and then replaced with neuronal maturation medium. The culture medium was refreshed every 3 d during the induction period.
The NDM comprised neurobasal medium/DMEM/F12 (1:1), 1% N2 (Gibco, 17502-048), 2% B27, 200 µM ascorbic acid (Sigma, A5960-25G), 10 ng/ml BDNF (PeproTech, 450-02-50), 10 ng/ml NGF (PeproTech, 450-34-20), 10 ng/ml NT-3 (PeproTech, 450-02-50), 10 ng/ml GDNF (PeproTech, 450-10-50), 3 mM VPA(Sigma, P4543-10G), 3 µM CHIR99021 (Selleck, S2924), 2 µM RO4929097 (Selleck, S1575), 10 µM SU5402 (Selleck, S7667).
Neuronal maturation medium: Neurobasal Medium (Gibco, 10888-022), 1% N2, 2% B27, 200 µM ascorbic acid, 10 ng/ml BDNF, 10 ng/ml NGF, 10 ng/ml NT-3, 10 ng/ml GDNF.
Immunofluorescence
Immunostaining of cultures was performed as described previously [24]. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, and then washed 3 times with phosphate-buffered saline (PBS) for 5 min each. Afterward, the slides were blocked with 0.2% Triton X-100 for 20 min and 5% BSA for 1 h at room temperature. Primary antibodies were incubated with the slides at 4 °C overnight. After washing 3 times with PBS, the appropriate secondary antibodies (Cy3/Alexa Fluor 488-labeled Goat Anti-Mouse/Rabbit IgG(H + L), 1: 500, Beyotime) were incubated with the slides for 2 h at room temperature. Cell nuclei were counterstained with Hoechst 33,258 (1:1000, Invitrogen). Fluorescent images were generated by Leica Microsystems LAS AF (Lecia AF6000 microscopy). The fraction of positive cells was determined by counting 4 nonoverlapping microscopic fields for each coverslip in at least three separate experiments using Image J software 1.4.3.67 (National Institutes of Health). The following primary antibodies were used: GS (1:400, Abcam, ab49873), Map2 (1:300, Cell Signaling Technology, 8707T), NeuN (1:500, Cell Signaling Technology, 24307T), Synapsin1 (1:200, Cell Signaling Technology, 5297T), GFAP (1:500, Cell Signaling Technology, 80788S), TrkA (1:200, Abcam, ab131472), S100 (1:200, Abcam, ab52642), Sox10 (1:400, Abcam, ab155279), p75 (1:300, Abcam, ab3125), Nestin (1:100, Abcam, ab6142), Sox2 (1:100, Abcam, ab79351), Tuj1 (1:500, Covance, MMS435P), Trpv1 (1:100, Santa Cruz, sc-398,417). Controls treated with nonspecific mouse/rabbit IgM or secondary antibodies alone showed no staining.
RT-PCR
RT-PCR was performed in CFX96 Real-Time PCR Detection System (Bio-Rad) and the quality and quantity of RNA was evaluated using NanoDrop 2000 (Thermo Fisher Scientific). Total RNA samples were collected using TRIzol reagents (Takara) according to the manufacturer’s instructions. Total RNA (1 µg) from each sample was reverse-transcribed into cDNA using a reverse transcription kit (Takara). Real-time RT-PCR reactions was performed in a 50-µl mixture containing cDNA, primers, and 1 × SYBR GREEN PCR Supermix (Bio-Rad). Reactions were carried out at 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The primers used are listed in Table S1, including three house-keeping genes (GAPDH, HPRT1, beta-actin). Each sample was run in duplicates, data were calculated using the 2ΔΔCT method. All gene expression changes were expressed as fold changes compared with Control, and were normalized to the expression of GAPDH, which was found to be stably expressed under our experimental conditions, a reliable reference gene with an M-value below 1 and coefficient of variation (CV) below 50% as suggested by Vandesompele et al. for a heterogeneous sample [25].
Electrophysiology
In order to detect the electrophysiological characteristics, the differentiated cells were processed as described previously [26, 27]. Briefly, cells were seeded onto PLL-coated coverslips as described for cell culture and were visualized in Nikon Ts2R-FL microscope. Whole cell patch clamp recordings were performed using a Multi-clamp 700 B amplifier (Molecular Devices) at room temperature (20–22 °C). Signals were sampled at 20 kHz. Patch pipettes (World Precision Instruments) were pulled from standard wall glass of 1.5-mm optical density OD using a flaming micropipette puller (P97, Sutter Instruments) and had ∼3 MΩ resistance when filled with internal solution. Voltage commands and digitization of membrane voltages were controlled using a Digidata 1440A interfaced with Clampex 10.2 of the pClamp software package (Molecular Devices). Series resistance (Rs) and capacitance (Cm) values were taken directly from readings of the amplifier after electronic subtraction of the capacitive transients. Series resistance was compensated to the maximum extent possible (at least 75%). For current-clamp and Na+ current recordings, the extracellular solution contained 128 mM NaCl, 2 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, and 25 mM HEPES, pH adjusted to 7.4 with NaOH (all from sigma). The intracellular solution contained 110 mM KCl, 10 mM NaCl, 2 mM EGTA, 25 mM HEPES, 4 mM MgATP, and 0.3 mM Na-GTP; the pH was adjusted to 7.4 with KOH (all from sigma). The voltage clamping potential was held at -60 mV. The injection current was − 20 pA, the step current was 50 pA, and the stimulation time was 1 s.
Calcium Imaging
The differentiated cells were subjected to calcium imaging as described previously [28, 29]. Briefly, differentiated cells were loaded with 2 µM Fluo-4 AM (Beyotime, S1060) for 30 min at 37 °C/5% CO2. Fluo-4 AM were added at in Tyrode’s solution (128 mM NaCl, 2 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, and 25 mM HEPES). Fluorescent images were generated by Leica Microsystems LAS AF (Lecia AF6000 microscopy). Calcium flux was monitored for 90 s, with 200 ms exposure time and 2 s delay between exposures. During the monitoring process, 10 × capsaicin (100 µM, MCE, HY-10,448) was added to the Tyrode’s solution. 25 mM KCl was added as a stimulation to confirm the neural identity of responsive cells. For experiments using the selective and competitive vanilloid receptor 1 (Trpv1) antagonist AMG9810 (5 µM, MCE, HY-101,736), compound was added to wells 10 minutes prior to calcium imaging, and then the protocol was followed as above.
Following data collection, image analysis was carried out to quantify calcium flux. Using Image J software 1.4.3.67 (National Institutes of Health), calcium responses were determined by calculating the change in fluorescence over the initial fluorescence at ~ 488 nm excitation wavelength (F - F0)/F0, where F = the fluorescence at a given time point and F0 = the mean basal, unstimulated fluorescence of each cell. At least 3 unstimulated time points were selected to calculate F0, and the fluorescence intensity of cells at each time point required background subtraction. 0.2 was applied as the threshold for a positive calcium response.
