Abstract
Derivation of induced Pluripotent Stem Cells (iPSCs) by reprogramming somatic cells to a pluripotent state has revolutionized stem cell research. Ensuing this, various groups have used genetic and non-genetic approaches to generate iPSCs from numerous cell types. However, achieving a pluripotent state in most of the reprogramming studies is marred by serious limitations such as low reprogramming efficiency and slow kinetics. These limitations are mainly due to the presence of potent barriers that exist during reprogramming when a mature cell is coaxed to achieve a pluripotent state. Several studies have revealed that intrinsic factors such as non-optimal stoichiometry of reprogramming factors, specific signaling pathways, cellular senescence, pluripotency-inhibiting transcription factors and microRNAs act as a roadblock. In addition, the epigenetic state of somatic cells and specific epigenetic modifications that occur during reprogramming also remarkably impede the generation of iPSCs. In this review, we present a comprehensive overview of the barriers that inhibit reprogramming and the understanding of which will pave the way to develop safe strategies for efficient reprogramming.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
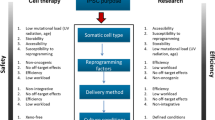
The groundbreaking discovery of induced Pluripotent Stem Cells (iPSCs) that resemble Embryonic Stem Cells (ESCs) has brought with it a wave of new possibilities for its suitability in various biomedical applications [1]. iPSCs have tremendous potential in developmental and disease biology, drug screening and toxicity testing, and personalized regenerative medicine [2,3,4,5]. To date, several groups have generated iPSCs by ectopic expression of reprogramming factors in numerous cell types isolated from healthy and diseased subjects using integrating and non-integrating approaches [3, 5,6,7,8,9,10,11,12]. Studies have also demonstrated that iPSCs can be derived successfully without the ectopic expression and integration of transgenes into the genome of target cells [2, 5, 8, 11]. However, the majority of these studies have not been able to address the issues related to poor reprogramming efficiency and/or prolonged kinetics. These major issues render cell fate manipulation highly inefficient and time-consuming [5, 10]. Here, we present a comprehensive insight into various reprogramming roadblocks that contribute to inefficient iPSC formation (Fig. 1).
Reprogramming roadblocks that prevent the efficient generation of iPSCs. The figure shows various reprogramming roadblocks that have been identified in mouse and human cells that restrict or impede the formation of iPSCs. Tcf3: T-cell factor 3; Bright: B-cell regulator of immunoglobulin heavy chain transcription; Tgf-β: transforming growth factor-β; Bmp: bone morphogenetic protein; CME: clathrin-mediated endocytosis; UPP: ubiquitin-proteasome pathway
Stoichiometry of Core Reprogramming Transcription Factors
Transcription factors recognize specific short DNA sequences and influence transcription either positively or negatively to regulate gene expression. Numerous studies have demonstrated that transcription factors Oct4 (O), Sox2 (S), Klf4 (K) and c-Myc (C) (collectively called OSKM; also known as Yamanaka factors) act as core reprogramming factors and are crucial regulators for induction and maintenance of pluripotent stem cell identity [1, 13]. Considering Yamanaka factor-based cell reprogramming, a balanced stoichiometric and temporal requirements are crucial for optimal expression of these four factors for efficient induction and maintenance of pluripotency [14,15,16]. Importantly, the stoichiometry of these transcription factors markedly influences the molecular characteristics [16], the epigenetic status [17], and the biological properties [16, 17] of iPSCs.
Among the Yamanaka factors, the dosage of master regulator Oct4 is critical during reprogramming as the expression level of this factor decides the fate of the somatic cell: either to drive the cells to a different lineage or to prepare it for induction of pluripotency [15, 16, 18]. To demonstrate this, an interesting study was performed to determine the optimal levels of Oct4 required for efficient reprogramming [15]. The study found that increasing the relative expression of Oct4 up to three-fold, while maintaining moderate expression levels of the other three transcription factors (Sox2, Klf4 and c-Myc), enhanced the reprogramming efficiency [15]. A similar observation was made where a stoichiometric ratio of high Oct4 levels over the other three factors resulted in the highest reprogramming efficiency [16]. Even though low expression levels of Oct4 is ideal to maintain a robust pluripotency in ESCs [19], its high expression is required to induce pluripotency in somatic cells [15, 16]. Further, the study also showed that high levels of Oct4 and c-Myc expression over the other two factors were also beneficial for efficient reprogramming [16]. The role of c-Myc could be to facilitate an ESC-like transcription profile and repression of the somatic cell gene expression program in the target cells during the initial phase of reprogramming [14]. Interestingly, more than a three-fold increase or relative decrease in Oct4 than moderate levels had a negative effect on reprogramming [15]. A significant increase in Oct4 expression above the threshold could have a detrimental effect on reprogramming due to the deviation in its function from the self-reinforcing autoregulatory loop (Oct4/Sox2/Klf4) to target specific promoters of other genes that promote cell differentiation [18, 20].
Recently, Wen and colleagues reported a significant increase in reprogramming efficiency by fine-tuning the expression of reprogramming factors [21]. In this study, the authors showed that equimolar expression of Oct4 and Sox2 and a higher and gradual increase in expression of c-Myc compared to Klf4 during reprogramming resulted in higher efficiency. Further, two independent studies demonstrated that high expression of Oct4 and Klf4 in combination with moderate-to-low expression of Sox2 and c-Myc could result in quality iPSCs [17, 22] with high reprogramming efficiency [22]. iPSCs generated with this stoichiometric ratio efficiently formed “all-iPSC mice” by tetraploid (4n) complementation, retained standard imprinting at the Dlk1-Dio3 locus and did not develop mice with tumors [17]. Moreover, another study reported that the optimal stoichiometric ratio having Oct4-high, Sox2-low, Klf4-high and c-Myc-high expression was the most efficient for the generation of iPSCs, primarily because this optimal ratio elevated expression of key members of the G protein-coupled receptor pathway (especially CCL2) and an epigenetic modifier, Whsc1l1 (variant 1) [23]. Interestingly, low Sox2 expression with moderate levels of Oct4, Klf4 and in the absence of c-Myc resulted in the generation of fully reprogrammed quality iPSCs [24]. Inclusion of c-Myc in this cocktail gave rise to numerous partially reprogrammed colonies, whereas its exclusion resulted in its conversion to fully reprogrammed iPSCs [24]. Cumulative results of this and the above studies clearly indicate that the stoichiometry of core reprogramming factors is critical for the efficient generation of quality iPSCs.
The differences in the studies mentioned above regarding the requirement of the expression levels of Yamanaka factors for efficient reprogramming may be due to different starting cell types, vectors and their design, and experimental conditions used for reprogramming (Table 1). Moreover, the continuous residual expression of reprogramming transgenes during the later stage averts iPSCs from attaining the transcriptional profile and the epigenetic signatures exhibited by ESCs [25]. Therefore, identifying and maintaining the optimal stoichiometric expression of the reprogramming factors during the entire process is critical to manipulate a somatic cell to attain and maintain their full pluripotency efficiently.
Transcription Factors
Transcription factors play a crucial role in the induction and maintenance of pluripotency in mouse and human cells [1, 13, 26]. On the contrary, expression of specific transcription factors can either decrease the reprogramming efficiency or even block the reprogramming process. Few major transcription factors are discussed below that negatively affect iPSC generation.
c-Jun
c-Jun is one of the important member of the AP-1 transcription factor complex. It was the first proto-oncogene to be identified as a transcription factor and plays a vital role in cell growth, proliferation, differentiation, apoptosis and cancer [27]. c-Jun-deficient mice do not develop and die at mid- to late-gestation, signifying that it is essential for proper development [28]. Unlike c-Myc, expression of c-Jun is absent in ESCs and therefore ESCs lacking c-Jun can self-renew and have a normal in vitro differentiation ability [28, 29]. Previous and recent studies have reported that c-Jun is expressed in fibroblasts and is essential for its proliferation by facilitating G1 to S phase progression [29, 30].
Despite being an oncogene and also reported to enhance proliferation of fibroblasts, c-Jun completely blocked, rather than promoted, reprogramming of mouse embryonic fibroblasts (MEFs) to iPSCs [29]. Mechanistically, c-Jun maintained repression of the pluripotency-associated genes, activated mesenchymal-related genes and inhibited the crucial mesenchymal-to-epithelial transition (MET) process [29], the latter is a primary step during the early stage of reprogramming [31, 32]. Further, c-Jun blocked the generation of mouse iPSCs by disrupting chromatin dynamics [33]. Moreover, its overexpression inhibited reprogramming [33], whereas, its downregulation resulted in enhanced efficiency and accelerated the reprogramming process [29]. These results clearly indicate that c-Jun safeguards somatic cell identity and act as a potent roadblock to iPSC generation.
T-cell Factor 3 (Tcf3)
T-cell factor (Tcf) family of proteins [Four mammalian isoforms: Tcf1 (Tcf7), Lef-1, Tcf3 (Tcf7L1) and Tcf4 (Tcf712)] are key constituents of the Wnt-canonical pathway [34]. Out of these four, transcription factor Tcf3 is the frequently expressed isoform in ESCs [35, 36], and functions as a transcriptional repressor in ESCs [35, 37, 38]. Recently, it has been reported that depletion of Tcf3 maintained the robust self-renewal of mouse ESCs and established naïve ground state pluripotency in these cells [39]. Expression of Tcf3 in ESCs prevented excessive transcription of pluripotency genes and maintained their steady state levels sufficient for self-renewal and pluripotency [35, 37, 38].
Earlier, Tcf3 has been identified as a negative regulator of reprogramming [36]. In this study, genetic ablation of this gene in neuronal precursor cells strongly augmented the reprogramming efficiency, attributing to the global epigenome modifications that resulted in an open and transcriptionally active chromatin structure [36]. These alterations in the epigenome was shown by an increase in acetylated histone 3 (AcH3), a slight increase in H3K4me3 and a decrease in H3K9me3 levels during the early stage of reprogramming [36]. Interestingly, this phenomenon was observed long before the initiation of expression of endogenous pluripotency genes. Furthermore, its expression in neuronal precursor cells repressed OK-induced reprogramming [36], most likely by inducing heterochromatin formation and global repression of transcription [35,36,37]. Interestingly, a new reprogramming study demonstrated the role of Tcf3 to be stage-specific [40]. The authors showed that depletion of endogenous Tcf3 inhibited the early stage but was essential for the later stage of reprogramming [40].
Bright/ARID3A
The gene Bright (B-cell regulator of immunoglobulin heavy chain transcription; human orthologue ARID3A) is a key member of ARID family of transcription factors implicated in chromatin remodeling [41]. This transcription factor is essential for development of B cell and hematopoietic stem cell differentiation during early embryonic development [42]. This study demonstrated that more than 99% Bright-deficient embryos did not survive due to failure in hematopoiesis, and the rare survivors exhibited hematopoietic stem cell deficiency in their bone-marrow and also developmental and functional deficiencies of the B-lineage. Moreover, depletion of Bright/ARID3A resulted in the expression of pluripotency-associated markers and promoted developmental plasticity in both mouse and human cells [43].
The observation that depletion of Bright/ARID3A exhibited developmental plasticity prompted Papowski and co-workers to investigate this in a reprogramming setup. The authors established that genetic ablation of Bright from MEFs spontaneously directed these cells towards a pluripotent-like state [44]. Further, they established that this gene was involved in transcriptional repression of crucial pluripotency-associated regulators Oct4, Sox2 and Nanog by directly binding to the promoter regions of these genes [44]. The authors put forward three reasons by which loss of Bright functions to induce pluripotency in MEF cells [44]. First, in the absence of this gene, the cells circumvented the senescence program, and this contributed to somatic self-renewal. Next, Bright-deficient MEFs and murine ESCs led to direct derepression of key pluripotency genes. Finally, deletion of this gene in MEFs might disrupt the signaling pathways that antagonize pluripotency and thereby induce dedifferentiation.
Other Transcription Factors
Similarly, Gata4 [45], Zfp281 [46] and Patz1 [47] were also reported to inhibit somatic cell reprogramming.
Gata4 blocked somatic cell reprogramming by binding to the conserved region located 9 kb upstream of the Nanog gene [45]. This resulted in downregulation of expression of this core pluripotency factor Nanog [45], vital for the generation bona fide iPSCs [48, 49]. On the other hand, downregulation of Gata4 by short hairpin RNAs in MEF cells resulted in enhanced reprogramming efficiency with faster kinetics due to the elevated expression levels of endogenous Nanog [45]. In contrast, a study recently showed that master regulator Oct4 can be replaced by any Gata member (Gata 1-6) to induce reprogramming by suppressing ectodermal lineage markers [50]. These Gata members promoted reprogramming by activating Sall4, an important member of the pluripotency network [50]. Therefore, further detailed investigation is required to explore the role of Gata members in iPSC formation.
Further, like Gata4 [45], a transcriptional repressor Zfp281 was also reported to repress Nanog [46]. Zfp281 recruited the Nucleosome remodeling and deacetylase (NuRD) complex onto the Nanog locus to impede somatic cell reprogramming [46]. The NuRD complex has histone deacetylase (HDAC) function [51, 52], whose inhibition by a HDAC inhibitor, greatly enhanced iPSC formation [53]. Therefore, the researchers then depleted Zfp281 and showed that its depletion facilitated the transition from pre-iPSCs to iPSCs through upregulation of endogenous Nanog [46], the upregulation of the latter is essential to attain a pluripotent ground state [54, 55].
One more transcription factor that has been reported to play an inhibitory role in iPSC formation is Patz1. Inclusion of this gene in the Yamanaka cocktail of transcription factors significantly decreased the reprogramming efficiency, whereas its depletion in MEFs resulted in enhanced efficiency [47]. The authors showed that heterozygous loss of Patz1 in MEFs can overcome the senescence roadblock of Ink4a/Arf locus to promote reprogramming. In addition, Patz1-depleted MEFs exhibited increased levels of active marks such as H3K4me2, H3K4me3, H3K36me3, acetylated histone H3, and decreased levels of heterochromatin protein 1α and a repressive mark H3K9me3, indicating that its depletion resulted in a globally open chromatin structure easily accessible for transcriptional activation [47].
