Abstract
Human hepatocytes display substantial functional inter-individual variation regarding drug metabolizing functions. In order to investigate if this diversity is mirrored in hepatocytes derived from different human pluripotent stem cell (hPSC) lines, we evaluated 25 hPSC lines originating from 24 different donors for hepatic differentiation and functionality. Homogenous hepatocyte cultures could be derived from all hPSC lines using one standardized differentiation procedure. To the best of our knowledge this is the first report of a standardized hepatic differentiation procedure that is generally applicable across a large panel of hPSC lines without any adaptations to individual lines. Importantly, with regard to functional aspects, such as Cytochrome P450 activities, we observed that hepatocytes derived from different hPSC lines displayed inter-individual variation characteristic for primary hepatocytes obtained from different donors, while these activities were highly reproducible between repeated experiments using the same line. Taken together, these data demonstrate the emerging possibility to compile panels of hPSC-derived hepatocytes of particular phenotypes/genotypes relevant for drug metabolism and toxicity studies. Moreover, these findings are of significance for applications within the regenerative medicine field, since our stringent differentiation procedure allows the derivation of homogenous hepatocyte cultures from multiple donors which is a prerequisite for the realization of future personalized stem cell based therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatotoxicity is a major concern in drug discovery and development and early detection of such problems would decrease costs and improve patient safety [1, 2]. Thus, for preclinical hepatotoxicity studies, physiologically relevant and robust human in vitro liver models are urgently needed. Currently, human primary hepatocytes (hphep), freshly isolated or cryopreserved (cryo), are considered the gold standard for evaluation of human hepatic drug transport, metabolism, clearance, and in vitro toxicity [3–5]. Hphep express a comprehensive set of enzymes and transporters involved in hepatic drug clearance [3–6]. However their rapid loss of functionality within a few days when cultured in vitro [6, 7], along with restricted availability, limits their utility. Other commonly used models are hepatoma cell lines such as HepG2 and HepaRG cells. However, these also have important shortcomings, e.g., low functionality (HepG2) or low predictivity of hepatotoxicity (HepaRG) [8].
Hepatocytes derived from human embryonic stem cells (hESC) [9] and human induced pluripotent stem cell (hiPSC) [10] represent an attractive alternative to hphep as in vitro liver models [11]. However, in order to be useful they must display sufficient expression levels of drug metabolizing enzymes and transporters as well as stable functionality over time in culture [12]. Another important feature is that hepatocytes derived from different hPSC lines should reflect the diversity of metabolic phenotypes present in the population [13]. Once achieved, defined panels of functionally diverse hepatocytes could be generated in a reproducible manner for drug discovery and toxicology studies. Such panels of hPSC-derived hepatocytes (hPSC-hep) could also be used for disease modeling, developmental studies, and cell-based therapies. For such applications, it is crucial to have a robust and efficient procedure for deriving homogenous hepatocyte cultures, in large scale, from many hPSC lines from multiple donors.
Several independent publications have described the derivation of hPSC-hep with varying differentiation efficiencies [14–31]. Some studies have also reported resistance of up to 20 % of hPSC lines to hepatic differentiation owing to inherent propensity of different lines to differentiate towards certain germ layers [30–36]. In order to compensate for this, adaptations of the protocol for individual hPSC lines have been required [30, 31] which makes it extremely time- and labor-intensive to use large panels of lines. Alternatively, researchers would be limited to use only a selection of hPSC lines with high intrinsic hepatic differentiation capacity.
Here, we present data from an evaluation of 25 hPSC lines derived from 24 separate donors using one standardized serum-and feeder-free differentiation protocol. In contrast to earlier reports [30–36], we could derive near-homogenous definitive endoderm (DE) and hepatocyte cultures from all 25 lines tested without any adaptations of the protocol to individual lines. To our knowledge, this is the first report of a virtually universal, highly efficient protocol towards DE and hepatocytes. Importantly, the hPSC-hep from different lines exhibit similar basic hepatic features. However, regarding drug metabolizing functions, they display metabolic diversity typically observed in hphep from different donors. This functional variation opens up the possibility to choose specific hPSC lines displaying certain phenotypes, e.g., with high or low Cytochrome P450 (CYP) activities or drug-induced liver injury (DILI)-sensitive phenotypes, for drug metabolism and toxicity studies. Also for applications within the field of regenerative medicine it is important to be able to derive hepatocyte cultures from multiple hPSC lines.