Statistical Analysis
All quantified data were statistically analyzed and presented as mean ± SD (standard deviation). Recording and analysis of whole cell membrane potential was performed using pCLAMP10.2 software (Molecular Devices). Statistical analysis was performed using GraphPad Prism 5 (GraphPad software). Two-sample analysis was performed using t-test analysis, and multi-sample analyses were performed using one-way analysis of variance (ANOVA) followed by Dunnett’s test. P values less than 0.05 (P < 0.05) were considered statistically significant.
Results
DRG-SGCs Have Stem Cell Properties
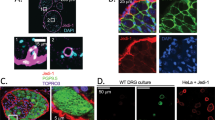
DRG-SGCs are the glial cells around the soma of sensory neurons in DRG, which have the functions of supporting, nourishing and protecting neurons [13, 18]. Evidence shows that after nerve injury, DRG-SGCs express NCC markers Nestin and Sox2, and can differentiate into sensory neurons [11, 21,22,23]. Thus, exploring the differentiation of DRG-SGCs into sensory neurons might help understand the persistent pathological stimulation, and contribute to reshape somatosensory function. To this end, we isolated, cultured and characterized DRG-SGCs using DRG explants from newborn SD rats. Upon 1 d of culture, we found that there were a small number of oval-shaped small cells around the DRG. After 6 d, a large number of cells migrated out of DRG (Fig. S1). At day 11, the DRG explants were removed and the remaining cells were passaged. After 24 h of incubation, immunofluorescence staining showed that the cultured cells expressed GS, GFAP and S100 (Fig. 1a, d), three markers for SGCs demonstrated by the in vitro and in vivo studies [12, 30].
DRG-SGCs express stem cell markers. a Immunofluorescence shows that the cells express SGCs markers GS, S100, GFAP. Scale bars, 250 µm. b Immunofluorescence shows that the cells express NCC markers Nestin, Sox2 and Sox10. Scale bars, 250 µm. c Double-immunostaining shows that most DRG-SGCs express NCC markers. d-f The positive rate of each marker. n = 5 batches. gThe RT-PCR results show that there was no significant difference between SGCs cultured using explants method and trypsinization approach. The cultured DRG-SGCs highly express Nestin, Sox10, and Sox2. n = 3 batches. Data are represented as mean ± SD. RDF: SD rat skin fibroblast; MSC: SD rat bone marrow mesenchymal stem cell; DT: DRG-SGCs cultured by trypsin digestion approach; DE: DRG-SGCs cultured by explants method
Furthermore, we showed that the cultured DRG-SGCs highly expressed NCC markers Nestin, Sox2, Sox10 and p75 (Fig. 1b-g, and Fig. S2), suggesting that these DRG-SGCs we obtained have the characteristics of stem cells [12, 21].
DRG-SGCs Differentiate Into Sensory Neurons
There is evidence showing that after peripheral nerve injury, SGCs around DRG neurons can differentiate into sensory neurons [12]. In the present study, we found that the cultured DRG-SGCs did not express sensory neuron markers Ngn1 (Neurogenin1), Ngn2 (Neurogenin2), Brn3a, Isl1, TrkA, TrkB, and TrkC (Fig. S3). We then treated these cells with CHIR99021, SU5402 and RO4929097 to activate Wnt pathway, and inhibit FGF and Notch pathway, a protocol that was reported to promote the production of sensory neurons [28, 31,32,33,34,35,36]. After 7 d, qRT-PCR results showed an increased expression of Brn3a, Runx3, TrkA, TrkB, and TrkC, the sensory neuron-related genes. However, immunofluorescence staining revealed the small proportion of neurons, as indicated by Tuj1-positive cells (Fig. 2).
CRS induces neuron differentiation. a After 7 d of differentiation, there was no significant difference in cell morphology between the groups. Control, continued DRG-SGCs medium culture; NDM, medium changed to neuron differentiation medium after passaged for 24 h; CRS, CHIR99021 (C), RO4929097 (R), SU5402 (S) were added to NDM. b Immunofluorescence results shows that a small number of Tuj1 positive cells were detected in the Control, NDM, and CRS. c The percentage of Tuj1+, Map2+ neuronal cells in total cells. n = 5 batches. d The RT-PCR results showed that the expression of sensory neuron genes was up-regulated after 7 d differentiation. Scale bars, 250 µm. n = 3 batches. Data are represented as mean ± SD
We then added the histone deacetylase inhibitor VPA to the medium and observed that the cell morphology changed significantly: cells had slender protrusions with good refractive properties after 3 d. On the 7th day, the number of Tuj1 and Map2 positive cells increased significantly (Fig. 3a-c), indicating that VPA can improve the neural differentiation efficiency of these cells. More importantly, cells were labeled with mature neuron marker NeuN after 21 d of differentiation (Fig. 3d), and these neuron-like cells survived in the normal culture medium (Fig. 3d, e, and Fig. S4). Furthermore, we found that after 3 d of differentiation, almost all Tuj1-positive cells expressed GS (Fig. S5), suggesting that these neurons we obtained were derived from SGCs.
VPA promotes CRS-induced neuronal differentiation efficiency. a-c After 7 d of VCRS treatment, the number of Tuj1 and Map2 positive cells increased significantly. a Control, continued DRG-SGCs medium culture; b VPA was added to the differentiation medium containing CRS; c the percentage of Tuj1+, Map2+ neuronal cells in total cells. n = 5 batches. d, e The results of immunofluorescence after 21 and 28 d of induction show that cells were Tuj1, Map2, and NeuN positive. f Quantitative analyses of sensory neuron related genes. n = 3 batches. Data are represented as mean ± SD. a, b, d Scale bars, 250 µm; e Scale bar, 25 µm
Neurons can be divided into motor neurons, intermediate neurons and sensory neurons according to their functions. In order to determine the types of neurons obtained, we used qRT-PCR to detect the expression of genes related to the three kinds of neurons at 3 d, 7 d, 14 d, 21 d and 28 d after differentiation. Results showed that the expression of sensory neuron-related genes Brn3a, Ngn1 and Isl1 were up-regulated during differentiation (Fig. 3f). In contrast, there was no significant change in the expression of motor neuron-related transcription factors (Lhx3 and Hb9) or intermediate neuron-related transcription factors (Prdm13 and SCL6A) (Fig. S6). These data reveal that DRG-SGCs differentiates into sensory neurons under the action of histone deacetylase inhibitor VPA, Wnt pathway activator CHIR99021, Notch pathway inhibitor RO4929097 and FGF pathway inhibitor SU5402.