In addition to these, other transcription factors that act as reprogramming roadblocks were also identified by performing knockdown experiments but their biological function from a reprogramming perspective is not yet investigated (Table 2). In conclusion, suppression of somatic and/or pluripotency-inhibiting transcription factors can eliminate a potent roadblock for an efficient iPSC generation.
Signaling Pathways
Numerous studies have shown that signaling pathways are critical for the induction and maintenance of stem cell pluripotency network. On the contrary, activation of specific signaling pathways during reprogramming is also reported to prevent induction and maintenance of pluripotency; and at a later stage, it may direct these cells from naïve to attain a primed state making them susceptible to differentiation. Therefore, inhibition of these signaling pathways is essential to promote cellular reprogramming.
Tgf-β Signaling Pathway
Tgf-β is a potent inducer of epithelial-to-mesenchymal transition (EMT) signals in specific cells [32]. Inhibition of the same promoted both early [31, 32, 61, 62] and late [63] phase of reprogramming. At an early stage, activation of Tgf-β signaling blocked the reprogramming by preventing the cells from undergoing MET and promoting pro-EMT signals [31, 32, 61, 62]. At a later stage, this signaling prevented the accomplishment of final pluripotency by trapping stable intermediate cells in a partially reprogrammed state [63]. Therefore, inhibition of this signaling at a later stage resulted in enhanced expression of Nanog that helped the cells to attain full pluripotency [63]. Further, Stadtfeld and colleagues established that inhibition of Tgf-β signaling with concurrent activation of Wnt/β-catenin in the presence of an antioxidant resulted in highly efficient (>80%) and non-stochastic acquisition of pluripotency in mouse fibroblasts [64]. Notably, blood progenitors were also reported to form iPSCs with a 100% efficiency by suppression of Tgf-β signaling or canonical Wnt activation [64]. Importantly, inhibition of this signaling pathway is sufficient to induce pluripotency in differentiated cells without the requirement of exogenous pluripotency-inducing factors, Sox2 [61, 63] or c-Myc [61]. Further, in combination with a protein arginine methyltransferase inhibitor AMI-5, it enabled reprogramming of MEF cells transduced with Oct4 alone [65]. Therefore, inhibition of Tgf-β signaling is essential to induce faithful reprogramming with minimal reprogramming factors.
Hippo Signaling
The Hippo signaling pathway function as a crossroad between cell specification and pluripotent reprogramming of somatic cells. This is a highly conserved signaling pathway that regulates growth with two main downstream effectors, Yap and its paralog Taz [66, 67]. Yap and Taz are transcriptional coactivators and are highly expressed during early embryonic development [66].
Interestingly, Yap and Taz have distinct functions in mouse and human cells. YAP plays a vital role in regulating the self-renewal of mouse ESCs [68] and enhance the formation of mouse iPSCs [69], but has no role in the self-renewal of human ESCs [70] or in the generation of human iPSCs [71]. On the contrary, TAZ regulates the self-renewal of human ESCs [72] and also promotes the generation of human iPSCs [71], playing no role in the self-renewal of mouse ESCs [72]. This could be because of the different signaling pathways involved in the regulation of pluripotency between mouse and human pluripotent stem cells.
Both these potent transcriptional coactivators, Yap and Taz, are negatively regulated by a tumor suppressor kinase Lats2 [71]. Lats2 is an important member of the Hippo signaling pathway and is expressed in primordial germ cells, but not in germ cell tumors and pluripotent stem cells [71]. Activated Lats2 was reported to phosphorylate both Yap and Taz, which resulted in their cytoplasmic retention and protein degradation [73]. Moreover, Lats2 was shown to antagonize reprogramming of human fibroblasts by suppression of Taz, whereas downregulation of Lats2 resulted in improvement in the generation of iPSCs [71]. This indicates that Lats2, an important member of the Hippo pathway, acts as a barrier to iPSC generation.
In addition, Hippo signaling pathway is shown to negatively regulate Wnt/β-catenin pathway [74, 75]. This is a common signaling pathway for self-renewal and maintenance of pluripotency in mouse and human pluripotent stem cells [76,77,78] as well as for the induction of pluripotency in somatic cells [79, 80]. To conclude, Hippo pathway is a negative regulator for the induction of pluripotency, and suppression of this pathway may alleviate a major roadblock to efficient reprogramming.
Protein Kinase Signaling
Different kinases have been reported to influence the reprogramming process negatively. Silva and colleagues demonstrated that iPSCs with an authentic naïve pluripotent state could be generated by reprogramming neural stem cells by using only two reprogramming factors, Oct4 and Klf4, and by dual inhibition of kinases namely, glycogen kinase 3 (GSK3) and mitogen-activated protein kinase/extracellular signal-regulated kinases 1 and 2 (MEK/ERK) [81]. Inhibition of GSK3 signaling was also shown to induce reprogramming in MEF cells transduced with only two factors, Oct4 and Klf4 [82]. Further, its inhibition in combination with Tranylcypromine, an inhibitor of lysine-specific demethylase 1 (LSD1) was reported to reprogram human primary keratinocytes transduced only with Oct4 and Klf4 [82]. Li and Rana performed a kinase inhibitor screen in MEF cells and identified kinases such as Aurora A kinase, inositol triphosphate 3-kinase, p38 mitogen-activated protein kinase and activin receptor-like kinase 4/5 that represented as a barrier to cell reprogramming, and showed that inhibition of these kinases led to improved iPSC formation [83]. On the contrary, another recent report by Lako and group showed that downregulation of p38 mitogen-activated protein kinase in human fibroblasts via small molecules or RNA interference led to shunting of cell cycle regulators followed by G1 arrest in the initial stage of reprogramming [84]. This caused significant decrease in pluripotency markers, abrogation of MET and increase in differentiation markers, halting the reprogramming process in a partially reprogrammed state. This discrepancy in the outcomes could be because of the differential role of mitogen-activated protein kinase signaling in the regulation of self-renewal and pluripotency in mouse [85] and human [86] pluripotent stem cells. Importantly, other studies also reported inhibition of kinases such as Rho-associated protein kinase [87], protein kinase C [88] and Src family tyrosine kinase [89] to demonstrate enhancement in iPSC formation. These studies suggest that the inclusion of various molecules targeting these barrier kinases will alleviate the roadblock to generate iPSCs with high efficiency.
BMP Signaling
BMP signaling plays a crucial role in early embryonic patterning [90] and lineage specification either towards trophoblast [91] or mesoderm [92, 93] in ESCs. Interestingly, this signaling has a controversial role during the reprogramming of fibroblasts to iPSCs [32, 94]. Hamasaki and co-workers tried to derive iPSCs from the human dermal fibroblasts of patients with fibrodysplasia ossificans progressiva that carried a missense mutation in ACVR1 [617G > A (R206H)] resulting in hyperactivation of the BMP signaling pathway [95]. However, the researchers achieved little success as they got only a few undifferentiated ESC-like colonies and many differentiated colonies due to incomplete reprogramming. This result suggested that BMP signaling negatively affects hiPSC derivation as well as their self-renewal. Surprisingly, a separate study reported a contradictory outcome showing a positive effect of BMP signaling during the initial phase of human iPSC formation by reprogramming human dermal fibroblasts from patients with the same disease [96]. Interestingly, Wrana lab demonstrated that BMP-dependent induction of microRNAs miR-205 and miR-200 family facilitated the generation of mouse iPSCs due to the promotion of MET process [32], which is an essential process during the early stage of reprogramming [31, 32]. However, it was also reported that during reprogramming many colonies having an ESC-like morphology, termed as “pre-iPSCs”, are devoid of the activation of the endogenous locus encoding Oct4 and Nanog [14, 56, 81, 94]. This is mainly due to BMP signaling that arrested reprogramming, and maintained intermediates at the pre-iPSC state [94, 97]. Moreover, it has also been reported that BMP4 promoted mesodermal commitment by inducing EMT [93], which is also in contradiction with its role during reprogramming [32]. This discrepancy could be due to the concentration- and time-dependent effects of BMPs on reprogramming resulting in different outcomes [97]. In general, these studies indicate that the role of BMP signaling in a cell reprogramming paradigm could be inhibitory, but is controversial due to conflicting reports and therefore requires further investigation.
Senescence
Senescence serves as another major barrier to cellular reprogramming as cells lose the capacity to proliferate and divide [98]. It occurs as a result of oxidative stress, DNA damage, telomere shortening and the derepression of Ink4a/Arf locus by chromatin remodeling in somatic cells (Fig. 2) [99].
Senescence as a critical barrier to prevent the efficient derivation of iPSCs. In young donor cells, DNA Damage Response (DDR) is triggered by oxidative stress via the generation of reactive oxygen species (ROS), reprogramming-induced senescence (RIS) and aberrant DNA replication; whereas in aged donor cells, DDR is triggered via mitochondrial dysfunction along with ROS generation, RIS, deficiency in DNA repair mechanism and replicative senescence (telomere shortening). DDR in both young and aged donor somatic cells are regulated through two major pathways, namely p14/p19Arf and p16Ink4a pathway. The p14/p19Arf leads to the activation of p53, followed by the activation of its downstream cyclin-dependent kinase inhibitor p21Cip/Waf1. This inhibits Cyclin E/Cdk2 complex, thus preventing the phosphorylation of retinoblastoma (Rb) protein, leading to its activation. Similarly, the p16Ink4a pathway inhibits Cyclin D/Cdk4/6 complex, following the downstream activation of Rb. Active Rb causes cell cycle arrest by prohibiting cells from entering S-phase, thus acting as a potent barrier. In addition, the aged donor cells are already in a pre-senescent or senescent stage with a functional senescent network. Upon induction of the reprogramming process, the expression of senescent regulators in aged donor cells is augmented, making the reprogramming even more difficult in comparison to young donor cells
Cells with short telomeres are indicative of damaged/uncapped telomeres; therefore, these cells cannot be efficiently reprogrammed into iPSCs despite having normal cell proliferation rates [100]. On the other hand, somatic cells having high replicative potential with longer telomeres (observed in low passage or young donor somatic cells) are easy to reprogram due to low expression of major senescence regulators [98]. The generation of oxidative stress is one of the leading causes of DNA damage and this leads to upregulation of p53 and p21Cip1 [99]. Additionally, introduction of reprogramming factors in fibroblasts promoted DNA damage, and the activation of the DNA damage response machinery resulted in the induction of p53 and its downstream target gene p21 during reprogramming [101,102,103,104], eventually leading to p53-p21-dependent cell cycle arrest or apoptosis [105]. Therefore, depletion or deletion of p53 facilitated efficient reprogramming of mouse and human iPSC formation [98, 101,102,103, 106,107,108,109], but the generated iPSCs sustained shorter telomeres and chromosomal aberrations [98, 101, 105]. Together, these studies conclude that the p53-p21 pathway is a determinant of reprogramming kinetics and efficiency. Interestingly, Jaenisch and co-workers showed that the depletion of this pathway resulted in enhanced proliferation which dramatically improved the reprogramming kinetics without having any effect on the efficiency in the model system they examined [110]. However, a permanent deletion or strong long-term inhibition of p53 will generate lower quality iPSCs due to severe genomic instability, and therefore will increase the chances of malignant transformation [98, 101,102,103, 107,108,109]. The presence of genomic instability has raised safety issues and can hinder the progress of iPSC-based therapeutic applications [111, 112].
To overcome this serious concern, employing temporary inhibition of p53 using a small molecule only for a specific duration during reprogramming [113] will be a more suitable strategy to derive genetically stable iPSCs. Further, inclusion of transcription factors such as Zscan4 or Zscan10 in a non-genetic form could also prevent genomic instability [104, 114, 115], inhibit p53 expression and maintain genetic stability of the derived iPSCs [104, 115]. Alternatively, overexpression of a physiological inhibitory p53 isoform Δ133p53 that lacks the N-terminal 132 amino acids promoted cellular reprogramming of human fibroblasts to a pluripotent state by regulating p53-inducible senescence genes [109]. Strikingly, Δ133p53 repressed the expression of p53-inducible senescence genes but did not alter the expression of p53-inducible apoptosis and DNA damage repair genes [109]. Therefore, overexpression of this isoform may overcome the p53 reprogramming barrier without affecting the genomic stability. Also, using specific target cell types having low protein expression levels of p53/p21 will be ideal for reprogramming. For example, keratinocytes exhibit reduced p53 and p21 protein expression levels than commonly used fibroblasts [101]. Therefore, this could be one major reason why this particular cell type gave a 100-fold higher reprogramming efficiency and two-fold faster reprogramming kinetics when compared to fibroblasts [116].
In addition, the other major senescence regulators namely the Ink4a/Arf locus [98, 103, 107] and its two constituents p19Arf [98, 107] and p16Ink4a [103, 107] have been reported to act as potent barriers of reprogramming. Notably, in mouse fibroblasts, p19Arf, rather than Ink4a, is the key roadblock to iPSC formation by activation of p53 and p21; whereas, it is p16Ink4a, rather than p19Arf, that acted as a reprogramming barrier in human fibroblasts [107]. In addition, diminishing or obliterating the constituents of senescence machinery can also reduce the number of reprogramming factors required for efficient reprogramming [98, 101, 102, 105, 107,108,109]. To summarize, senescence acts as a roadblock to reprogramming, preventing efficient iPSC generation. Identifying the precise timing for temporary inhibition of vital constituent(s) of senescence machinery using small interfering RNAs or small molecules or inclusion of genomic stabilizers in the reprogramming cocktail would be the next logical step for efficient formation of genetically stable iPSCs.
MicroRNAs
MicroRNAs (miRNAs) are widely known for its regulation of gene expression post-transcriptionally by targeting one or more messenger RNA (mRNA) to favor degradation or inhibit translation. They function via full or partial base-pairing with complementary sequences with the 3’-untranslated region (UTR) or within the target mRNA. This novel class of naturally occurring, short, non-coding RNA sequences plays a vital role in various biological processes such as embryogenesis, differentiation, apoptosis, proliferation, autophagy, immune responses and human diseases [117, 118]. In pluripotent stem cells, loss of key genes involved in miRNA biogenesis namely DGCR8 [119] and Dicer [120, 121] affected the proliferation and differentiation of these cells. Studies have also reported that specific miRNAs can induce cellular reprogramming without [122, 123] or substituting the reprogramming factors [124, 125]. Numerous miRNAs have been reported to play a crucial role in the induction of pluripotency [8]. On the contrary, there are specific miRNAs that impede the reprogramming process (Fig. 3).