Materials and Methods
Human Pluripotent Stem Cell Lines
The hESC lines SA121, SA167, SA181, and SA461 (Takara Bio Europe AB, former Cellartis AB, Göteborg, Sweden; www.cellartis.com; www.clontech.com) were derived as previously described [37–39]. The Val9 hESC line was derived as previously described [40] and obtained from the National Stem Cell Bank of Spain (Banco Nacional de Lineas Celulares). The hiPSC line ChiPSC4 (Takara Bio Europe) was derived as described before using human dermal fibroblasts [29]. The hiPSC line ChiPSC6b was derived from fibroblast line P11031 (Ectycell/Cellectis SA, Paris, France) by transfecting with episomal vectors encoding OCT4, SOX2, KLF4, LIN28, and L-MYC using electroporation (Neon transfection system; Invitrogen). All other hiPSC lines were obtained from Cellectis SA and were derived from either peripheral blood cells (P11021) or adult dermal fibroblasts (all others) using either episomal (P11012, P11021, P11025) or retroviral reprogramming (all others) with OCT4, SOX2, KLF4, LIN28, and L-MYC. P11021 and ChiPSC20 were derived from the same donor. These hiPSC lines were initially established in a Matrigel-based culture system and subsequently transferred to the Cellartis DEF-CS Culture System. All hPSC lines were cultured under standard conditions in the Cellartis DEF-CS [41] with continuous passaging twice a week and were immuno-positive for OCT4, TRA1-60, TRA1-81, and SSEA-4, and immuno-negative for SSEA-1. Pluripotency was confirmed by in vitro differentiation. Karyotyping as described in [39] showed a normal chromosomal profile. The generation time was calculated as following: Generation time = (ln (harvested cells/seeded cells)) /ln2.
Hepatic Differentiation of hPSC
For hepatic differentiation, we applied a serum- and feeder-free procedure recapitulating liver development. First, hPSC are guided to differentiate into DE using the Cellartis DE Diff Kit (Takara Bio Europe AB; Y30030) which contains frozen complete media and frozen coating solution for the differentiation of hPSC to DE cells in 2D culture. The kit was used according to the manufacturer’s instructions. In short, on day 0, cell culture vessels were coated using the ready-to-use coating solution. hPSC were dissociated into a single cell suspension, re-suspended in warm DE diff Day 0 medium and seeded with a density of 30–40 K cells/cm2 (in 0.2 ml medium/cm2). On day 1, 2, 3, 4 and 6, medium changes were performed using warm DE diff Day 1, 2, 3, 4 and 6 medium, respectively. On day 7, the DE cells were enzymatically dissociated and re-seeded in the appropriate cell culture vessels. First, cell culture vessels were coated. To this end, Fibronectin (Sigma; F0895) was diluted 1:20 in D-PBS +/+ (Life Tech; 14080-048) to a final concentration of 50 μg/ml, added to cell culture vessels (0.15 ml/cm2), and incubated for >60 min at RT. Then, the DE cells were washed with D-PBS −/− (Life Tech; 14200-067), warm TrypLE™ Select (Life Tech; 12563-011) was added (0.1 ml/cm2) and incubated for 3–5 min at 37 °C until the cells had detached. Then 10 % KO-SR (Life Tech; 10828-028) in D-PBS −/− was added (0.1 ml/cm2) to achieve a 1:1 dilution of the cell suspension. Next, the cell suspension was centrifuged for 5 min at 300 g at RT, the supernatant was removed, and the cell pellet was re-suspended in warm Hepatocyte Thawing and Seeding medium (Cellartis Hepatocyte Diff Kit; Takara Bio Europe AB; Y30050). Then, excess coating was removed and 120–130 K DE cells/cm2 were seeded in 0.5 ml medium/cm2. On days 9 and 11 (counted from the start of hPSC differentiation), medium changes were performed using Hepatocyte Progenitor Medium (Cellartis Hepatocyte Diff Kit). On day 14 and onwards, medium changes were performed every second or third day using warm Williams Medium E (Life Tech; 32551-087) supplemented with 0.1 % PEST, HCM Single Quots (Lonza; CC-4182; GA-1000 was omitted), 10 ng/ml Oncostatin M (PromoKine; C-65020), 40 ng/ml Hepatocyte Growth Factor (PromoKine; C-64530), 0.1 μM Dexamethasone (Sigma; D8893), and 1.4 μM BIO (Sigma; B1686). If medium was changed every second day (preferred) 0.5 ml/cm2 was added and for medium change every third day 0.75 ml/cm2 was added.
Human Primary Hepatocytes
Cryoplateable hphep from four donors (BioreclamationIVT, Brussels, Belgium) were thawed and plated in InVitroGRO CP Medium (BioreclamationIVT) on rat tail Collagen I-coating (Invitrogen) according to the manufacturer’s instructions. CYP activity assays were started 4 and 52 h after plating, respectively. Samples for RNA extractions were collected directly post-thawing. Fresh hphep were isolated from excess liver tissue from seven patients who have undergone liver surgery (Sahlgrenska Hospital, Gothenburg, Sweden; derived with donor consent), and RNA was prepared directly after isolation.