DRG-SGCs Differentiate Into Functional Nociceptive Neurons
According to the types of stimuli afferent to the central nervous system, sensory neurons can be divided into three types: nociceptive neurons that respond to pain or itching, mechanical sensory neurons that respond to mechanical stimuli, and proprioceptive neurons that respond to limb and muscle movement, which selectively express TrkA, TrkB, and TrkC [37, 38]. Our results showed that the sensory neurons we obtained highly expressed TrkA, Trpv1, Synapsin, SP, sodium channel SCN9A, SCN10A, and TrpA1 (Fig. 4a-d, g) in response to harmful stimuli, which are consistent with the characteristics of nociceptive sensory neurons reported [3, 28, 29, 39,40,41], implying that we get nociceptive sensory neurons. Indeed, in the process of obtaining sensory neurons, there was no change in the expression of mechanical sensory neuron-related genes TrkB and Shox2 (Fig. 4a, e). Although TrkC expression was up-regulated, the expression of Runx3 and Etv1, which are proprioceptive neuron-specific transcription factors [37, 42], was down-regulated in the late stage of differentiation (Fig. 4a, e, f), further confirming that DRG-SGCs differentiate into nociceptive sensory neurons.
Transformation of DRG-SGCs into nociceptive sensory neurons. a The expression of the sensory neuron-associated genes TrkA and TrkC was continuously upregulated during the transformation of DRG-SGCs into neurons. However, there was no change in TrkB expression. b The expression of the sodium channels SCN9A and SCN10A was continuously upregulated during the conversion of DRG-SGCs into neurons. c After 28 d of induction, the expression of the nociceptive neurons related genes SP, Trpv1, and TrpA1 were detected. Control refers to undifferentiated cells. d Immunofluorescence showed that Tuj1+ cells were TrkA+ and Synapsin+. e The expression of Runx1, a specific sensory neuron transcription factor, was downregulated, and the expression of Shox2 was unchanged. The expression of Runx3 was upregulated during VCRS treatment, but it was downregulated after switching to neuronal maturation medium. f The expression of Etv1 was downregulated in the late stage of differentiation. g Trpv1 expression was detected by immunofluorescence staining after 28 d of induction. a, b, e, f The undifferentiated cells were used as controls at the corresponding time points. n = 3 batches. Data are represented as mean ± SD. d, g Scale bars, 25 µm
By performing a whole-cell patch-clamp recording of 11 cells with neuronal morphology, we found that the resting membrane potential was between − 39 and − 53 mV, with an average of -45.6 mV (SEM = 4.051324, n = 11), and that the cells could generate action potential (Fig. 5a). Using a current-voltage protocol, voltage-gated Na+ currents were observed in all cells with neuronal morphology (n = 15, 361.67 ± 215.14 pA) (Fig. 5b). These data indicate that the neurons transformed by DRG-SGCs have the same electrophysiological characteristics as mature neurons.
Functional characterization of the induced nociceptive neurons. a, b After 28 d of induction, neuron-specific action potentials (a) and Na+ currents (b) were detected. c Representative calcium responses for 10 µM capsaicin. Calcium transients were measured using Fluo-4 AM. Calcium responses were calculated as the change in fluorescence (∆F) over the initial fluorescence (F0). d ∆F/F0 intensity plot showing the response of individual cells to KCl and capsaicin. e Calcium flux images of the induced nociceptive neurons in response to the addition of 25 mM KCl and 10 µM capsaicin. f-h Calcium flux images of the induced nociceptive neurons in response to the addition of 10 µM capsaicin. To confirm that capsaicin was indeed activating the Trpv1 vanilloid receptor, we carried out calcium flux analysis with the selective Trpv1 antagonist AMG9810. Pretreatment with AMG9810 significantly decreased calcium flux response induced by capsaicin (f, g); withdrawal of Trpv1 antagonist AMG9810 led to rapid calcium transients in response to capsaicin (h). n = 3 batches. e Scale bars, 50 µm; f-h Scale bars, 100 µm
We further evaluated calcium flux of the nociceptive neurons in response to 10 µM capsaicin, known to activate a subset of nociceptive neurons through binding to the Trpv1 vanilloid receptor [28, 43], and observed that ~ 12% of the cells exhibited rapid calcium transients in response to capsaicin (Fig. 5c-e). To confirm that capsaicin was indeed activating the Trpv1 vanilloid receptor, we carried out calcium flux with the selective Trpv1 antagonist AMG9810 and observed that pretreatment with AMG9810 significantly decreased calcium flux response induced by capsaicin (Fig. 5f, g, Movie S1, 2); withdrawal of Trpv1 antagonist AMG9810 led to rapid calcium transients in response to capsaicin (Fig. 5h, Movie S3), demonstrating that these cells have the functional properties of nociceptive neurons.
(MP4 24529 kb)
(MP4 24367 kb)
(MP4 24813 kb)
VPA Promotes DRG-SGCs Differentiation of Sensory Neurons, While Wnt, FGF and Notch Signaling Pathways are Involved in the Formation of Specific Sensory Neurons
In the process of DRG-SGCs differentiation into nociceptive sensory neurons, we detected the expression of histone deacetylase HDAC1 by immunofluorescence staining, and found that HDAC1 expression was continuously down-regulated (Fig. S7). Through qRT-PCR detection, it was found that the expression of the target gene Nedd9 downstream of the Wnt signaling pathway was continuously up-regulated, while the expression of the target gene Hes5 downstream of the Notch signaling pathway and the cMyc downstream of the FGF signaling pathway were continuously down-regulated (Fig. S7). The above results indicate that the acetylation state, continuous activation of the Wnt signaling pathway, and continuous inhibition of the FGF and Notch signaling pathways help DRG-SGCs transform into nociceptive sensory neurons.
Next, we investigated the roles of VPA, CHIR99021, RO4929097 and SU5402 in the differentiation of DRG-SGCs into nociceptive sensory neurons. The above results showed that treatment with CHIR99021, RO4929097, and SU5402 increased the expression of Brn3a, Runx3, TrkA, TrkB, and TrkC (Fig. 2d); VPA addition resulted in the up-regulation of Ngn2 expression (Fig. 2d, and Fig. S8). It has been reported that Ngn2 alone or in combination with Brn3a can transform astrocytes or fibroblasts into neurons or sensory neurons [29, 44, 45], suggesting that VPA may improve the neural differentiation efficiency by up-regulating the expression of Ngn2. Further, qRT-PCR testing revealed that VPA treatment up-regulated the expression of early sensory neuron-related genes Brn3a, Isl1 and Ngn2, and of the genes related to nociceptive, mechanical and proprioceptive neurons such as Runx1, Runx3, TrkA, TrkB and TrkC (Fig. 6a). Immunofluorescence staining showed that the rate of Tuj1 positive cells reached more than 85% (Fig. 6b-e), indicating that VPA can promote the differentiation of DRG-SGCs into multiple types of sensory neurons.