An overview of various microRNAs that act as reprogramming barriers. The schematic shows the list and function of various microRNAs that have been identified as reprogramming roadblocks in mouse and human cells. MBD3: methyl-CpG binding domain protein 3; ERK1/2: extracellular signal-regulated kinase 1/2; Let-7: lethal-7
One of the miRNAs that act as a barrier to cellular reprogramming is the miR-34 family [126], which comprises of the evolutionarily conserved mammalian non-coding RNAs namely, miR-34a, miR-34b and miR-34c [127]. The expression of these members were induced by oncogenic stress and DNA damage response in a p53-dependent manner and their overexpression caused cellular senescence or apoptosis [127]. Due to this observation, the same group further investigated the role of this miR-34 family in a cell reprogramming paradigm [126]. Expectedly, the authors found that members of this family act as a roadblock to cell reprogramming since depletion of any member of this family resulted in enhanced reprogramming efficiency with faster kinetics. Among these three mammalian homologs, miR-34a depletion exhibited the highest increase in reprogramming efficiency. miR-34a-depleted MEFs infected with OSK showed a 4.5-fold increase in reprogramming efficiency, whereas a 4-fold increase was reported with OSKM transduction [126]. This enhancement in efficiency was mainly due to posttranscriptional derepression of pluripotency-associated genes, Nanog, Sox2 and N-Myc. Importantly, miR-34a exhibited p53-dependent induction and cooperated with p21 during reprogramming to prevent the formation of iPSCs. A similar observation was made in a separate study which showed inhibition of miR-34a resulted in improved iPSC formation, whereas, expression of this miRNA resulted in inhibition of reprogramming [128]. The study reported that the role of this miRNA is mainly during the early stage of reprogramming, most likely by downregulation of Sirtuin 1 expression. Mechanistically, expression of Sirtuin 1 induced Nanog expression and repressed p53 expression and thereby the expression of its downstream target p21; and this action of Sirtuin 1 was markedly inhibited by miR-34a resulting in poor reprogramming efficiency in mouse fibroblasts [128].
Various miRNAs highly expressed in specialized cells post-transcriptionally regulate proteins that serve as reprogramming barriers. Therefore, depletion of these miRNAs in specialized cells in a reprogramming set-up can result in improvement in reprogramming efficiency and kinetics. Employing this strategy, Blelloch and co-workers depleted let-7 family of microRNAs in MEF cells to investigate its effect on reprogramming [129]. This family of miRNAs is highly expressed in MEF cells [130]. Depletion of the let-7 family dramatically increased the reprogramming efficiency, especially when a reprogramming factor c-Myc was not included in the cocktail [129]. A similar conclusion was drawn, indicating that let-7 acts as a reprogramming barrier to human iPSC generation by stimulating the expression of pro-differentiation genes [131]. Interestingly, inhibition of let-7 in human fibroblasts transduced with OSK yielded a similar reprogramming efficiency as that of OSKM cocktail [131]. Using the same strategy, Rana and colleagues depleted MEF-enriched miRNAs, miR-21 and miR-29a, in MEF cells to determine their roles in reprogramming [132]. The authors established that both these miRNAs regulate p53 and ERK1/2 signaling pathways to act as reprogramming barriers. Depletion of these miRNAs in MEF cells resulted in two- to three-fold enhancement in iPSC generation [132]. During reprogramming, these miRNAs are inhibited by c-Myc to promote reprogramming by increasing CDC42 and p85α expression [132], the latter proteins are targets of miR-29a as reported earlier [133]. Later, the same group identified two other MEF-enriched miRNAs, miR-223 and miR-495, to have inhibitory roles in iPSC formation [134]. Further, Wu and co-workers identified miR-199a-3p and miR-363 as novel reprogramming barriers [135]. miR-199a-3p is induced and regulated by p53 in MEF cells [135]. Expression of this MEF-enriched miR-199a-3p resulted in a significant reduction in reprogramming by imposing G1 cell cycle arrest via the upregulation of p21 gene, whereas inhibition of the same resulted in improvement due to enhanced cell proliferation. Depletion of another set of miRNAs, miR-132 and miR-212, resulted in improved reprogramming efficacy [136]. These miRNAs exerted their function by targeting two epigenetic regulators, the H3K4 demethylase Jarid1a (KDM5a) and the histone acetyltransferase p300, to act as an endogenous roadblock of somatic cell reprogramming. In addition, a miRNA, miR-134, is highly expressed during neural differentiation but downregulated in human ESCs and iPSCs [137]. Inhibition of miR-134 in neural progenitor cells enhanced the reprogramming efficiency and maturation of pre-iPSCs, whereas its overexpression inhibited iPSC induction and generation. Upon expression, this miRNA targeted the 3’-UTR of Methyl-CpG-binding domain protein 3 (Mdb3) to downregulate its expression to suppress iPSC generation and maturation [137]. In human cells, depletion of the miR-29a in fibroblasts resulted in global DNA demethylation during the early stages of reprogramming eventually leading to enhanced reprogramming efficiency [138]. The iPSCs generated in this study were also reported to be epigenetically closer to ESCs. Another miRNA, miR-145 is highly expressed in human fibroblasts and low in pluripotent cells [139]. In human ESCs, Oct4 repressed miR-145, thereby blocking its function [139]. However, overexpression of this miRNA in human ESCs inhibited ESC self-renewal by targeting 3’-UTR of pluripotency factors Oct4, Sox2 and Klf4 and induced lineage-specific differentiation, thus indicating the existence of a double-negative feedback loop involving Oct4, Sox2, Klf4 and miR-145 [139]. Depletion of miR-145 in human dermal fibroblasts resulted in low expression of mesenchymal markers and miRNA let-7b, upregulation of epithelial markers and expression of pluripotency-associated genes, thereby facilitating and enhancing iPSC formation [140].
To summarize, various miRNAs act as reprogramming barriers by regulating different downstream genes to prevent faithful reprogramming, and inhibition of these miRNAs can overcome these roadblocks to improve not only reprogramming kinetics and efficiency but also the quality of iPSCs.
Epigenetic State and Modifications
Epigenetics is the study of phenotypic alterations due to changes in gene expression without any change in the original DNA sequence [141]. Epigenetic modifications, such as DNA methylation and histone modifications, are vital for chromatin organization and the regulation of gene expression [142, 143]. The conversion of a differentiated cell to a pluripotent cell involves resetting of the global epigenome. The epigenetic state in somatic cells and the specific epigenetic modifications that occur during reprogramming specify lineage-specific programs rather than the induction of pluripotency, and thereby act as a roadblock to efficient reprogramming [144].
DNA Methylation
DNA methylation involves the covalent addition of a methyl group onto the 5th carbon of cytosine residue in the DNA strand resulting in the establishment of 5-methylcytosine (5mC). The family of enzymes called DNA methyltransferases (DNMTs) catalyzes this epigenetic modification. DNMT1 methylates hemimethylated CpG (5’-Cytosine-phosphate-Guanine-3’) sites thus referred to as “maintenance” DNMT; whereas DNMT3a and DNMT3b methylate new CpG sites, therefore called as “de novo” DNMT [145, 146]. Inhibition of DNMTs by DNMT inhibitors, 5-Azacitidine and RG108, improved the reprogramming efficiency or can generate iPSCs with minimal reprogramming factors [53, 147, 148]. In a study by Mikkelson and colleagues, transient inhibition of DNMT1 in partially reprogrammed cells with specific shRNA/siRNA or using DNMT inhibitors (such as 5-Azacitidine) for 48 hours rapidly enhanced the complete transition from partially to fully reprogrammed iPSCs [56]. Concurrently, a histone methyltransferase enzyme G9a was reported to recruit Dnmt3a and Dnmt3b to promote de novo DNA methylation at crucial pluripotency genes thus prevented reprogramming to a pluripotent state [149]. This function was independent of its histone methyltransferase activity.
Conversely, DNA demethylation is the removal of a methyl group from 5-methylcytosine via the sequential modification of cytosine bases. Newly discovered Ten-eleven translocation (Tet) family of dioxygenases is believed to have a role in active DNA demethylation by binding to CpG rich regions to avoid undesirable DNA methyltransferase activity, and by successively oxidizing 5mC to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine through hydroxylase activity [150]. Three independent groups reported that Tet enzymes reactivated Oct4 promoter [151, 152] and facilitated the generation of iPSCs [151,152,153]. Interestingly, Tet-deficient MEFs could not be reprogrammed [152, 153], due to block in MET [152]. One of the Tet enzymes, Tet1, enhanced reprogramming by promoting Oct4 demethylation and reactivation, and could also substitute this master regulator Oct4 in the reprogramming process [151]. Simultaneously, Tet enzymes and Nanog were identified as interaction partners and synergistically improved the reprogramming efficiency by enhancing the expression of crucial pluripotency genes [153]. Furthermore, two epigenetic modification factors namely Tet2 [154, 155] and poly(ADP-ribose) polymerase-1 (Parp1) [154] were reportedly recruited to the Esrrb and Nanog loci during the early stage of reprogramming to facilitate iPSC formation [154]. Tet2 was also reported to interact with C/EBPa, Klf4 and Tfcp2l1, and these factors recruited Tet2 to specific DNA sites, leading to enhancer demethylation and activation of pluripotency-associated genes [155]. Previously, a popular water-soluble antioxidant vitamin C was reported to enhance the reprogramming of somatic cells [156], in part by mitigating cellular senescence [156] and perhaps by promoting DNA demethylation [157, 158]. It was shown that vitamin C enhanced Tet-dependent demethylation activity in mouse ESCs [159] and modulated Tet function during somatic cell reprogramming [160]. Additionally, MEFs deficient in Thymine DNA Glycosylase, an enzyme reported to efficiently excise 5-formylcytosine and 5-carboxylcytosine, also showed similar impaired reprogramming ability and was attributed to the defects in the activation of critical miRNAs [152].
Remarkably, Activation-Induced cytidine Deaminase (AID/AICDA), a DNA demethylase enzyme responsible for secondary antibody diversification, was also reported to be essential for the reprogramming process [161]. This enzyme is expressed in oocytes, B lymphocytes, and in pluripotent cells such as ESCs and embryonic germ cells [162]. In a study generating inter-species heterokaryons (fusion of human skin cells and mouse ESCs), the researchers demonstrated that AID was involved in demethylation of promoter regions of key pluripotency genes Oct4 and Nanog and enabled reprogramming with higher efficiency and faster kinetics [161]. Further, siRNA mediated silencing of AID showed that it is essential for promoter demethylation and induction of expression of Oct4 and Nanog, thus highlighting its putative demethylating activity [161]. Hence, all these studies corroborate the fact that DNA methylation act as a barrier and DNA demethylation of pluripotency-related genes is indispensable for the generation of iPSCs (Fig. 4).
In addition to DNA methylation, specific chromatin modifications disturb the ability of the reprogramming factors to bind to their target sites and affect iPSC formation (Fig. 5). Few prominent histone modification marks that act as a reprogramming barrier are discussed below.
An overview of histone modifications acting as epigenetic barriers during the reprogramming process to prevent efficient iPSC formation. me: methylation; vit. C: vitamin C; Kdm: lysine demethylase; LSD1: lysine-specific demethylase 1; Suv: suppressor of variegation; Setdb1: SET domain bifurcated histone lysine methyltransferase 1; Ehmt: euchromatin histone methyltransferase; Glp: G9a like protein; Trim 28: tripartite motif-containing 28; Jhdm: jumonji-domain containing histone demethylase; DOT1L: disruptor of telomeric silencing; shRNA: short hairpin RNA; Utx: ubiquitously transcribed tetratricopeptide repeat X chromosome; ac: acetylation; VPA: valproic acid; NaB: sodium butyrate; SAHA: suberoylanilide hydroxamic acid; TSA: trichostatin A
Histone Methylation
The major histone modification responsible for heterochromatin assembly and gene silencing is the methylation at lysine 9 of histone H3 (H3K9me), a mark conserved from yeast to human [143, 163]. Trimethylation of H3K9 (H3K9me3) is mediated by a histone methyltransferase enzyme, G9a, to generate a heterochromatin structure by recruiting heterochromatin protein 1 [149, 164]. This repressive histone mark is introduced to silence the crucial pluripotency genes through the SET domain of G9a to prevent cellular reprogramming [149, 164].
In line with these observations, the study by Pei lab further established that methylation of H3K9 attenuated somatic cell reprogramming, and this epigenetic roadblock was sensitive to vitamin C [94]. To demonstrate this, the authors showed that conversion of reprogramming intermediate cells, termed as “pre-iPSCs”, to iPSCs was possible only after the supplementation with vitamin C into the ESC medium, and these quality iPSCs could contribute to chimeric mice. Interestingly, these pre-iPSCs cultured in ESC medium devoid of vitamin C failed to activate the endogenous locus encoding for Oct4 and could not contribute to chimeric mice. Deducing the intrinsic mechanism through which vitamin C promoted the formation of quality iPSCs, the researchers revealed that vitamin C remarkably decreased H3K9 methylation, and knockdown of H3K9 methyltransferases further enhanced reprogramming of pre-iPSCs to iPSCs in a medium containing fetal bovine serum and vitamin C [94]. This observation was reinforced in different studies where the researchers further demonstrated that expression of H3K9 methyltransferases (Suv39h1, Suv39h2, Setdb1, Ehmt1/Glp and Ehmt2/G9a) inhibited whereas H3K9 demethylases (Kdm3a, Kdm3b, Kdm4b and Kdm4c) promoted the reprogramming efficiency[94, 165,166,167,168]. It could be that H3K9 methylation may have recruited heterochromatin protein 1 to establish a heterochromatin state [164, 169], which possibly hampered reprogramming. This probably resulted in impairment in the activation of pluripotency-inducing genes by OKSM, due to the inaccessibility of the latter to its target sites [94, 165, 166]. Interestingly, removal of H3K9me3 marks during reprogramming could also be influenced by the culture conditions used to derive iPSCs [94, 170]. Also, it was reported that the downregulation of heterochromatin protein-1γ/Chromobox homolog 3 (or Cbx3), a protein known to identify H3K9 methylation, improved the reprogramming efficiency [171].