Effects of VPA, CHIR99021, RO4929097 and SU5402 in the differentiation of DRG-SGCs into nociceptive neurons. a After VPA treatment for 7 d, RT-PCR showed that the expression of related genes in sensory neurons was up-regulated. b-d After passage for 24 h, the cells were treated with V, VC, VR, VS, VCR, VCS, VRS and VCRS for 7 d. immunofluorescence results showed that all groups were Tuj1 positive. Scale bars, 250 µm. V, VC, VR, VS, VRS, VCR, VCS, and VCRS refer to VPA, CHIR99021, RO4929097, and SU5402 added separately or in combination in neuron differentiation culture (NDM). e The percentage of Tuj1+ neuronal cells in total cells. n = 5 batches. f, g In the presence of VPA, CHIR99021, RO4929097 and SU5402 regulated specific sensory neuron transcription factors Runx1. *P < 0.05; **P < 0.01; ***P < 0.001; unpaired t-test. h After 7 d of VCRS treatment, removal of S, R or the simultaneous withdrawal of SR lead to the down-regulation of neuron-related gene expression, especially the sodium channels SCN9A, n = 3 batches. Data are represented as mean ± SD. ***P < 0.001; one-way ANOVA followed with Dunnett’s multiple comparison test
In the presence of VPA, we examined by qRT-PCR the effects of CHIR99021, RO4929097, and SU5402 on the expression of Runx1, which acts as a transcriptional activator and is necessary for the differentiation of nociceptive sensory neurons [46,47,48,49]. The results showed that Runx1 expression was down-regulated (Fig. 6f, g) in all groups containing CHIR99021, while no changes were observed upon treatment with RO4929097 or SU5402 (Fig. 6f, g), suggesting that CHIR99021, an activator of Wnt signaling pathway, regulates Runx1 expression, and that inhibiting Notch or FGF signaling had no effect on the expression of Runx1 in the presence of VPA. These results indicate that Wnt signaling pathway plays an important role in the formation of nociceptive sensory neurons. Although inhibition of Notch and FGF signaling pathways had no effect on Runx1 expression, withdrawal of SU5402 or RO4929097 during sensory neuron differentiation led to the down-regulation of neuron-related genes NeuN (Fig. 6c, e, h), implying that Notch and FGF pathways play a key role in the differentiation of DRG-SGCs into sensory neurons.
Subsequently, we investigated the effects of CHIR99021, RO4929097, and SU5402 on the expression of proprioceptive neuron-specific transcription factor Runx3 [50, 51]. It was found that on the basis of histone deacetylase inhibitor VPA, the inhibition of FGF signaling caused an increase in Runx3 expression, the inhibition of Notch signal pathway caused a decrease in Runx3 expression, while the Wnt signaling pathway activator CHIR99021 had no effect on Runx3 expression (Fig. 7), suggesting that Notch and FGF signaling pathways are involved in the formation of proprioceptive neurons.
The regulatory effect of CHIR99021, RO4929097 and SU5402 on Runx3 and Shox2 in the presence of VPA. a, c, d, g In the presence of VPA, CHIR99021 had no effect on the expression of Runx3. (a-c, e) when SU5402 was added to the medium, Runx3 expression was up-regulated. (a-f) the simultaneous addition of RO4929097 and SU5402 increased the expression of Shox2. *P < 0.05; **P < 0.01; ***P < 0.001; unpaired t-test. n = 3 batches. Data are represented as mean ± SD
Finally, we analyzed the effects of CHIR99021, RO4929097, and SU5402 on the expression of Shox2, the conditional deletion of which in NCCs was reported to cause a 60∼65% reduction in the number of mechanical sensory neurons [52]. Our results revealed that the addition of CHIR99021 to the medium had no effect on Shox2 expression (Fig. 7a, c, d, g), and the simultaneous addition of RO4929097 and SU5402 increased the expression of Shox2 (Fig. 7a-f), suggesting that Notch and FGF signaling pathways participate in mechanical sensory neuron formation.
Discussion
Damage or loss of sensory neurons in DRG causes somatosensory dysfunction, which seriously affects patients’ quality of life. Therefore, it is hopeful to reshape the somatosensory function by inducing DRG cells into sensory neurons. Studies have found that after nerve injury, SGCs around DRG sensory neurons begin to express Nestin and Sox2 [9, 11, 22, 23] and can differentiate into sensory neurons and glial cells [12, 30]. A group of cells expressing Nestin and p75 will migrate from adult rat DRG cultured in vitro, which can differentiate into glial cells and neurons in the presence of serum [21], suggesting that DRG-SGCs may have differentiation plasticity. In this study, we found that DRG-SGCs isolated and cultured from newborn rat DRGs highly express Nestin, p75, Sox10, Sox2, the NCC markers, and can differentiate into nociceptive sensory neurons. Using the same induced differentiation conditions, SGCs derived from trigeminal ganglia and adult rat DRG can be transformed into neurons (data not shown). These data indicate that SGCs have the characteristics of stem cells.
DRG sensory neurons originate from NCCs, a transient population in the embryo that migrate to specific locations to form brain ganglia, DRG, autonomic ganglia, and these NCCs become progressively restricted in developmental potential and rapidly lose their multipotency, and differentiates into different types of cells, including SGCs, Schwann cells, melanocytes, and sensory neurons [53,54,55,56,57]. Under the action of multiple signaling pathways such as Wnt, FGF, Notch, NCCs express early sensory neuron-specific transcription factors Ngn1 and Ngn2, and become sensory neurons, and subsequently differentiate into nociceptive sensory neurons, mechanical sensory neurons, and proprioceptive neurons through expressing specific sensory neuronal transcription factors Runx1, Shox2 and Runx3. Consistent with previous results [9, 11, 22, 23], we clearly show that SGCs we obtained express Nestin, Sox10 and Sox2, the NCC markers. Moreover, these cells can give rise to nociceptive sensory neurons. Immunofluorescence staining showed that the nociceptive neurons we obtained expressed synapsin, which has also been reported previously in the nociceptors derived from mouse embryonic fibroblasts and human induced pluripotent stem cells [29, 41], suggesting that synaptogenesis occurred in these neurons. The nociceptive neurons we obtained also expressed Trpv1. Using calcium-imaging assay, we observed that the cells exhibited rapid calcium transients in response to capsaicin, known to activate a subset of nociceptive neurons through binding to the Trpv1 vanilloid receptor [28, 43]. To confirm that capsaicin was indeed activating the Trpv1 vanilloid receptor, we carried out calcium flux analysis with the selective Trpv1 antagonist AMG9810. Results showed that pretreatment with AMG9810 significantly decreased calcium flux response induced by capsaicin and that withdrawal of Trpv1 antagonist AMG9810 resulted in the rapid calcium transients in response to capsaicin, confirming that the cells have the functional properties of nociceptive sensory neurons. In addition, we found that after 3 days of differentiation, almost all Tuj1-positive cells expressed GS, suggesting that the neurons we obtained were derived from SGCs.
Our results show that in the presence of the histone deacetylase inhibitor VPA, Wnt pathway activator CHIR99021, FGF pathway inhibitor SU5402, and Notch pathway inhibitor RO4929097, DRG-SGCs differentiate into nociceptive sensory neurons, during which VPA increases the efficiency of sensory neuron differentiation, whereas Wnt signaling via Runx1, the specific transcription factor necessary for the differentiation of nociceptive sensory neurons [46,47,48,49], FGF and Notch signals via SCN9A, a functionally-related gene in nociceptive sensory neurons [58,59,60,61], regulate the formation of these cells from DRG-SGCs. We also found that the addition of SU5402 up-regulated the expression of Runx3, and the addition of RO4929097 and SU5402 up-regulated the expression of Shox2, suggesting that FGF and Notch signal pathways might be involved in the formation of proprioceptive and mechanical sensory neurons. Further studies are required to explore the conditions for the differentiation of these cells.
In addition, our results show that after 14 d of DRG-SGCs differentiation, the expression of proprioceptive neuron-related genes decreases, suggesting that the time required for proprioceptive neuron differentiation is relatively shorter than that of nociceptive neurons. Similar to our findings, during the conversion of human pluripotent stem cells into sensory neurons, prolonged activation of the Wnt pathway leads to down-regulation of proprioceptive and mechanical sensory neuron-related genes [28], indicating that the time required for different types of sensory neurons may be different.
Reversible acetylation and deacetylation of histone lysine residues is one of the most widely studied epigenetic regulation. Histone acetyltransferase adds acetyl groups to lysine, resulting in more open chromatin. Histone deacetylases (HDAC) remove acetyl groups, leading to chromatin concentration, which is associated with gene silencing [62, 63]. Studies have shown that histone deacetylase inhibitor VPA can promote neural progenitor cells to differentiate into neurons [64,65,66]. Our results show that histone deacetylase inhibition can significantly increase the expression of sensory neuron-related genes in DRG-SGCs, promoting the differentiation of DRG-SGCs into sensory neurons. There is evidence showing that HDAC inhibitors can prevents and treats multiple symptoms of cisplatin-induced peripheral neuropathy, such as spontaneous pain and numbness, and reverse loss of intra-epidermal nerve fibers (IENFs) [67], suggesting that histone acetylation plays an important role in treating peripheral neuropathy. In addition to restoring mitochondrial function in peripheral nerve and DRG [67], our results provide another potential explanation for HDAC inhibitors to effectively reverse IENF loss and numbness by the production of new nociceptive sensory neurons from DRG-SGCs, a new theoretical basis for clinical treatment of sensory neuron-related diseases.
Peripheral neurological diseases or injuries can lead to reduction of dorsal root ganglion neurons, disordered connections between neurons, formation of traumatic neuromas, leading to the abnormal sensory conduction and neuropathic pain. Neuropathic pain caused by the nervous system injury is a kind of chronic pain which is difficult to cure. At present, the mechanism of neuropathic pain is not fully understood. Intrathecal use of Wnt/β-catenin signaling inhibitor XAV939 can effectively reduce neuropathic pain [68]. In a rat model of the sciatic nerves chronic constriction injury (CCI) and a mouse model of bone cancer pain induced by tumor cell implantation, the expression of Wnt3a in DRG and spinal cord increases before pain and continues to be expressed during the onset of pain, and Wnt inhibitors can effectively alleviate mechanical allodynia and thermal hyperalgesia in neuropathic pain models, suggesting that Wnt plays a key role in inducing neuropathic pain [69]. Intrathecal injection of Wnt/β-catenin inhibitors IWR-1-endo and TCF4 siRNA reduced CCI-induced mechanical allodynia and thermal hyperalgesia, suggesting that TCF4 in DRG and spinal cord may be involved in the maintenance of CCI-induced neuropathic pain through Wnt/β-catenin signaling [70]. The above results indicate that the Wnt signaling pathway plays an important role in the generation of neuropathic pain. Our results show that during the differentiation of DRG-SGCs into nociceptive sensory neurons, the Wnt signaling regulates the expression of transcription factor Runx1, which is necessary for the differentiation of nociceptive sensory neurons. The runt domain protein Runx1 is initially expressed in most nociceptive sensory neurons during embryonic development, and then undergoes dynamic changes during perinatal and postnatal development [46, 71]. Persistent expression of Runx1 is required for proper development of non-peptidergic nociceptive sensory neurons [46,47,48]. However, the consequence of Runx1 persistence is the loss of several peptidergic markers, including CGRP, TRPV1, and TRPA. As a result, Runx1 inactivation is essential in the development of peptidergic nociceptive sensory neurons [49, 71]. These results demonstrate the importance of Wnt signaling pathway in the formation of nociceptive sensory neurons. Therefore, we speculate that after peripheral nerve injury, the erroneous production of some nociceptive neurons may be one of the causes of neuropathic pain.
Conclusions
Our results in the present study show that DRG-SGCs highly express NCC markers Nestin, p75, Sox10, and Sox2, and differentiate into nociceptive sensory neurons in the presence of histone deacetylase inhibitor VPA, Wnt pathway activator CHIR99021, Notch pathway inhibitor RO4929097, and FGF pathway inhibitor SU5402. The nociceptive sensory neurons express multiple functionally-related genes (SCN9A, SCN10A, SP, Trpv1, and TrpA1) and are able to generate action potentials and voltage-gated Na+ currents. Moreover, these cells exhibit rapid calcium transients in response to capsaicin through binding to the Trpv1 vanilloid receptor, confirming that the DRG-SGC-derived cells are nociceptive sensory neurons. Further, we show that Wnt signaling promotes the differentiation of DRG-SGCs into nociceptive sensory neurons by regulating the expression of specific transcription factor Runx1, while Notch and FGF signaling pathways are involved in the expression of SCN9A. These results demonstrate that DRG-SGCs have stem cell characteristics and can efficiently differentiate into functional nociceptive sensory neurons, shedding light on the clinical treatment of sensory neuron-related diseases such as CIPA or chronic pain.
Data Availability
All data generated or analysed during this study are included in this published article.
References
Basbaum, A. I., Bautista, D. M., Scherrer, G., & Julius, D. (2009). Cellular and molecular mechanisms of pain. Cell, 139(2), 267–284. https://doi.org/10.1016/j.cell.2009.09.028.
Meyer, K., & Kaspar, B. K. (2014). Making sense of pain: are pluripotent stem cell-derived sensory neurons a new tool for studying pain mechanisms? Molecular Therapy, 22(8), 1403–1405. https://doi.org/10.1038/mt.2014.123.
Woolf, C. J., & Ma, Q. (2007). Nociceptors–noxious stimulus detectors. Neuron, 55(3), 353–364. https://doi.org/10.1016/j.neuron.2007.07.016.
Jackson, T., Thomas, S., Stabile, V., Han, X., Shotwell, M., & McQueen, K. (2015). Prevalence of chronic pain in low-income and middle-income countries: a systematic review and meta-analysis. Lancet, 385(Suppl 2), S10. https://doi.org/10.1016/S0140-6736(15)60805-4.