Previously, it was reported that Trim28 and Setdb1 interacted to establish the repressive H3K9me3 mark [172], and recently, a group of researchers established that the loss of Trim28 resulted in the upregulation of genes involved in maintenance of an open chromatin configuration, ideal for efficient reprogramming [173]. These genes were located in the proximity of repressed chromatin regions associated with high levels of H3K9me3 [173]. In contrast to other H3K9 demethylases that promoted reprogramming, suppression of the lysine-specific histone demethylase LSD1 (KDM1A) has been reported to enhance iPSC generation[82, 174,175,176], primarily through epithelialization (elevation of a set of genes enriched in epithelial markers, particularly E-cadherin (CDH1), CLDN10, EPCAM and KRT19) during the early stage of reprogramming [175]. Also, improvement in the formation of iPSCs due to LSD1 inhibition could be by regulating both H3K4 and H3K9 demethylation [176]. On the whole, the elimination of this modification is necessary for the efficient conversion of pre-iPSCs towards iPSCs to overcome this epigenetic barrier.
Additionally, vitamin C was also reported to reduce the levels of H3K36me2/3 during reprogramming via downstream vitamin C-dependent H3K36 demethylases Jhdm1a (Kdm2a) and Jhdm1b (Kdm2b) [177]. Removal of these modifications by Jhdm1b accelerated cell cycle progression and prevented senescence by repressing the Ink4a/Arf locus [177]. Also, Oct4 in cooperation with Jhdm1b triggered the stimulation of a microRNA cluster 302/367 [177], a vital cluster that promoted reprogramming by inhibiting Tgf-β to accelerate MET [62, 122, 178]. This significantly promoted the reprogramming process by overcoming the senescence roadblock. It was in accordance with the previously reported studies that Jhdm1b enhanced proliferation of fibroblasts through suppression of the Ink4a/Arf locus by eliminating H3K36me2/3 modifications [179, 180]. Further, the demethylation of H3K36 by Jhdm1b (Kdm2b) enzyme has been reported to enhance iPSC formation [181]. This enzyme is an H3K36me2-specific demethylase and is crucial for demethylation of promoters of genes involved in the early stage of reprogramming [181]. Therefore, H3K36me2/3 demethylation is also crucial for efficient iPSC generation.
Similar to H3K9 and H3K36, one more histone modification that has been implicated to act as a reprogramming roadblock is H3K79me2 which facilitated EMT [165]. In this study, the researchers identified a histone methyltransferase, Disruptor of telomeric silencing 1-like (DOT1L), that act on H3K79 as a reprogramming barrier [165]. The DOT1L is vital for establishing a heterochromatin structure and mammalian development [182]. In contrast, where H3K79 methylation represents transcriptional activation [183, 184], Onder and co-workers observed that inhibition of DOT1L, SUV39H1 (an H3K9 methyltransferase) and YY1 (a context-dependent repressor or activator of transcription), improved reprogramming of fibroblasts. The authors observed that suppression of DOT1L in the early stage of reprogramming was concomitant with a marked increase in the expression of pluripotency factors, namely Lin28 and Nanog. Further, the researchers performed global genome-wide analysis to determine the distribution of H3K79me2 upon inhibition of DOT1L, and this experiment revealed that the lineage-specific genes associated with the EMT lose H3K79me2 during the early stage of reprogramming.
Concurrently, Hanna lab revealed that demethylation of H3K27 during reprogramming contributed towards the ground state of pluripotency [185]. This study demonstrated that Utx (Ubiquitously transcribed tetratricopeptide repeat, X chromosome; also known as Kdm6a), an H3K27me3 demethylase, is responsible for properly timed H3K27me3 demethylation during reprogramming. If aberrant (in cells lacking Utx), it would negatively impact the activation of pluripotency-inducing genes namely Utf1, Sall1 and Sall4, leading to a block in iPSC formation [185]. Similarly, the two isoforms of macroH2A (macroH2A1 and macroH2A2) along with H3K27me3 co-occupy the crucial pluripotency-associated genes providing a redundant silencing layer on these genes [186]. Removal of these variants resulted in enhanced reprogramming efficiency [186,187,188], possibly due to an effective epigenetic remodeling or loss of trimethylation mark on H3K27 [186].
Histone Deacetylation
Histone acetylation and deacetylation are the crucial epigenetic mechanisms that controls the expression of specific genes. These mechanisms are mainly regulated by the activity of two classes of enzymes: the histone acetyltransferases (HATs) that acetylate conserved lysines on histone proteins, and histone deacetylases (HDACs) that remove the acetyl groups from the histone tails [189]. The chromatin structure marked by acetylated lysine represents transcriptional activation, whereas deacetylated lysine represents transcriptional repression [190, 191].
Mammalian genomes have four classes of HDACs [189]. Histone deacetylation by HDACs promote chromatin compaction and generates a heterochromatin structure that makes the chromatin inaccessible for transcription [189, 192]. Numerous studies have reported that inhibition of histone deacetylation significantly enhanced the reprogramming of somatic cells to iPSCs [53, 122, 147, 148, 193,194,195,196,197,198]. This is accomplished by inhibiting expression of HDACs using small molecules such as Valproic acid [53, 122, 193, 198], Sodium butyrate [147, 194, 196], Suberoylanilide hydroxamic acid [53, 195] and Trichostatin A [53]. Enhancement in the reprogramming efficiency by inhibiting HDACs during the early stage of reprogramming could be due to one or more of these reasons: either by induction of miR302/367 cluster [122, 196], or epigenome remodeling and activation of pluripotency-related genes [147, 194, 195, 197], or by suppression of reprogramming-induced senescence stress [198].
Interestingly, the role of HDACs in the conversion of pre-iPSCs to iPSCs is still debatable due to the conflicting reports [197, 199]. While one study claims that HDAC2 acts as a barrier to the reprogramming process during the maturation stage [197], whereas the other study claims that it could be beneficial during the maturation stage in the reprogramming [199]. Therefore, the exact mechanisms by which HDAC inhibitors enhance reprogramming efficiency and the role of HDACs during the different stages of reprogramming is still controversial and requires detailed investigation. Altogether, these studies indicate that HDACs function as epigenetic roadblocks to reprogramming by establishing a chromatin structure which restricts the activation of a transcriptional network that regulates pluripotency.
Histone Variants
Histone variants are non-canonical proteins that replace the octamer made up of core histones (H2A, H2B, H3, H4) in eukaryotic chromatin and have specific functional and structural characteristics. In a reprogramming context, the first study to establish the role of histone variants carried out nuclear transfer experiments and demonstrated that macroH2A prevented the X-chromosome reactivation and activation of pluripotency-related genes to confer resistance to nuclear reprogramming [200]. Depletion of macroH2A variants, macroH2A.1 and macroH2A.2, resulted in enhanced reprogramming efficiency [186,187,188, 201], and its overexpression prevented efficient reprogramming [187, 188, 201], of which, macroH2A.2 is the major roadblock towards reprogramming [186]. The negative role of macroH2A in a cell reprogramming paradigm could be due to its ability to inhibit the key MET process during the early stage of reprogramming [201]. Genome-wide analysis revealed that the macroH2A variants occupied the promoters of bivalent developmental regulators and pluripotency-related genes, and showed substantial overlap with the genes repressed by H3K27me3 in differentiated cells [186, 188, 201]. The presence of macroH2A.1 at these silenced genes precludes the introduction of H3K4me2 at these sites in the course of reprogramming, imposing an extra layer of repression that maintains somatic cell identity [188].
One more histone variant that has been implicated in the maintenance of somatic cell identity is H3.3 [202]. Removal of this histone variant during the early stage of reprogramming improved the reprogramming ability of differentiated cells, whereas, it was found to be essential for the acquisition of pluripotency during the later stage of reprogramming [202]. This histone variant is essential for supporting heterochromatic features and thereby maintaining genome integrity [203], explaining its role in the generation of stable iPSCs during the later stage of reprogramming. Thus, various histone variants play a crucial role in the maintenance of the somatic cell identity to prevent efficient reprogramming.
Chromatin Remodelers
NuRD/MBD Complexes
NuRD complexes are chromatin remodeling factors that are mainly associated with a repressive chromatin structure [52, 204]. Out of the seven constituents, one of the vital constituents of the NuRD complex is either Methyl-CpG Binding Domain Protein 2 (MBD2) or Methyl-CpG Binding Domain Protein 3 (MBD3) (hereafter MBD2/NuRD and MBD3/NuRD), and thus forms two separate complexes having different biochemical and functional characteristics [205]. The major distinction between MBD2/NuRD and MBD3/NuRD is that the latter is devoid of four conserved amino acids in the MBD domain, and therefore, it does not bind to methylated DNA [206]. Data from different studies have revealed that MBD2/NuRD only or MBD3/NuRD has a major effect on the induction of pluripotency; however, whether these complexes affect positively or negatively is still a matter of debate.
Demonstrating the negative effect of NuRD repressor complex on reprogramming, Fidalgo and co-workers established that recruitment of the NuRD complex by transcription factor Zfp281 onto the Nanog locus blocked reprogramming through suppression of Nanog expression [46]. Further, this study showed that suppression of Zfp281 resulted in enhanced reprogramming efficiency, thereby supporting an inhibitory role of MBD3/NuRD in iPSC generation. In accordance with the previous reports, another study also reported that overexpression of MBD3/NuRD resulted in the establishment of heterochromatic features and silencing of key ESC-specific genes (Nanog and Oct4) [207]. Additionally, the researchers showed that depletion of MBD3/NuRD resulted in the expression of pluripotency genes and gave rise to fully reprogrammed iPSCs, even in the absence of core reprogramming factors, Sox2 or c-Myc [207]. In line with this study, Hanna lab reprogrammed mouse and human cells within seven days in naïve pluripotency promoting conditions resulting in deterministic reprogramming with nearly 100% efficiency [208]. They also demonstrated that the chromatin in MBD3/NuRD-depleted cells was more open and active and displayed high Oct4 binding with an increased H3K27ac and H3K4me3 marks as well as reduced levels of H3K27me3 modifications [208]. Similarly, they further provided additional facts to support their earlier claim [208] and reconfirmed that MBD3/NuRD pathway is a potent inhibitor of induction and maintenance of pluripotency [209].
In contrast to these observations, dos Santos and colleagues performed reprogramming of multipotent neural stem cells, pre-iPSCs and epiblast stem cells and showed reduced reprogramming efficiency on depletion of Mbd3 [210], however, no obvious effect was observed for MEF cells [168, 210]. Overexpression of the isoform MBD3b/NuRD only or MBD3b/NuRD in combination with Nanog facilitated reprogramming with accelerated kinetics [210], which was also in contrast to the previous studies [46, 207]. These differences could be attributed to the use of different reprogramming cassettes, culture conditions, cell types or altered reprogramming factor stoichiometry.
In addition to this, two independent studies indicated that subunit MBD2/NuRD also has a substantial effect on reprogramming [211, 212]. Elucidating the molecular mechanism of reprogramming, Lee and colleagues showed that MBD2 binds to the Nanog promoter to suppress its transcriptional activation and thereby giving rise to partially reprogrammed cells [211]. Moreover, overexpression of miR-302 (miR-302 cluster without miR-367) ensured the generation of fully reprogrammed iPSCs as this relieved MBD2-mediated transcriptional repression of Nanog [211]. However, these results [211, 212] were not in line with the other studies [165, 168] where no substantial increase in iPSC colonies was established on silencing of NuRD/MBD. Explaining this disparity, Marto lab presumed an isoform-specific function of MBD2 (MBD2 has two isoforms: MBD2a and MBD2c) on reprogramming [212]. This study revealed that MBD2a preferentially cooperated with the repressive NuRD complex and directed the human iPSCs towards lineage specification, whereas, overexpression of MBD2c promoted reprogramming of fibroblasts to iPSCs [212]. Further, this study showed that the 3’-UTR of MBD2a was a direct target of miR-302 and its expression was under the control of miR-302 [212], which resulted in downregulation of MBD2 and upregulation of Nanog expression [211, 212]. These studies highlighted the importance of specific isoforms in differentiation or reprogramming, and further studies will reveal the epigenetic and transcriptional alterations in response to deletion of these isoforms to identify downstream targets that play a crucial role in lineage-specification or in induction of pluripotency.
Recently, one more key constituent of the NuRD complex Methyl-CpG-binding protein 2 (MeCP2) has been implicated in cell reprogramming [213]. MeCP2 interacts with co-repressor complexes and acts as a transcriptional repressor [214]. Downregulation of MeCP2 resulted in improved reprogramming efficiency via activation of cell cycle genes by ribosomal proteins and stimulation of IGF1/AKT/mTOR signaling in the initial stage of reprogramming [213]. Therefore, the investigation of other members of the NuRD complex should also be carried out in the context of reprogramming to understand their molecular function.
Histone Chaperones
Chromatin assembly factor (CAF-1) is a highly conserved protein complex comprising of p40, p60 and p150 subunits which enable de novo nucleosome assembly onto newly replicated DNA in vitro [215, 216]. Depletion of this histone chaperone CAF-1 in ESCs resulted in altered epigenetic histone methylation marks at the pericentric heterochromatin, indicating its role in heterochromatin organization [217]. To identify the reprogramming barriers, a comprehensive study performed two independent RNAi screens and found CAF-1 (Chaf1a and Chaf1b subunits of CAF-1), Ube2i and Setdb1 as the most prominent hits [168]. Investigating specifically the role of CAF-1 in reprogramming, the study showed that CAF-1 depletion resulted in a decrease in H3K9me3 marks at specific somatic heterochromatin areas called as ‘reprogramming-resistant regions’ to make the chromatin more accessible for efficient transcriptional activation by core reprogramming factors [168]. The decrease in H3K9me3 modifications at specific sites partly removed this epigenetic barrier [218], and resulted in an improved efficiency in a short span of only four days [168]. Together, this study indicates that CAF-1 contribute to the maintenance of somatic cell identity and act as a potent reprogramming barrier.