Indo, Y. (2018). NGF-dependent neurons and neurobiology of emotions and feelings: Lessons from congenital insensitivity to pain with anhidrosis. Neuroscience and Biobehavioral Reviews, 87, 1–16. https://doi.org/10.1016/j.neubiorev.2018.01.013.
Indo, Y. (2012). Nerve growth factor and the physiology of pain: lessons from congenital insensitivity to pain with anhidrosis. Clinical Genetics, 82(4), 341–350. https://doi.org/10.1111/j.1399-0004.2012.01943.x.
Kim, Y. S., Anderson, M., Park, K., Zheng, Q., Agarwal, A., Gong, C., Saijilafu, Young, L., He, S., LaVinka, P. C., Zhou, F., Bergles, D., Hanani, M., Guan, Y., Spray, D. C., & Dong, X. (2016). Coupled activation of primary sensory neurons contributes to chronic pain. Neuron, 91(5), 1085–1096. https://doi.org/10.1016/j.neuron.2016.07.044.
Sikandar, S., Minett, M. S., Millet, Q., Santana-Varela, S., Lau, J., Wood, J. N., & Zhao, J. (2018). Brain-derived neurotrophic factor derived from sensory neurons plays a critical role in chronic pain. Brain, 141(4), 1028–1039. https://doi.org/10.1093/brain/awy009.
Muratori, L., Ronchi, G., Raimondo, S., Geuna, S., Giacobini-Robecchi, M. G., & Fornaro, M. (2015). Generation of new neurons in dorsal root Ganglia in adult rats after peripheral nerve crush injury. Neural Plasticity, 2015, 860546. https://doi.org/10.1155/2015/860546.
Hu, P., & McLachlan, E. M. (2003). Selective reactions of cutaneous and muscle afferent neurons to peripheral nerve transection in rats. The Journal of Neuroscience, 23(33), 10559–10567. https://doi.org/10.1155/2015/860546.
Gallaher, Z. R., Johnston, S. T., & Czaja, K. (2014). Neural proliferation in the dorsal root ganglia of the adult rat following capsaicin-induced neuronal death. The Journal of Comparative Neurology, 522(14), 3295–3307. https://doi.org/10.1002/cne.23598.
Zhang, L., Xie, R., Yang, J., Zhao, Y., Qi, C., Bian, G., Wang, M., Shan, J., Wang, C., Wang, D., Luo, C., Wang, Y., & Wu, S. (2019). Chronic pain induces nociceptive neurogenesis in dorsal root ganglia from Sox2-positive satellite cells. Glia, 67(6), 1062–1075. https://doi.org/10.1002/glia.23588.
Hanani, M. (2005). Satellite glial cells in sensory ganglia: from form to function. Brain Research. Brain Research Reviews, 48(3), 457–476. https://doi.org/10.1016/j.brainresrev.2004.09.001.
Pannese, E. (2010). The structure of the perineuronal sheath of satellite glial cells (SGCs) in sensory ganglia. Neuron Glia Biology, 6(1), 3–10. https://doi.org/10.1017/S1740925X10000037.
Spray, D. C., Iglesias, R., Shraer, N., Suadicani, S. O., Belzer, V., Hanstein, R., & Hanani, M. (2019). Gap junction mediated signaling between satellite glia and neurons in trigeminal ganglia. Glia, 67(5), 791–801. https://doi.org/10.1002/glia.23554.
Belzer, V., & Hanani, M. (2019). Nitric oxide as a messenger between neurons and satellite glial cells in dorsal root ganglia. Glia, 67(7), 1296–1307. https://doi.org/10.1002/glia.23603.
Zhang, X., Chen, Y., Wang, C., & Huang, L. Y. (2007). Neuronal somatic ATP release triggers neuron-satellite glial cell communication in dorsal root ganglia. Proceedings of the National Academy of Sciences of the United States of America, 104(23), 9864–9869. https://doi.org/10.1073/pnas.0611048104.
Huang, L. Y., Gu, Y., & Chen, Y. (2013). Communication between neuronal somata and satellite glial cells in sensory ganglia. Glia, 61(10), 1571–1581. https://doi.org/10.1002/glia.22541.
Verkhratsky, A., Parpura, V., & Rodriguez, J. J. (2011). Where the thoughts dwell: the physiology of neuronal-glial “diffuse neural net". Brain Research Reviews, 66(1–2), 133–151. https://doi.org/10.1016/j.brainresrev.2010.05.002.
Lin, G., Bella, A. J., Lue, T. F., & Lin, C. S. (2006). Brain-derived neurotrophic factor (BDNF) acts primarily via the JAK/STAT pathway to promote neurite growth in the major pelvic ganglion of the rat: part 2. The Journal of Sexual Medicine, 3(5), 821–829. https://doi.org/10.1111/j.1743-6109.2006.00292.x.
Li, H. Y., Say, E. H., & Zhou, X. F. (2007). Isolation and characterization of neural crest progenitors from adult dorsal root ganglia. Stem Cells, 25(8), 2053–2065. https://doi.org/10.1634/stemcells.2007-0080.
Koike, T., Wakabayashi, T., Mori, T., Takamori, Y., Hirahara, Y., & Yamada, H. (2014). Sox2 in the adult rat sensory nervous system. Histochemistry and Cell Biology, 141(3), 301–309. https://doi.org/10.1007/s00418-013-1158-x.
Krishnan, A., Bhavanam, S., & Zochodne, D. (2018). An intimate role for adult dorsal root ganglia resident cycling cells in the generation of local macrophages and satellite glial cells. Journal of Neuropathology and Experimental Neurology, 77(10), 929–941. https://doi.org/10.1093/jnen/nly072.
Zhang, H., Vutskits, L., Pepper, M. S., & Kiss, J. Z. (2003). VEGF is a chemoattractant for FGF-2-stimulated neural progenitors. The Journal of Cell Biology, 163(6), 1375–1384. https://doi.org/10.1083/jcb.200308040.
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., & Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology, 8(2), R19. https://doi.org/10.1186/gb-2007-8-2-r19.
Zhang, Y., Jiang, D., Zhang, Y., Jiang, X., Wang, F., & Tao, J. (2012). Neuromedin U type 1 receptor stimulation of A-type K + current requires the betagamma subunits of Go protein, protein kinase A, and extracellular signal-regulated kinase 1/2 (ERK1/2) in sensory neurons. The Journal of Biological Chemistry, 287(22), 18562–18572. https://doi.org/10.1074/jbc.M111.322271.
Mitric, M., Seewald, A., Moschetti, G., Sacerdote, P., Ferraguti, F., Kummer, K. K., & Kress, M. (2019). Layer- and subregion-specific electrophysiological and morphological changes of the medial prefrontal cortex in a mouse model of neuropathic pain. Scientific Reports, 9(1), 9479. https://doi.org/10.1038/s41598-019-45677-z.
Chambers, S. M., Qi, Y., Mica, Y., Lee, G., Zhang, X. J., Niu, L., Bilsland, J., Cao, L., Stevens, E., Whiting, P., Shi, S. H., & Studer, L. (2012). Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nature Biotechnology, 30(7), 715–720. https://doi.org/10.1038/nbt.2249.