Recently, one more histone chaperone FACT (facilitates chromatin transcription; comprising of Spt16/SUPT16H and Pob3/SSRP1) was identified as a reprogramming barrier in Caenorhabditis elegans and human cells [219]. Unlike other reprogramming barriers that repress gene expression, this chromatin regulator remodels the tetranucleosomal unit of chromatin fibres to facilitate gene expression [220, 221]. FACT collaborates with other histone chaperones, CAF-1 and Rtt106, to deposit new histones onto replicating DNA and participate in replication-coupled nucleosome assembly [222]. Subsequently, it was shown that the intact human FACT complex performs destabilization of the nucleosome and preserve its integrity at the single nucleosome level during DNA replication and transcription [223]. From a reprogramming perspective, depletion of FACT leads to reduced expression of previously identified reprogramming barriers [219], such as repressive chromatin regulators, SUV39H1/2 and NR2F1 [165], CAF1 [168], PTPN11 [57], PRRX1 [224] and SUMO2 [225]. Consequently, downregulation of FACT resulted in a more accessible chromatin structure that enabled the transcription of reprogramming-inducing factors, demonstrating the role of FACT to safeguard somatic cell identity [219].
Other Reprogramming Barriers
Cell Adhesion and Motility
A Disintegrin And Metalloproteinase (ADAM) family members are multidomain transmembrane proteins crucial for numerous biological processes such as cell adhesion, migration, signaling and proteolysis [226]. This family of proteins has various domains: a pro-domain, a metalloproteinase, a disintegrin, a cysteine-rich, an epidermal-growth factor like, a transmembrane domain and a C-terminal cytoplasmic tail [227]. Recently, Ramalho-Santos and co-workers demonstrated that the knockdown of ADAM7, ADAM21 or ADAM29 significantly improved the reprogramming efficiency of human fibroblasts to iPSCs, with the most prominent effect observed for ADAM29 [57]. To identify the domain(s) in the ADAM family responsible for this inhibitory effect on reprogramming, the authors mutated the key domains and found that mutation in the disintegrin domain obliterates the adverse effect of ADAM29 on reprogramming efficiency [57]. These results established that it is the disintegrin domain in the ADAM family that serve as a reprogramming barrier.
Clathrin-Mediated Endocytosis
Endocytosis is a cellular process in all eukaryotic cells that involves the internalization of proteins (such as transporters, receptors and channels), nutrients and membrane-associated molecules [228, 229]. The main endocytic pathway, clathrin-mediated endocytosis, regulate the uptake of many cell surface proteins with the help of a clathrin coat [230, 231]. Recently, a study reported the negative influence of clathrin-mediated endocytosis on cellular reprogramming [57]. Downregulation of the endosomal surface proteins such as DRAM1, SLC17A5 or ARSD dramatically improved the reprogramming efficiency [57]. In addition, the inclusion of clathrin-specific inhibitors, Pitstop1 and Pitstop2, hampers clathrin-mediated endocytosis exhibiting the same effect, i.e. a significant increase in the reprogramming efficiency.
Earlier, it has been shown that clathrin-mediated endocytosis positively regulated the activation of Tgf-β signaling via recycling the receptors [228, 232]. Also, Tgf-β signaling is a well-known barrier of reprogramming that blocks MET [32, 62]. Thus, clathrin-mediated endocytosis impedes reprogramming by preventing MET at an early phase via activation of Tgf-β signaling. These outcomes establish that clathrin-mediated endocytosis is a potent reprogramming barrier, and inhibition of endocytosis is likely to enhance reprogramming efficiency.
The Ubiquitin-Proteasome Pathway
The Ubiquitin-Proteasome Pathway (UPP) is a key mechanism that mediates the rapid intracellular protein degradation in the mammalian cytosol and nucleus [233, 234]. This pathway involves two distinct sequential steps to carry out the degradation of the protein: first, the covalent attachment of ubiquitin molecules to a target protein; second, the degradation of the ubiquitinated target protein by an ATP-dependent large supramolecular proteolytic complex, the 26S proteasome [235, 236]. The initial observation was that the proteasome restricted non-specific transcriptional initiation through sequestration of transcription factors from their target sequences [237]. Further studies claimed that core pluripotency regulators that control self-renewal and differentiation of ESCs were ubiquitinated by molecular mechanisms that are still unidentified [238, 239].
To investigate the role of UPP in a reprogramming perspective, Buckley and colleagues demonstrated that silencing a member of the UPP, an E3 ubiquitin ligase Fbxw7, promoted the reprogramming efficiency from MEFs to iPSCs through stabilization of c-Myc and a subsequent rise in the levels of the pluripotency factors Oct4, Nanog and Sox2 [240]. Concurrently, Fbxw7, and another E3 ubiquitin ligase Wwp2, were also reported to be repressed by a microRNA miR-25, and therefore, overexpression of miR-25 in MEF cells resulted in the enhanced formation of iPSCs [241]. Earlier, both Fbxw7 and Wwp7 were reported to promote degradation of pluripotency-associated genes Oct4 [139, 238, 242], c-Myc [243] and Klf5 [244]. As a result, enhancement in the formation of iPSCs due to overexpression of miR-25 could be due to prevention of the degradation of Oct4, c-Myc and Klf5 by Fbxw7 and Wwp2 [241]. These studies concluded that Fbxw7 and Wwp2 acts as a barrier to cell reprogramming.
In line with these studies, a subsequent study identified multiple members of the UPP that either delay or affect the reprogramming efficiency [57]. The researchers demonstrated that individual knockdown of conjugating enzymes, UBE2D3 and UBE2E3, or ubiquitin ligase RNF40 significantly enhanced human iPSC generation. Consequently, the data from all these studies indicated that various constituents of the UPP act as negative regulators of reprogramming. This necessitates further studies on individual players of the UPP to identify their role in iPSC formation.
Sumoylation
Sumoylation, similar to ubiquitination, is a post-translational modification that involves the addition of SUMOs (small ubiquitin-like modifiers) to modify the function of a protein. In humans, there are three SUMO isoforms: SUMO 1–3 [245]. Among the other hits, a recent study identified SUMO2 as a top-scoring hit and showed its depletion resulted in enhanced and accelerated reprogramming [225]. Thus, SUMO2 acts as a potent barrier to reprogramming during the early-to-mid stage of iPSC generation [225]. Also, suppression of SUMO2 in combination with reprogramming enhancing small molecules generated iPSCs in less than 38 hours. In addition, an earlier study from the same group found an upstream SUMO-conjugating enzyme, Ubc9 (UBE2i) as a novel inhibitor of iPSC generation [168]. On the contrary, Yang lab established that Ubc9 is essential for efficient reprogramming as its depletion resulted in marked inhibition of iPSC induction [246]. Therefore, a further detailed investigation is required to unravel the importance of SUMO isoforms and other enzymes involved in sumoylation in the context of reprogramming.
Conclusion
iPSCs hold great promise for biomedical applications such as drug screening, disease modeling and personalized medicine. Various genetic and non-genetic approaches have been explored to derive integration-free iPSCs from numerous specialized cell types [11, 12]. Despite these advancements, in general, the generation of iPSCs from adult somatic cells is known to be an inefficient process. The poor efficiency and delayed kinetics of somatic cell reprogramming are serious concerns that restrict the generation of quality iPSCs for regenerative medicine. As discussed above, this is mainly due to the presence of numerous barriers that act as a roadblock that prevents efficient reprogramming. In addition to the aforementioned studies, other genes have also been identified as potent reprogramming roadblocks that prevent efficient iPSC generation (Table 3), but their biological function from a reprogramming perspective is not yet investigated. Also, all these players may act as a barrier during a specific stage or in a specific cell type but may be essential for reprogramming in the other stage or in a different cell type so detailed investigations are required to understand their role in different cell types and at different stages of the reprogramming. The better understanding of all these barriers and their consequences will pave the way to improve the reprogramming efficiency and kinetics. This knowledge will further help us develop safe strategies for efficient reprogramming that could generate genetically stable iPSCs. Eventually, this will result in the successful translation of this promising technology for various biomedical applications.
References
Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676.
Seifinejad, A., Tabebordbar, M., Baharvand, H., Boyer, L. A., & Hosseini Salekdeh, G. (2010). Progress and promise towards safe induced pluripotent stem cells for therapy. Stem Cell Reviews and Reports, 6(2), 297–306.
Young, W. (2012). Patient-specific induced pluripotent stem cells as a platform for disease modeling, drug discovery and precision personalized medicine. Journal of Stem Cell Research & Therapy, 01(S10), 2.
Singh, V. K., Kalsan, M., Kumar, N., Saini, A., & Chandra, R. (2015). Induced pluripotent stem cells: Applications in regenerative medicine, disease modeling, and drug discovery. Frontiers in Cell and Developmental Biology, 3, 2.
Omole, A. E., & Fakoya, A. O. J. (2018). Ten years of progress and promise of induced pluripotent stem cells: Historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ, 6, e4370.
Patel, M., & Yang, S. (2010). Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Reviews and Reports, 6(3), 367–380.
Walia, B., Satija, N., Tripathi, R. P., & Gangenahalli, G. U. (2012). Induced pluripotent stem cells: Fundamentals and applications of the reprogramming process and its ramifications on regenerative medicine. Stem Cell Reviews and Reports, 8(1), 100–115.
Hu, K. (2014). All roads lead to induced pluripotent stem cells: The technologies of iPSC generation. Stem Cells and Development, 23(12), 1285–1300.
Chhabra, A. (2017). Derivation of human induced pluripotent stem cell (iPSC) lines and mechanism of pluripotency: Historical perspective and recent advances. Stem Cell Reviews and Reports, 13(6), 757–773.
Saha, B., Borgohain, M. P., Dey, C., & Thummer, R. P. (2018). iPS cell generation: Current and future challenges. Annals of Stem Cell Research and Therapy, 1(2), 1007.
Borgohain, M. P., Haridhasapavalan, K. K., Dey, C., Adhikari, P., & Thummer, R. P. (2019). An insight into DNA-free reprogramming approaches to generate integration-free induced pluripotent stem cells for prospective biomedical applications. Stem Cell Reviews and Reports, 15(2), 286–313.
Haridhasapavalan, K. K., Borgohain, M. P., Dey, C., Saha, B., Narayan, G., Kumar, S., & Thummer, R. P. (2018). An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene, 686, 146–159.
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., & Yamanaka, S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131(5), 861–872.
Sridharan, R., Tchieu, J., Mason, M. J., Yachechko, R., & Kuoy, E. (2009). Resource role of the Murine reprogramming factors in the induction of pluripotency. Cell, 136(2), 364–377.
Papapetrou, E. P., Tomishima, M. J., Chambers, S. M., Mica, Y., Reed, E., & Menon, J. (2009). Stoichiometric and temporal requirements of Oct4 , Sox2 , Klf4 , and c-Myc expression for efficient human iPSC induction and differentiation. Proceedings of the National Academy of Sciences USA, 106(31), 12759–12764.
Tiemann, U., Sgodda, M., Warlich, E., Ballmaier, M., Schöler, H. R., Schambach, A., & Cantz, T. (2011). Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry Part A, 79(6), 426–435.
Carey, B. W., Markoulaki, S., Hanna, J. H., Faddah, D. A., Buganim, Y., Kim, J., et al. (2011). Short article reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Stem Cells, 9(6), 588–598.
Stefanovic, S., & Pucéat, M. (2007). Oct-3/4: Not just a gatekeeper of pluripotency for embryonic stem cell, a cell fate instructor through a gene dosage effect. Cell Cycle, 6(1), 8–10.
Karwacki-Neisius, V., Göke, J., Osorno, R., Halbritter, F., Ng, J. H., Weiße, A. Y., et al. (2013). Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell, 12(5), 531–545.
Stefanovic, S., Abboud, N., Désilets, S., Nury, D., Cowan, C., & Pucéat, M. (2009). Interplay of Oct4 with Sox2 and Sox17: A molecular switch from stem cell pluripotency to specifying a cardiac fate. Journal of Cell Biology, 186(5), 665–673.
Wen, W., Zhang, J.-P., Xu, J., Su, R. J., Neises, A., Ji, G.-Z., et al. (2016). Enhanced generation of integration-free iPSCs from human adult peripheral blood mononuclear cells with an optimal combination of episomal vectors. Stem Cell Reports, 6(6), 873–884.
Schmitt, C. E., Morales, B. M., Schmitz, E. M. H., Hawkins, J. S., Lizama, C. O., Zape, J. P., et al. (2017). Fluorescent tagged episomals for stoichiometric induced pluripotent stem cell reprogramming. Stem Cell Research and Therapy, 8(1), 132.
Nagamatsu, G., Saito, S., Kosaka, T., Takubo, K., Kinoshita, T., Oya, M., et al. (2012). Optimal ratio of transcription factors for somatic cell reprogramming. Journal of Biological Chemistry, 287(43), 36273–36282.
Yamaguchi, S., Hirano, K., Nagata, S., & Tada, T. (2011). Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cell Research, 6(2), 177–186.
Sommer, C. A., Christodoulou, C., Gianotti-Sommer, A., Shen, S. S., Sailaja, B. S., Hezroni, H., et al. (2012). Residual expression of reprogramming factors affects the transcriptional program and epigenetic signatures of induced pluripotent stem cells. PLOS ONE, 7(12), 1–10.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318(5858), 1917–1920.
Vogt, P. K. (2002). Fortuitous convergences: The beginnings of JUN. Nature Reviews Cancer, 2(6), 465–469.
Hilberg, F., Aguzzi, A., Howells, N., & Wagner, E. F. (1993). c-Jun is essential for normal mouse development and hepatogenesis. Nature, 365(6442), 179.
Liu, J., Han, Q., Peng, T., Peng, M., Wei, B., Li, D., et al. (2015). The oncogene c-Jun impedes somatic cell reprogramming. Nature Cell Biology, 17(7), 856–867.
Schreiber, M., Kolbus, A., Piu, F., Szabowski, A., Möhle-Steinlein, U., Tian, J., et al. (1999). Control of cell cycle progression by c-Jun is p53 dependent. Genes and Development, 13(5), 607–619.
Li, R., Liang, J., Ni, S., Zhou, T., Qing, X., Li, H., et al. (2010). A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell, 7(1), 51–63.
Samavarchi-Tehrani, P., Golipour, A., David, L., Sung, H. K., Beyer, T. A., Datti, A., et al. (2010). Functional genomics reveals a BMP-Driven mesenchymal-to-Epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell, 7(1), 64–77.
Li, D., Liu, J., Yang, X., Zhou, C., Guo, J., Wu, C., et al. (2017). Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell, 21(6), 819–833.e6.