Blanchard, J. W., Eade, K. T., Szucs, A., Lo Sardo, V., Tsunemoto, R. K., Williams, D., Sanna, P. P., & Baldwin, K. K. (2015). Selective conversion of fibroblasts into peripheral sensory neurons. Nature Neuroscience, 18(1), 25–35. https://doi.org/10.1038/nn.3887.
George, D., Ahrens, P., & Lambert, S. (2018). Satellite glial cells represent a population of developmentally arrested Schwann cells. Glia, 66(7), 1496–1506. https://doi.org/10.1002/glia.23320.
Hari, L., Brault, V., Kleber, M., Lee, H. Y., Ille, F., Leimeroth, R., Paratore, C., Suter, U., Kemler, R., & Sommer, L. (2002). Lineage-specific requirements of beta-catenin in neural crest development. The Journal of Cell Biology, 159(5), 867–880. https://doi.org/10.1083/jcb.200209039.
Lee, H. Y., Kleber, M., Hari, L., Brault, V., Suter, U., Taketo, M. M., Kemler, R., & Sommer, L. (2004). Instructive role of Wnt/beta-catenin in sensory fate specification in neural crest stem cells. Science, 303(5660), 1020–1023. https://doi.org/10.1126/science.1091611.
Murphy, M., Reid, K., Ford, M., Furness, J. B., & Bartlett, P. F. (1994). FGF2 regulates proliferation of neural crest cells, with subsequent neuronal differentiation regulated by LIF or related factors. Development, 120(12), 3519–3528.
Israsena, N., Hu, M., Fu, W., Kan, L., & Kessler, J. A. (2004). The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Developmental Biology, 268(1), 220–231. https://doi.org/10.1016/j.ydbio.2003.12.024.
Ota, M., & Ito, K. (2006). BMP and FGF-2 regulate neurogenin-2 expression and the differentiation of sensory neurons and glia. Developmental Dynamics, 235(3), 646–655. https://doi.org/10.1002/dvdy.20673.
Hu, Z. L., Shi, M., Huang, Y., Zheng, M. H., Pei, Z., Chen, J. Y., Han, H., & Ding, Y. Q. (2011). The role of the transcription factor Rbpj in the development of dorsal root ganglia. Neural Development, 6, 14. https://doi.org/10.1186/1749-8104-6-14.
Marmigere, F., & Carroll, P. (2014). Neurotrophin signalling and transcription programmes interactions in the development of somatosensory neurons. Handbook of Experimental Pharmacology, 220, 329–353. https://doi.org/10.1007/978-3-642-45106-5_13.
Lallemend, F., & Ernfors, P. (2012). Molecular interactions underlying the specification of sensory neurons. Trends in Neurosciences, 35(6), 373–381. https://doi.org/10.1016/j.tins.2012.03.006.
Renganathan, M., Cummins, T. R., & Waxman, S. G. (2001). Contribution of Na(v)1.8 sodium channels to action potential electrogenesis in DRG neurons. Journal of Neurophysiology, 86(2), 629–640. https://doi.org/10.1152/jn.2001.86.2.629.
Liu, Y., & Ma, Q. (2011). Generation of somatic sensory neuron diversity and implications on sensory coding. Current Opinion in Neurobiology, 21(1), 52–60. https://doi.org/10.1016/j.conb.2010.09.003.
Schoepf, C. L., Zeidler, M., Spiecker, L., Kern, G., Lechner, J., Kummer, K. K., & Kress, M. (2020). Selected ionotropic receptors and voltage-gated ion channels: more functional competence for human induced pluripotent stem cell (iPSC)-derived nociceptors. Brain Sciences, 10(6). https://doi.org/10.3390/brainsci10060344.
Sun, Y., Dykes, I. M., Liang, X., Eng, S. R., Evans, S. M., & Turner, E. E. (2008). A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nature Neuroscience, 11(11), 1283–1293. https://doi.org/10.1038/nn.2209.
Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D., & Julius, D. (1997). The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature, 389(6653), 816–824. https://doi.org/10.1038/39807.
Masserdotti, G., Gillotin, S., Sutor, B., Drechsel, D., Irmler, M., Jorgensen, H. F., Sass, S., Theis, F. J., Beckers, J., Berninger, B., Guillemot, F., & Gotz, M. (2015). Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell, 17(1), 74–88. https://doi.org/10.1016/j.stem.2015.05.014.
Liu, M. L., Zang, T., Zou, Y., Chang, J. C., Gibson, J. R., Huber, K. M., & Zhang, C. L. (2013). Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nature Communications, 4, 2183. https://doi.org/10.1038/ncomms3183.
Chen, C. L., Broom, D. C., Liu, Y., de Nooij, J. C., Li, Z., Cen, C., Samad, O. A., Jessell, T. M., Woolf, C. J., & Ma, Q. (2006). Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron, 49(3), 365–377. https://doi.org/10.1016/j.neuron.2005.10.036.
Yoshikawa, M., Senzaki, K., Yokomizo, T., Takahashi, S., Ozaki, S., & Shiga, T. (2007). Runx1 selectively regulates cell fate specification and axonal projections of dorsal root ganglion neurons. Developmental Biology, 303(2), 663–674. https://doi.org/10.1016/j.ydbio.2006.12.007.
Marmigere, F., Montelius, A., Wegner, M., Groner, Y., Reichardt, L. F., & Ernfors, P. (2006). The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nature Neuroscience, 9(2), 180–187. https://doi.org/10.1038/nn1631.
Gascon, E., Gaillard, S., Malapert, P., Liu, Y., Rodat-Despoix, L., Samokhvalov, I. M., ., Delmas, P., Helmbacher, F., Maina, F., & Moqrich, A. (2010). Hepatocyte growth factor-Met signaling is required for Runx1 extinction and peptidergic differentiation in primary nociceptive neurons. The Journal of Neuroscience, 30(37), 12414–12423. https://doi.org/10.1523/JNEUROSCI.3135-10.2010.
Levanon, D., Bettoun, D., Harris-Cerruti, C., Woolf, E., Negreanu, V., Eilam, R., Bernstein, Y., Goldenberg, D., Xiao, C., Fliegauf, M., Kremer, E., Otto, F., Brenner, O., Lev-Tov, A., & Groner, Y. (2002). The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. The EMBO Journal, 21(13), 3454–3463. https://doi.org/10.1093/emboj/cdf370.
Kramer, I., Sigrist, M., de Nooij, J. C., Taniuchi, I., Jessell, T. M., & Arber, S. (2006). A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron, 49(3), 379–393. https://doi.org/10.1016/j.neuron.2006.01.008.
Abdo, H., Li, L., Lallemend, F., Bachy, I., Xu, X. J., Rice, F. L., & Ernfors, P. (2011). Dependence on the transcription factor Shox2 for specification of sensory neurons conveying discriminative touch. The European Journal of Neuroscience, 34(10), 1529–1541. https://doi.org/10.1111/j.1460-9568.2011.07883.x.
Pavan, W. J., & Raible, D. W. (2012). Specification of neural crest into sensory neuron and melanocyte lineages. Developmental Biology, 366(1), 55–63. https://doi.org/10.1016/j.ydbio.2012.02.038.
Marmigere, F., & Ernfors, P. (2007). Specification and connectivity of neuronal subtypes in the sensory lineage. Nature Reviews. Neuroscience, 8(2), 114–127. https://doi.org/10.1038/nrn2057.
Pla, P., & Monsoro-Burq, A. H. (2018). The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Developmental Biology, 444(1), S36–S46. https://doi.org/10.1016/j.ydbio.2018.05.018.
Vandamme, N., & Berx, G. (2019). From neural crest cells to melanocytes: cellular plasticity during development and beyond. Cellular and Molecular Life Sciences, 76(10), 1919–1934. https://doi.org/10.1007/s00018-019-03049-w.
Mehrotra, P., Tseropoulos, G., Bronner, M. E., & Andreadis, S. T. (2020). Adult tissue-derived neural crest-like stem cells: Sources, regulatory networks, and translational potential. Stem Cells Translational Medicine, 9(3), 328–341. https://doi.org/10.1002/sctm.19-0173.
McDermott, L. A., Weir, G. A., Themistocleous, A. C., Segerdahl, A. R., Blesneac, I., Baskozos, G., Clark, A. J., Millar, V., Peck, L. J., Ebner, D., Tracey, I., Serra, J., & Bennett, D. L. (2019). Defining the functional role of NaV1.7 in human nociception. Neuron, 101(5), 905-919 e908. https://doi.org/10.1016/j.neuron.2019.01.047.
Drenth, J. P., & Waxman, S. G. (2007). Mutations in sodium-channel gene SCN9A cause a spectrum of human genetic pain disorders. The Journal of Clinical Investigation, 117(12), 3603–3609. https://doi.org/10.1172/JCI33297.
Nahorski, M. S., Chen, Y. C., & Woods, C. G. (2015). New mendelian disorders of painlessness. Trends in Neurosciences, 38(11), 712–724. https://doi.org/10.1016/j.tins.2015.08.010.
Minett, M. S., Falk, S., Santana-Varela, S., Bogdanov, Y. D., Nassar, M. A., Heegaard, A. M., & Wood, J. N. (2014). Pain without nociceptors? Nav1.7-independent pain mechanisms. Cell Reports, 6(2), 301–312. https://doi.org/10.1016/j.celrep.2013.12.033.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693–705. https://doi.org/10.1016/j.cell.2007.02.005.
Hu, X. L., Wang, Y., & Shen, Q. (2012). Epigenetic control on cell fate choice in neural stem cells. Protein and Cell, 3(4), 278–290. https://doi.org/10.1007/s13238-012-2916-6.
Hsieh, J., Nakashima, K., Kuwabara, T., Mejia, E., & Gage, F. H. (2004). Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proceedings of the National Academy of Sciences of the United States of America, 101(47), 16659–16664. https://doi.org/10.1073/pnas.0407643101.
Zhang, X., He, X., Li, Q., Kong, X., Ou, Z., Zhang, L., Gong, Z., Long, D., Li, J., Zhang, M., Ji, W., Zhang, W., Xu, L., & Xuan, A. (2017). PI3K/AKT/mTOR signaling mediates valproic acid-induced neuronal differentiation of neural stem cells through epigenetic modifications. Stem Cell Reports, 8(5), 1256–1269. https://doi.org/10.1016/j.stemcr.2017.04.006.
Yu, I. T., Park, J. Y., Kim, S. H., Lee, J. S., Kim, Y. S., & Son, H. (2009). Valproic acid promotes neuronal differentiation by induction of proneural factors in association with H4 acetylation. Neuropharmacology, 56(2), 473–480. https://doi.org/10.1016/j.neuropharm.2008.09.019.
Krukowski, K., Ma, J., Golonzhka, O., Laumet, G. O., Gutti, T., van Duzer, J. H., Mazitschek, R., Jarpe, M. B., Heijnen, C. J., & Kavelaars, A. (2017). HDAC6 inhibition effectively reverses chemotherapy-induced peripheral neuropathy. Pain, 158(6), 1126–1137. https://doi.org/10.1097/j.pain.0000000000000893.
Yang, Q. O., Yang, W. J., Li, J., Liu, F. T., Yuan, H., & Ou Yang, Y. P. (2017). Ryk receptors on unmyelinated nerve fibers mediate excitatory synaptic transmission and CCL2 release during neuropathic pain induced by peripheral nerve injury. Molecular Pain, 13, 1744806917709372. https://doi.org/10.1177/1744806917709372.
Zhang, Y. K., Huang, Z. J., Liu, S., Liu, Y. P., Song, A. A., & Song, X. J. (2013). WNT signaling underlies the pathogenesis of neuropathic pain in rodents. The Journal of Clinical Investigation, 123(5), 2268–2286. https://doi.org/10.1172/JCI65364.
Xu, Z., Chen, Y., Yu, J., Yin, D., Liu, C., Chen, X., & Zhang, D. (2015). TCF4 mediates the maintenance of neuropathic pain through Wnt/beta-catenin signaling following peripheral nerve injury in rats. Journal of Molecular Neuroscience, 56(2), 397–408. https://doi.org/10.1007/s12031-015-0565-y.
Lopes, C., Liu, Z., Xu, Y., & Ma, Q. (2012). Tlx3 and Runx1 act in combination to coordinate the development of a cohort of nociceptors, thermoceptors, and pruriceptors. The Journal of Neuroscience, 32(28), 9706–9715. https://doi.org/10.1523/JNEUROSCI.1109-12.2012.
Funding
This work was supported by National Natural Science Foundation of China (Grant no. 31970705, 31771509, 31371407), the Priority Academic Program Development of Jiangsu Higher Education Institutions, Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_2523), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (NO. 2017PT31042), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (NO. 2018PT31048).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Dongyan Wang, Junhou Lu, Xiaojing Xu, Ye Yuan, and Yu Zhang. The first draft of the manuscript was written by Dongyan Wang, Junhou Lu, Xiaojing Xu, Ye Yuan, Huanhuan Chen, Jinming Liu, and Huanxiang Zhang and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethical Approval
All animal experimental protocols were approved by the guidelines of the Institutional Medical Experimental Animal Use Committee of Soochow University.
Consent to Participate
Not applicable.
Consent to Publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 19658 kb)
Rights and permissions
About this article
Cite this article
Wang, D., Lu, J., Xu, X. et al. Satellite Glial Cells Give Rise to Nociceptive Sensory Neurons. Stem Cell Rev and Rep 17, 999–1013 (2021). https://doi.org/10.1007/s12015-020-10102-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-020-10102-w