Cadigan, K. M., & Waterman, M. L. (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harbor Perspectives in Biology, 4(11), a007906.
Pereira, L., Yi, F., & Merrill, B. J. (2006). Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Molecular and Cellular Biology, 26(20), 7479–7491.
Lluis, F., Ombrato, L., Pedone, E., Pepe, S., Merrill, B. J., & Cosma, M. P. (2011). T-cell factor 3 (Tcf3) deletion increases somatic cell reprogramming by inducing epigenome modifications. Proceedings of the National Academy of Sciences USA, 108(29), 11912–11917.
Cole, M. F., Johnstone, S. E., Newman, J. J., Kagey, M. H., & Young, R. A. (2008). Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes and Development, 22(6), 746–755.
Tam, W.-L., Lim, C. Y., Han, J., Zhang, J., Ang, Y.-S., Ng, H.-H., et al. (2008). T-Cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells, 26(8), 2019–2031.
Ye, S., Zhang, T., Tong, C., Zhou, X., He, K., Ban, Q., et al. (2017). Depletion of Tcf3 and Lef1 maintains mouse embryonic stem cell self-renewal. Biology Open, 6(4), 511–517.
Ho, R., Papp, B., Hoffman, J. A., Merrill, B. J., & Plath, K. (2013). Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Reports, 3(6), 2113–2126.
Lin, D., Ippolito, G. C., Zong, R. T., Bryant, J., Koslovsky, J., & Tucker, P. (2007). Bright/ARID3A contributes to chromatin accessibility of the immunoglobulin heavy chain enhancer. Molecular Cancer, 6(1), 23.
Webb, C. F., Bryant, J., Popowski, M., Allred, L., Kim, D., Harriss, J., et al. (2011). The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Molecular and Cellular Biology, 31(5), 1041–1053.
An, G., Miner, C. A., Nixon, J. C., Kincade, P. W., Bryant, J., Tucker, P. W., & Webb, C. F. (2010). Loss of bright/ARID3a function promotes developmental plasticity. Stem Cells, 28(9), 1560–1567.
Popowski, M., Templeton, T. D., Lee, B. K., Rhee, C., Li, H., Miner, C., et al. (2014). Bright/Arid3A acts as a barrier to somatic cell reprogramming through direct regulation of Oct4, Sox2, and Nanog. Stem Cell Reports, 2(1), 26–35.
Serrano, F., Calatayud, C. F., Blazquez, M., Torres, J., Castell, J. V., & Bort, R. (2013). Gata4 blocks somatic cell reprogramming by directly repressing Nanog. Stem Cells, 31(1), 71–82.
Fidalgo, M., Faiola, F., Pereira, C.-F., Ding, J., Saunders, A., Gingold, J., et al. (2012). Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proceedings of the National Academy of Sciences USA, 109(40), 16202–16207.
Ma, H., Ow, J. R., Tan, B. C. P., Goh, Z., Feng, B., Loh, Y. H., et al. (2014). The dosage of Patz1 modulates reprogramming process. Scientific Reports, 4, 7519.
Okita, K., Ichisaka, T., & Yamanaka, S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature, 448(7151), 313.
Wernig, M., Meissner, A., Foreman, R., Brambrink, T., Ku, M., Hochedlinger, K., et al. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature, 448(7151), 318.
Shu, J., Zhang, K., Zhang, M., Yao, A., Shao, S., Du, F., et al. (2015). GATA family members as inducers for cellular reprogramming to pluripotency. Cell Research, 25(2), 169–180.
Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Côté, J., & Wang, W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molecular Cell, 2(6), 851–861.
Zhang, W., Aubert, A., Gomez de Segura, J. M., Karuppasamy, M., Basu, S., Murthy, A. S., et al. (2016). The nucleosome remodeling and deacetylase complex NuRD is built from preformed catalytically active sub-modules. Journal of Molecular Biology, 428(14), 2931–2942.
Huangfu, D., Maehr, R., Guo, W., Eijkelenboom, A., Snitow, M., Chen, A. E., & Melton, D. A. (2008). Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nature Biotechnology, 26(7), 795–797.
Silva, J., Nichols, J., Theunissen, T. W., Guo, G., van Oosten, A. L., Barrandon, O., et al. (2009). Nanog is the gateway to the pluripotent ground state. Cell, 138(4), 722–737.
Allouba, M. H., ElGuindy, A. M., Krishnamoorthy, N., Yacoub, M. H., & Aguib, Y. E. (2015). NaNog: A pluripotency homeobox (master) molecule. Global Cardiology Science & Practice, 2015(3), 36.
Mikkelsen, T. S., Hanna, J., Zhang, X., Ku, M., Wernig, M., Schorderet, P., et al. (2008). Dissecting direct reprogramming through integrative genomic analysis. Nature, 454(7200), 49–55.
Qin, H., Diaz, A., Blouin, L., Lebbink, R. J., Patena, W., Tanbun, P., et al. (2014). Systematic identification of barriers to human iPSC generation. Cell, 158(2), 449–461.
Chronis, C., Fiziev, P., Papp, B., Butz, S., Bonora, G., Sabri, S., et al. (2017). Cooperative binding of transcription factors orchestrates reprogramming. Cell, 168(3), 442–459.e20.
Knaupp, A. S., Buckberry, S., Pflueger, J., Lim, S. M., Ford, E., Larcombe, M. R., et al. (2017). Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell, 21(6), 834–845.e6.
Meissner, A., Wernig, M., & Jaenisch, R. (2007). Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature Biotechnology, 25(10), 1177–1181.
Maherali, N., & Hochedlinger, K. (2009). Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Current Biology, 19(20), 1718–1723.
Subramanyam, D., Lamouille, S., Judson, R. L., Liu, J. Y., Bucay, N., Derynck, R., & Blelloch, R. (2011). Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnology, 29(5), 443–448.
Ichida, J. K., Blanchard, J., Lam, K., Son, E. Y., Chung, J. E., Egli, D., et al. (2009). A small-molecule inhibitor of Tgf-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell, 5(5), 491–503.
Vidal, S. E., Amlani, B., Chen, T., Tsirigos, A., & Stadtfeld, M. (2014). Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem Cell Reports, 3(4), 574–584.
Yuan, X., Wan, H., Zhao, X., Zhu, S., Zhou, Q., & Ding, S. (2011). Brief report: Combined chemical treatment enables Oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells, 29(3), 549–553.
Varelas, X. (2014). The hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development (Cambridge), 141(8), 1614–1626.
Ramos, A., & Camargo, F. D. (2012). The Hippo signaling pathway and stem cell biology. Trends in Cell Biology, 22(7), 339–346.
Tamm, C., Böwer, N., & Annerén, C. (2011). Regulation of mouse embryonic stem cell self-renewal by a Yes–YAP–TEAD2 signaling pathway downstream of LIF. Journal of Cell Science, 124(7), 1136–1144.
Lian, I., Kim, J., Okazawa, H., Zhao, J., Zhao, B., Yu, J., et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes and Development, 24(11), 1106–1118.
Chia, N. Y., Chan, Y. S., Feng, B., Lu, X., Orlov, Y. L., Moreau, D., et al. (2010). A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature, 468(7321), 316–320.
Qin, H., Blaschke, K., Wei, G., Ohi, Y., Blouin, L., Qi, Z., et al. (2012). Transcriptional analysis of pluripotency reveals the hippo pathway as a barrier to reprogramming. Human Molecular Genetics, 21(9), 2054–2067.
Varelas, X., Sakuma, R., Samavarchi-Tehrani, P., Peerani, R., Rao, B. M., Dembowy, J., et al. (2008). TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nature Cell Biology, 10(7), 837–848.
Zhao, B., Li, L., Tumaneng, K., Wang, C.-Y., & Guan, K.-L. (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes and Development, 24(1), 72–85.
Varelas, X., Miller, B. W., Sopko, R., Song, S., Gregorieff, A., Fellouse, F. A., et al. (2010). The Hippo pathway regulates Wnt/β-catenin signaling. Developmental Cell, 18(4), 579–591.
Heallen, T., Zhang, M., Wang, J., Bonilla-Claudio, M., Klysik, E., Johnson, R. L., & Martin, J. F. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science (New York, N.Y.), 332(6028), 458–461.
Sato, N., Meijer, L., Skaltsounis, L., Greengard, P., & Brivanlou, A. H. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nature Medicine, 10(1), 55–63.
Hao, J., Li, T. G., Qi, X., Zhao, D. F., & Zhao, G. Q. (2006). WNT/β-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Developmental Biology, 290(1), 81–91.
Xu, Z., Robitaille, A. M., Berndt, J. D., Davidson, K. C., Fischer, K. A., Mathieu, J., et al. (2016). Wnt/β-catenin signaling promotes self-renewal and inhibits the primed state transition in naïve human embryonic stem cells. Proceedings of the National Academy of Sciences USA, 113(42), E6382–E6390.
Lluis, F., Pedone, E., Pepe, S., & Cosma, M. P. (2008). Periodic activation of Wnt/β-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell, 3(5), 493–507.
Marson, A., Foreman, R., Chevalier, B., Bilodeau, S., Kahn, M., Young, R. A., & Jaenisch, R. (2008). Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell, 3(2), 132–135.
Silva, J., Barrandon, O., Nichols, J., Kawaguchi, J., Theunissen, T. W., & Smith, A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLOS Biology, 6(10), 2237–2247.
Li, W., Zhou, H., Abujarour, R., Zhu, S., Young Joo, J., Lin, T., et al. (2009). Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells, 27(12), 2992–3000.
Li, Z., & Rana, T. M. (2012). A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nature Communications, 3, 1011–1085.
Neganova, I., Shmeleva, E., Munkley, J., Chichagova, V., Anyfantis, G., Anderson, R., et al. (2016). JNK/SAPK signaling is essential for efficient reprogramming of human fibroblasts to induced pluripotent stem cells. Stem Cells, 34(5), 1198–1212.
Qi, X., Li, T.-G., Hao, J., Hu, J., Wang, J., Simmons, H., et al. (2004). BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proceedings of the National Academy of Sciences USA, 101(16), 6027–6032.
Li, J., Wang, G., Wang, C., Zhao, Y., Zhang, H., Tan, Z., et al. (2007). MEK/ERK signaling contributes to the maintenance of human embryonic stem cell self-renewal. Differentiation, 75(4), 299–307.
Lai, W.-H., Ho, J. C.-Y., Lee, Y.-K., Ng, K.-M., Au, K.-W., Chan, Y.-C., et al. (2010). ROCK inhibition facilitates the generation of human-induced pluripotent stem cells in a defined, feeder-, and serum-free system. Cellular Reprogramming, 12(6), 641–653.
Lin, Z., Liu, F., Shi, P., Song, A., Huang, Z., Zou, D., et al. (2018). Fatty acid oxidation promotes reprogramming by enhancing oxidative phosphorylation and inhibiting protein kinase C. Stem Cell Research and Therapy, 9(1), 47.
Staerk, J., Lyssiotis, C. A., Medeiro, L. A., Bollong, M., Foreman, R. K., Zhu, S., et al. (2011). Pan-Src family kinase inhibitors replace Sox2 during the direct reprogramming of somatic cells. Angewandte Chemie International Edition (English), 50(25), 5734–5736.
Kishigami, S., & Mishina, Y. (2005). BMP signaling and early embryonic patterning. Cytokine and Growth Factor Reviews, 16(3), 265–278.
Xu, R.-H., Chen, X., Li, D. S., Li, R., Addicks, G. C., Glennon, C., et al. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature Biotechnology, 20(12), 1261–1264.
Zhang, P., Li, J., Tan, Z., Wang, C., Liu, T., Chen, L., et al. (2008). Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood, 111(4), 1933–1941.
Richter, A., Valdimarsdottir, L., Hrafnkelsdottir, H. E., Runarsson, J. F., Omarsdottir, A. R., Oostwaard, D. W., et al. (2014). BMP4 promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells, 32(3), 636–648.
Chen, J. J., Liu, H., Liu, J., Qi, J., Wei, B., Yang, J., et al. (2012). H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nature Genetics, 45(1), 34.
Hamasaki, M., Hashizume, Y., Yamada, Y., Katayama, T., Hohjoh, H., Fusaki, N., et al. (2012). Pathogenic mutation of ALK2 inhibits induced pluripotent stem cell reprogramming and maintenance: Mechanisms of reprogramming and strategy for drug identification. Stem Cells, 30(11), 2437–2449.
Hayashi, Y., Hsiao, E. C., Sami, S., Lancero, M., Schlieve, C. R., Nguyen, T., et al. (2016). BMP-SMAD-ID promotes reprogramming to pluripotency by inhibiting p16/INK4A-dependent senescence. Proceedings of the National Academy of Sciences USA, 113(46), 13057–13062.
Lin, L., Liang, L., Yang, X., Sun, H., Li, Y., Pei, D., & Zheng, H. (2018). The homeobox transcription factor MSX2 partially mediates the effects of bone morphogenetic protein 4 (BMP4) on somatic cell reprogramming. Journal of Biological Chemistry, 293(38), 14905–14915.
Utikal, J., Polo, J. M., Stadtfeld, M., Maherali, N., Kulalert, W., Walsh, R. M., et al. (2009). Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature, 460(7259), 1145–1148.
Collado, M., Blasco, M. A., & Serrano, M. (2007). Cellular senescence in cancer and aging. Cell, 130(2), 223–233.
Marion, R. M., Strati, K., Li, H., Tejera, A., Schoeftner, S., Ortega, S., et al. (2009). Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell, 4(2), 141–154.
Kawamura, T., Suzuki, J., Wang, Y. V., Menendez, S., Morera, L. B., Raya, A., et al. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature, 460(7259), 1140–1144.
Hong, H., Takahashi, K., Ichisaka, T., Aoi, T., Kanagawa, O., Nakagawa, M., et al. (2009). Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature, 460(7259), 1132–1135.
Banito, A., Rashid, S. T., Acosta, J. C., De Li, S., Pereira, C. F., Geti, I., et al. (2009). Senescence impairs successful reprogramming to pluripotent stem cells. Genes and Development, 23(18), 2134–2139.
Jiang, J., Lv, W., Ye, X., Wang, L., Zhang, M., Yang, H., et al. (2012). Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Research, 23(1), 92–106.
Marión, R. M., Strati, K., Li, H., Murga, M., Blanco, R., Ortega, S., et al. (2009). A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature, 460(7259), 1149–1153.
Zhao, Y., Yin, X., Qin, H., Zhu, F., Liu, H., Yang, W., et al. (2008). Two supporting factors greatly improve the efficiency of human iPSC generation. Stem Cells, 3(5), 475–479.
Li, H., Collado, M., Villasante, A., Strati, K., Ortega, S., Cãamero, M., et al. (2009). The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature, 460(7259), 1136–1139.
Sarig, R., Rivlin, N., Brosh, R., Bornstein, C., Kamer, I., Ezra, O., et al. (2010). Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. Journal of Experimental Medicine, 207(10), 2127–2140.
Horikawa, I., Park, K. Y., Isogaya, K., Hiyoshi, Y., Li, H., Anami, K., et al. (2017). Δ133P53 represses P53-inducible senescence genes and enhances the generation of human induced pluripotent stem cells. Cell Death and Differentiation, 24(6), 1017–1028.
Hanna, J., Saha, K., Pando, B., Van Zon, J., Lengner, C. J., Creyghton, M. P., et al. (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature, 462(7273), 595–601.
Yoshihara, M., Hayashizaki, Y., & Murakawa, Y. (2017). Genomic instability of iPSCs: Challenges towards their clinical applications. Stem Cell Reviews and Reports, 13(1), 7–16.
Attwood, S., & Edel, M. (2019). iPS-cell technology and the problem of genetic instability—Can it ever be safe for clinical use? Journal of Clinical Medicine, 8(3), 288.
Abdelalim, E. M., & Tooyama, I. (2012). The p53 inhibitor, pifithrin-α, suppresses self-renewal of embryonic stem cells. Biochemical and Biophysical Research Communications, 420(3), 605–610.
Zalzman, M., Falco, G., Sharova, L. V., Nishiyama, A., Thomas, M., Lee, S. L., et al. (2010). Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature, 464(7290), 858–863.
Skamagki, M., Correia, C., Yeung, P., Baslan, T., Beck, S., Zhang, C., et al. (2017). ZSCAN10 expression corrects the genomic instability of iPSCs from aged donors. Nature Cell Biology, 19(9), 1037.
Aasen, T., Raya, A., Barrero, M. J., Garreta, E., Consiglio, A., Gonzalez, F., et al. (2008). Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnology, 26(11), 1276–1284.
Tüfekci, K. U., Öner, M. G., Meuwissen, R. L. J., & Genç, Ş. (2014). The role of MicroRNAs in human diseases BT. In M. Yousef & J. Allmer (Eds.), miRNomics: MicroRNA biology and computational analysis (pp. 33–50). Totowa: Humana Press.
Zeng, Z.-L., Lin, X., Tan, L.-L., Liu, Y.-M., Qu, K., & Wang, Z. (2018). MicroRNAs: Important regulators of induced pluripotent stem cell generation and differentiation. Stem Cell Reviews and Reports, 14(1), 71–81.
Wang, Y., Medvid, R., Melton, C., Jaenisch, R., & Blelloch, R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nature Genetics, 39(3), 380–385.
Kanellopoulou, C., Muljo, S. A., Kung, A. L., Ganesan, S., Drapkin, R., Jenuwein, T., et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes and Development, 19(4), 489–501.
Murchison, E. P., Partridge, J. F., Tam, O. H., Cheloufi, S., & Hannon, G. J. (2005). Characterization of Dicer-deficient murine embryonic stem cells. Proceedings of the National Academy of Sciences USA, 102(34), 12135–12140.
Anokye-Danso, F., Trivedi, C. M., Juhr, D., Gupta, M., Cui, Z., Tian, Y., et al. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell, 8(4), 376–388.
Miyoshi, N., Ishii, H., Nagano, H., Haraguchi, N., Dewi, D. L., Kano, Y., et al. (2011). Reprogramming of mouse and human cells to pluripotency using mature MicroRNAs. Cell Stem Cell, 8(6), 633–638.
Judson, R. L., Greve, T. S., Parchem, R. J., & Blelloch, R. (2013). MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nature Structural and Molecular Biology, 20(10), 1227–1237.
Pfaff, N., Fiedler, J., Holzmann, A., Schambach, A., Moritz, T., Cantz, T., & Thum, T. (2011). miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO reports, 12(11), 1153–1159.
Choi, Y. J., Lin, C., Ho, J. J., He, X., Okada, N., Bu, P., et al. (2011). miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nature Cell Biology, 13(11), 1353–1360.
He, X., He, L., & Hannon, G. J. (2007). The guardian’s little helper: MicroRNAs in the p53 tumor suppressor network. Cancer Research, 67(23), 11099–11101.
Lee, Y. L., Peng, Q., Fong, S. W., Chen, A. C. H., Lee, K. F., Ng, E. H. Y., et al. (2012). Sirtuin 1 facilitates generation of induced pluripotent stem cells from mouse embryonic fibroblasts through the miR-34a and p53 pathways. PLOS ONE, 7(9), e45633.
Melton, C., Judson, R. L., & Blelloch, R. (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature, 463(7281), 621–626.
Marson, A., Levine, S. S., Cole, M. F., Frampton, G. M., Brambrink, T., Johnstone, S., et al. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell, 134(3), 521–533.
Worringer, K. A., Rand, T. A., Hayashi, Y., Sami, S., Takahashi, K., Tanabe, K., et al. (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell, 14(1), 40–52.
Yang, C. S., Li, Z., & Rana, T. M. (2011). microRNAs modulate iPS cell generation. RNA, 17(8), 1451–1460.
Park, S. Y., Lee, J. H., Ha, M., Nam, J. W., & Kim, V. N. (2009). miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nature Structural and Molecular Biology, 16(1), 23–29.
Li, Z., Dang, J., Chang, K. Y., & Rana, T. M. (2014). MicroRNA-mediated regulation of extracellular matrix formation modulates Somatic cell reprogramming. RNA, 20(12), 1900–1915.
Wang, J., He, Q., Han, C., Gu, H., Jin, L., Li, Q., et al. (2012). P53-facilitated Mir-199a-3P regulates somatic cell reprogramming. Stem Cells, 30(7), 1405–1413.
Pfaff, N., Liebhaber, S., Möbus, S., Beh-Pajooh, A., Fiedler, J., Pfanne, A., et al. (2017). Inhibition of miRNA-212/132 improves the reprogramming of fibroblasts into induced pluripotent stem cells by de-repressing important epigenetic remodelling factors. Stem Cell Research, 20, 70–75.
Zhang, L., Zheng, Y., Sun, Y., Zhang, Y., Yan, J., Chen, Z., & Jiang, H. (2016). MiR-134-Mbd3 axis regulates the induction of pluripotency. Journal of Cellular and Molecular Medicine, 20(6), 1150–1158.
Hysolli, E., Tanaka, Y., Su, J., Kim, K. Y., Zhong, T., Janknecht, R., et al. (2016). Regulation of the DNA methylation landscape in human somatic cell reprogramming by the miR-29 family. Stem Cell Reports, 7(1), 43–54.
Xu, N., Papagiannakopoulos, T., Pan, G., Thomson, J. A., & Kosik, K. S. (2009). MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell, 137(4), 647–658.
Barta, T., Peskova, L., Collin, J., Montaner, D., Neganova, I., Armstrong, L., & Lako, M. (2016). Brief report: Inhibition of miR-145 enhances reprogramming of human dermal fibroblasts to induced pluripotent stem cells. Stem Cells, 34(1), 246–251.
Weinhold, B. (2006). Epigenetics: The science of change, A160–A167.
Armstrong, L. (2012). Epigenetic control of embryonic stem cell differentiation. Stem Cell Reviews and Reports, 8(1), 67–77.
Gonzalez, M., & Li, F. (2012). DNA replication, RNAi and epigenetic inheritance. Epigenetics, 7(1), 14–19.
Hochedlinger, K., & Jaenisch, R. (2015). Induced pluripotency and epigenetic reprogramming. Cold Spring Harbor Perspectives in Biology, 7(12), a019448.
Li, E., Bestor, T. H., & Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69(6), 915–926.
Okano, M., Bell, D. W., Haber, D. A., & Li, E. (1999). DNA Methyltransferases Dnmt3a and Dnmt3b are essential for De Novo methylation and mammalian development. Cell, 99(3), 247–257.
Mali, P., Chou, B. K., Yen, J., Ye, Z., Zou, J., Dowey, S., et al. (2010). Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells, 28(4), 713–720.
Pasha, Z., Haider, H. K., & Ashraf, M. (2011). Efficient non-viral reprogramming of myoblasts to stemness with a single small molecule to generate cardiac progenitor cells. PLOS ONE, 6(8), e23667–e23667.
Epsztejn-Litman, S., Feldman, N., Abu-Remaileh, M., Shufaro, Y., Gerson, A., Ueda, J., et al. (2008). De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nature Structural & Molecular Biology, 15(11), 1176.
Li, D., Guo, B., Wu, H., Tan, L., & Lu, Q. (2015). TET family of dioxygenases: Crucial roles and underlying mechanisms. Cytogenetic and Genome Research, 146(3), 171–180.
Gao, Y., Chen, J., Li, K., Wu, T., Huang, B., Liu, W., et al. (2013). Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell, 12(4), 453–469.
Hu, X., Zhang, L., Mao, S. Q., Li, Z., Chen, J., Zhang, R. R., et al. (2014). Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell, 14(4), 512–522.
Costa, Y., Ding, J., Theunissen, T. W., Faiola, F., Hore, T. A., Shliaha, P. V., et al. (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature, 495(7441), 370–374.
Doege, C. A., Inoue, K., Yamashita, T., Rhee, D. B., Travis, S., Fujita, R., et al. (2012). Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature, 488(7413), 652–655.
Sardina, J. L., Collombet, S., Tian, T. V., Gómez, A., Di Stefano, B., Berenguer, C., et al. (2018). Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell, 23(5), 727–741.
Esteban, M. A., Wang, T., Qin, B., Yang, J., Qin, D., Cai, J., et al. (2010). Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell, 6(1), 71–79.
Chung, T., Brena, R. M., Kolle, G., Grimmond, S. M., Berman, B. P., Laird, P. W., et al. (2010). Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells, 28(10), 1848–1855.
Gao, Y., Han, Z., Li, Q., Wu, Y., Shi, X., Ai, Z., et al. (2015). Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating micro RNA expression. The FEBS journal, 282(4), 685–699.
Blaschke, K., Ebata, K. T., Karimi, M. M., Zepeda-Martínez, J. A., Goyal, P., Mahapatra, S., et al. (2013). Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature, 500(7461), 222–226.
Chen, J., Guo, L., Zhang, L., Wu, H., Yang, J., Liu, H., et al. (2013). Vitamin C modulates TET1 function during somatic cell reprogramming. Nature Genetics, 45(12), 1504.
Bhutani, N., Brady, J. J., Damian, M., Sacco, A., Corbel, S. Y., & Blau, H. M. (2010). Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature, 463(7284), 1042–1047.
Morgan, H. D., Dean, W., Coker, H. A., Reik, W., & Petersen-Mahrt, S. K. (2004). Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. Journal of Biological Chemistry, 279(50), 52353–52360.
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D., & Grewal, S. I. S. (2001). Role of histone H3 Lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292(2001), 110–113.
Feldman, N., Gerson, A., Fang, J., Li, E., Zhang, Y., Shinkai, Y., et al. (2006). G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nature Cell Biology, 8(2), 188–194.
Onder, T. T., Kara, N., Cherry, A., Sinha, A. U., Zhu, N., Bernt, K. M., et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature, 483(7391), 598–602.
Soufi, A., Donahue, G., & Zaret, K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell, 151(5), 994–1004.
Sridharan, R., Gonzales-Cope, M., Chronis, C., Bonora, G., McKee, R., Huang, C., et al. (2013). Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nature Cell Biology, 15(7), 872–882.
Cheloufi, S., Elling, U., Hopfgartner, B., Jung, Y. L., Murn, J., Ninova, M., et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature, 528(7581), 218–224.
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K., & Jenuwein, T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410(6824), 116–120.
Zhu, J., Adli, M., Zou, J. Y., Verstappen, G., Coyne, M., Zhang, X., et al. (2013). Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell, 152(3), 642–654.
Sridharan, R., Gonzales-Cope, M., Chronis, C., Bonora, G., McKee, R., Huang, C., et al. (2013). Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nature Cell Biology, 15(7), 872–882.
Schultz, D. C., Ayyanathan, K., Negorev, D., Maul, G. G., & Rauscher, F. J. (2002). SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes and Development, 16(8), 919–932.
Miles, D. C., de Vries, N. A., Gisler, S., Lieftink, C., Akhtar, W., Gogola, E., et al. (2017). TRIM28 is an epigenetic barrier to induced pluripotent stem cell reprogramming. Stem Cells, 35(1), 147–157.
Wang, Q., Xu, X., Li, J., Liu, J., Gu, H., Zhang, R., et al. (2011). Lithium, an anti-psychotic drug, greatly enhances the generation of induced pluripotent stem cells. Cell Research, 21(10), 1424.
Cacchiarelli, D., Trapnell, C., Ziller, M. J., Soumillon, M., Cesana, M., Karnik, R., et al. (2015). Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell, 162(2), 412–424.
Sun, H., Liang, L., Li, Y., Feng, C., Li, L., Zhang, Y., et al. (2016). Lysine-specific histone demethylase 1 inhibition promotes reprogramming by facilitating the expression of exogenous transcriptional factors and metabolic switch. Scientific Reports, 6, 30903.
Wang, T., Chen, K., Zeng, X., Yang, J., Wu, Y., Shi, X., et al. (2011). The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell, 9(6), 575–587.
Liao, B., Bao, X., Liu, L., Feng, S., Zovoilis, A., Liu, W., et al. (2011). MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. Journal of Biological Chemistry, 286(19), 17359–17364.
He, J., Kallin, E. M., Tsukada, Y. I., & Zhang, Y. (2008). The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15Ink4b. Nature Structural and Molecular Biology, 15(11), 1169–1175.
Tzatsos, A., Pfau, R., Kampranis, S. C., & Tsichlis, P. N. (2009). Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the lnk4a/Arf locus. Proceedings of the National Academy of Sciences USA, 106(8), 2641–2646. https://doi.org/10.1073/pnas.0813139106.
Liang, G., He, J., & Zhang, Y. (2012). Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nature Cell Biology, 14(5), 457–466.
Jones, B., Su, H., Bhat, A., Lei, H., Bajko, J., Hevi, S., et al. (2008). The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLOS Genetics, 4(9), e1000190.
Ng, H. H., Ciccone, D. N., Morshead, K. B., Oettinger, M. A., & Struhl, K. (2003). Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proceedings of the National Academy of Sciences USA, 100(4), 1820–1825.
Wood, K., Tellier, M., & Murphy, S. (2018). DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules, 8(1), 11.
Mansour, A. A., Gafni, O., Weinberger, L., Zviran, A., Ayyash, M., Rais, Y., et al. (2012). The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature, 488(7411), 409–413.
Gaspar-Maia, A., Qadeer, Z. A., Hasson, D., Ratnakumar, K., Adrian Leu, N., Leroy, G., et al. (2013). MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nature Communications, 4, 1512–1565.
Pasque, V., Radzisheuskaya, A., Gillich, A., Halley-Stott, R. P., Panamarova, M., Zernicka-Goetz, M., et al. (2012). Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. Journal of Cell Science, 125(24), 6094–6104.
Barrero, M. J., Sese, B., Kuebler, B., Bilic, J., Boue, S., Martí, M., & Izpisua Belmonte, J. C. (2013). Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell Reports, 3(4), 1005–1011.
Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harbor Perspectives in Biology, 6(4), 1–26.
Kretsovali, A., Hadjimichael, C., & Charmpilas, N. (2012). Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells International, 2012, 1–10.
Shahbazian, M. D., & Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annual Review of Biochemistry, 76(1), 75–100.
Huynh, N. C.-N., Everts, V., & Ampornaramveth, R. S. (2017). Histone deacetylases and their roles in mineralized tissue regeneration. Bone Reports, 7, 33–40.
Huangfu, D., Osafune, K., Maehr, R., Guo, W., Eijkelenboom, A., Chen, S., et al. (2008). Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nature Biotechnology, 26(11), 1269–1275.
Liang, G., Taranova, O., Xia, K., & Zhang, Y. (2010). Butyrate promotes induced pluripotent stem cell generation. Journal of Biological Chemistry, 285(33), 25516–25521.
Pandian, G. N., Sato, S., Anandhakumar, C., Taniguchi, J., Takashima, K., Syed, J., et al. (2014). Identification of a small molecule that turns on the pluripotency gene circuitry in human fibroblasts. ACS Chemical Biology, 9(12), 2729–2736.
Zhang, Z., & Wu, W. S. (2013). Sodium butyrate promotes generation of human induced pluripotent stem cells through induction of the miR302/367 cluster. Stem Cells and Development, 22(16), 2268–2277.
Wei, T., Chen, W., Wang, X., Zhang, M., Chen, J., Zhu, S., et al. (2015). An HDAC2-TET1 switch at distinct chromatin regions significantly promotes the maturation of pre-iPS to iPS cells. Nucleic Acids Research, 43(11), 5409–5422.
Zhai, Y., Chen, X., Yu, D., Li, T., Cui, J., Wang, G., et al. (2015). Histone deacetylase inhibitor valproic acid promotes the induction of pluripotency in mouse fibroblasts by suppressing reprogramming-induced senescence stress. Experimental Cell Research, 337(1), 61–67.
Saunders, A., Huang, X., Fidalgo, M., Reimer, M. H., Faiola, F., Ding, J., et al. (2017). The SIN3A/HDAC corepressor complex functionally cooperates with NANOG to promote pluripotency. Cell Reports, 18(7), 1713–1726.
Pasque, V., Gillich, A., Garrett, N., & Gurdon, J. B. (2011). Histone variant macroH2A confers resistance to nuclear reprogramming. The EMBO Journal, 30(12), 2373–2387.
Pliatska, M., Kapasa, M., Kokkalis, A., Polyzos, A., & Thanos, D. (2018). The histone variant macroH2A blocks cellular reprogramming by inhibiting mesenchymal-to-epithelial transition. Molecular and Cellular Biology, 38(10), e00669–e00617.
Fang, H. T., El Farran, C. A., Xing, Q. R., Zhang, L. F., Li, H., Lim, B., & Loh, Y. H. (2018). Global H3.3 dynamic deposition defines its bimodal role in cell fate transition. Nature Communications, 9(1), 1537.
Jang, C. W., Shibata, Y., Starmer, J., Yee, D., & Magnuson, T. (2015). Histone H3.3 maintains genome integrity during mammalian development. Genes and Development, 29(13), 1377–1393.
Denslow, S. A., & Wade, P. A. (2007). The human Mi-2/NuRD complex and gene regulation. Oncogene, 26(37), 5433–5438.
Le Guezennec, X., Vermeulen, M., Brinkman, A. B., Hoeijmakers, W. A. M., Cohen, A., Lasonder, E., & Stunnenberg, H. G. (2006). MBD2 / NuRD and MBD3 / NuRD, two distinct complexes with different biochemical and functional properties. Molecular and Cellular Biology, 26(3), 843–851.
Menafra, R., & Stunnenberg, H. G. (2014). MBD2 and MBD3: Elusive functions and mechanisms. Frontiers in Genetics, 5, 428.
Luo, M., Ling, T., Xie, W., Sun, H., Zhou, Y., Zhu, Q., et al. (2013). NuRD blocks reprogramming of mouse somatic cells into Pluripotent stem cells. Stem Cells, 31(7), 1278–1286.
Rais, Y., Zviran, A., Geula, S., Gafni, O., Chomsky, E., Viukov, S., et al. (2013). Deterministic direct reprogramming of somatic cells to pluripotency. Nature, 502(7469), 65–70.
Zviran, A., Rais, Y., Mor, N., Novershtern, N., & Hanna, J. (2015). Mbd3/NuRD is a key inhibitory module during the induction and maintenance of naïve pluripotency. bioRxiv, 013961.
Dos Santos, R. L., Tosti, L., Radzisheuskaya, A., Caballero, I. M., Kaji, K., Hendrich, B., & Silva, J. C. R. (2014). MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell, 15(1), 102–110.
Lee, M. R., Prasain, N., Chae, H. D., Kim, Y. J., Mantel, C., Yoder, M. C., & Broxmeyer, H. E. (2013). Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cells, 31(4), 666–681.
Lu, Y., Loh, Y. H., Li, H., Cesana, M., Ficarro, S. B., Parikh, J. R., et al. (2014). Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell, 15(1), 92–101.
Zhang, W., Feng, G., Wang, L., Teng, F., Wang, L., Li, W., et al. (2018). MeCP2 deficiency promotes cell reprogramming by stimulating IGF1/AKT/mTOR signaling and activating ribosomal protein-mediated cell cycle gene translation. Journal of Molecular Cell Biology, 10(6), 515–526.
Jones, P. L., Veenstra, G. C. J., Wade, P. A., Vermaak, D., Kass, S. U., Landsberger, N., et al. (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genetics, 19(2), 187.
Takami, Y., Ono, T., Fukagawa, T., Shibahara, K., & Nakayama, T. (2007). Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Molecular Biology of the Cell, 18(1), 129–141.
Sauer, P. V, Gu, Y., Liu, W. H., Mattiroli, F., Panne, D., Luger, K., & Churchill, M. E. (2018). Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Research, 46(19), 9907–9917.
Houlard, M., Berlivet, S., Probst, A. V., Quivy, J. P., Héry, P., Almouzni, G., & Gérard, M. (2006). CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLOS Genetics, 2(11), 1686–1696.
Chen, J., Liu, H., Liu, J., Qi, J., Wei, B., Yang, J., et al. (2013). H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nature Genetics, 45(1), 34–42.
Kolundzic, E., Ofenbauer, A., Bulut, S. I., Uyar, B., Baytek, G., Sommermeier, A., et al. (2018). FACT sets a barrier for cell fate reprogramming in caenorhabditis elegans and human cells. Developmental Cell, 46(5), 611–626.
Orphanides, G., LeRoy, G., Chang, C.-H., Luse, D. S., & Reinberg, D. (1998). FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92(1), 105–116.
Li, W., Chen, P., Yu, J., Dong, L., Liang, D., Feng, J., et al. (2016). FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Molecular Cell, 64(1), 120–133.
Yang, J., Zhang, X., Feng, J., Leng, H., Li, S., Xiao, J., et al. (2016). The histone chaperone FACT contributes to dna replication-coupled nucleosome assembly. Cell Reports, 14(5), 1128–1141.
Chen, P., Dong, L., Hu, M., Wang, Y. Z., Xiao, X., Zhao, Z., et al. (2018). Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Molecular Cell, 71(2), 284–293.
Yang, C., Lopez, C. G., & Rana, T. M. (2011). Discovery of nonsteroidal anti-inflammatory drug and anticancer drug enhancing reprogramming and induced pluripotent stem cell generation. Stem Cells, 29(10), 1528–1536.
Borkent, M., Bennett, B. D., Lackford, B., Bar-Nur, O., Brumbaugh, J., Wang, L., et al. (2016). A serial shRNA screen for roadblocks to reprogramming identifies the protein modifier SUMO2. Stem Cell Reports, 6(5), 704–716.
Duffy, M. J., Mullooly, M., O’Donovan, N., Sukor, S., Crown, J., Pierce, A., & McGowan, P. M. (2011). The ADAMs family of proteases: New biomarkers and therapeutic targets for cancer? Clinical Proteomics, 8(1), 1–13.
Edwards, D. R., Handsley, M. M., & Pennington, C. J. (2009). The ADAM metalloproteinases. Molecular Aspects of Medicine, 29(5), 258–289.
Scita, G., & Di Fiore, P. P. (2010). The endocytic matrix. Nature, 463(7280), 464–473.
Watanabe, S., & Boucrot, E. (2017). Fast and ultrafast endocytosis. Current Opinion in Cell Biology, 47, 64–71.
Doherty, G. J., & McMahon, H. T. (2009). Mechanisms of endocytosis. Annual Review of Biochemistry, 78(1), 857–902.
McMahon, H. T., & Boucrot, E. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nature Reviews Molecular Cell Biology, 12(8), 517–533.
Di Guglielmo, G. M., Le Roy, C., Goodfellow, A. F., & Wrana, J. L. (2003). Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nature Cell Biology, 5(5), 410.
Reits, E. A. J., Benham, A. M., Plougastel, B., Neefjes, J., & Trowsdale, J. (1997). Dynamics of proteasome distribution in living cells. The EMBO journal, 16(20), 6087–6094.
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annual Review of Biochemistry, 70(1), 503–533.
Tu, Y., Chen, C., Pan, J., Xu, J., Zhou, Z.-G. G., & Wang, C.-Y. Y. (2012). The Ubiquitin Proteasome Pathway (UPP) in the regulation of cell cycle control and DNA damage repair and its implication in tumorigenesis. International Journal of Clinical and Experimental Pathology, 5(8), 726–738.
Okita, Y., & Nakayama, K. I. (2012). UPS delivers pluripotency. Cell Stem Cell, 11(6), 728–730.
Szutorisz, H., Georgiou, A., Tora, L., & Dillon, N. (2006). The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell, 127(7), 1375–1388.
Liao, B., & Jin, Y. (2010). Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Research, 20(3), 332.
Ramakrishna, S., Suresh, B., Lim, K.-H., Cha, B.-H., Lee, S.-H., Kim, K.-S., & Baek, K.-H. (2011). PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells and Development, 20(9), 1511–1519.
Buckley, S. M., Aranda-Orgilles, B., Strikoudis, A., Apostolou, E., Loizou, E., Moran-Crusio, K., et al. (2012). Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell, 11(6), 783–798.
Lu, D., Davis, M. P. A., Abreu-Goodger, C., Wang, W., Campos, L. S., Siede, J., et al. (2012). MiR-25 regulates Wwp2 and Fbxw7 and promotes reprogramming of mouse fibroblast cells to iPSCs. PLOS ONE, 7(8), e40938.
Xu, H. M., Liao, B., Zhang, Q. J., Wang, B. B., Li, H., Zhong, X. M., et al. (2004). Wwp2, An E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. Journal of Biological Chemistry, 279(22), 23495–23503.
Welcker, M., Orian, A., Grim, J. A., Eisenman, R. N., & Clurman, B. E. (2004). A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Current Biology, 14(20), 1852–1857.
Liu, N., Li, H., Li, S., Shen, M., Xiao, N., Chen, Y., et al. (2010). The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. Journal of Biological Chemistry, 285(24), 18858–18867.
Hay, R. T. (2005). SUMO: A history of modification. Molecular Cell, 18(1), 1–12.
Tahmasebi, S., Ghorbani, M., Savage, P., Gocevski, G., & Yang, X. J. (2014). The SUMO conjugating enzyme Ubc9 is required for inducing and maintaining stem cell pluripotency. Stem Cells, 32(4), 1012–1020.
Liao, J., Marumoto, T., Yamaguchi, S., Okano, S., Takeda, N., Sakamoto, C., et al. (2013). Inhibition of PTEN tumor suppressor promotes the generation of induced pluripotent stem cells. Molecular Therapy, 21(6), 1242–1250.
Yang, C. S., Chang, K. Y., & Rana, T. M. (2014). Genome-wide functional analysis reveals factors needed at the transition steps of induced reprogramming. Cell Reports, 8(2), 327–337.
Acknowledgments
We thank all the members of the Laboratory for Stem Cell Engineering and Regenerative Medicine (SCERM) for their excellent support. This work was supported by North Eastern Region – Biotechnology Programme Management Cell (NERBPMC), Department of Biotechnology, Government of India (BT/PR16655/NER/95/132/2015) and also by IIT Guwahati Institutional Top-Up on Start-Up Grant. The authors sincerely apologize to all scientists whose research could not be cited in this review due to space restrictions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no potential conflicts of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Haridhasapavalan, K.K., Raina, K., Dey, C. et al. An Insight into Reprogramming Barriers to iPSC Generation. Stem Cell Rev and Rep 16, 56–81 (2020). https://doi.org/10.1007/s12015-019-09931-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-019-09931-1