CYP Activity Assay
CYP1A, 2C9, 2D6, and 3A activities were evaluated in hPSC-hep on day 29 (i.e., 29 days after start of differentiation) and in cryo hphep (4 and 52 h after plating) by the following assay. First, cells were washed twice with warm WME without phenol red (LifeTechnologies). Then, the assay was started by adding 110 μl/cm2 culture area of warm WME (without phenol red) supplemented with 0.1 % PEST, 25 mM HEPES, 2 mM L-glutamine, 26 μM phenacetin, 10 μM bufuralol, 9 μM diclofenac, and 3 μM midazolam. After 16 h, 100 μl of supernatant was collected and kept at −80 °C until LC/MS analysis of paracetamol, 4′-hydroxydiclofenac, hydroxybufuralol, and 3′-hydroxymidazolam was performed at Pharmacelsus (Saarbrücken, Germany). Samples from Val9 were analyzed at Advancell (Valencia, Spain) using an equivalent LC/MS method. Protein amount per well was quantified using the Pierce BCA Protein Assay Kit (ThermoScientific, Rockford, IL) according to the manufacturer’s instructions. Metabolite concentrations were normalized to protein amount per well and assay duration.
Immunocytochemistry
Cells were stained and examined as described previously [29]. Primary antibodies used were: rabbit anti-Albumin (1:1000, A0001, DAKO), rabbit anti-α1-Antitrypsin (1:200, A0012, DAKO), mouse anti-CK18 (1:100, M7010, DAKO), rabbit anti-HNF4α (1:300, sc-8987, SantaCruz Biotechnology), mouse anti-Oct3/4 (1:200, C-10, sc-5279, SantaCruz), and mouse anti-SSEA-4 (1:200, sc-21704, SantaCruz). Secondary antibodies used were: donkey anti-rabbit Alexa Fluor 488 IgG (1:1000, A21206), donkey anti-rabbit Alexa Fluor 594 IgG (1:1000, A21207), goat-anti-mouse Alexa Fluor 488 (1:1000, A11029), and donkey anti-mouse Alexa Fluor 488 IgG (1:500, A11029, all LifeTech). For OCT4 quantification of hPSC, representative DAPI and OCT4 pictures were analyzed using the Macro Process method for automated counting of nuclei (Image J). For SOX17 and OCT4 quantification of DE, representative SOX17 and OCT4 pictures were merged with the respective DAPI stainings, and the number of SOX17, DAPI and OCT4-stained nuclei was calculated using the Plug-in “Cell counter” method (Image J). Similarly, for HNF4α quantification of hPSC-hep, representative HNF4α pictures were merged with the respective DAPI stainings, and the number of HNF4α-DAPI double-stained nuclei versus HNF4α-negative and DAPI-positive nuclei was evaluated.
RNA Preparation, cDNA Synthesis, and Quantitative Real-Time PCR (qPCR)
RNA preparation, cDNA synthesis, and qPCR were performed as described previously [29]. Gene expression was analyzed using the following TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA): AAT (alpha-1 antitrypsin; Hs00165475_m1), Albumin (Hs009102225_m1), CEBPα (CCAAT/enhancer-binding protein α; Hs00269972_s1), CREBBP (CREB binding protein; Hs00231733_m1), CYP1A1 (Hs00153120_m1), CYP1A2 (Hs00167927_m1), CYP2C9 (Hs004260376_m1), CYP3A4 (Hs00430021_m1), CYP3A5 (Hs00241417_m1), CYP3A7 (Hs00426361_m1), GSTA1-1 (Glutathione S-Transferase A1; Hs00275575_m1), HNF4α (hepatocyte nuclear factor α; Hs00230853_m1), NTCP (sodium-taurocholate co-transporting polypeptide; Hs01081543_m1), OATP1B1 (organic anion-transporting polypeptide; OATP2; SLCO1B1; Hs00272374_m1), and UGT2B7 (UDP glucuronosyl-transferase 2B7; Hs00426592_m1). Expression levels were calculated using the ∆∆Ct method and normalized against a calibrator mix consisting of cDNA from hPSC, hPSC-derived embryoid bodies, hPSC-derived DE, hPSC-derived cardiomyocytes, hphep, HepG2, and HEK29c cells. For hPSC-derived DE, each sample was further normalized to CREBBP expression and presented as relative quantification. For hPSC-hep, samples were normalized to CEBPα expression which showed equivalent expression levels in hphep and hPSC-hep (which was not the case for CREBBP; data not shown) and presented as relative quantification. ∆∆Ct was transformed into fold change by the formula: fold change = 2-∆∆Ct.
Results
The DEF-CS Culture System is a Robust Culturing System for Multiple hPSC Lines
All hPSC lines in this study were maintained in DEF-CS, a defined, feeder- and serum-free culture system. Two lines (ChiPSC4, ChiPSC6b) were established in the DEF-CS, whereas all others were established in Matrigel- or feeder-based culture systems and subsequently transferred into the DEF-CS. Already after one or two passages, all transferred hPSC lines had adapted to the DEF-CS and were cultured under standard culture conditions thereafter. Since a highly homogenous hPSC population is crucial when starting differentiation experiments, we investigated the homogeneity of the hPSC cultures in the DEF-CS. All lines displayed typical hPSC morphology (representative pictures in Fig. 1A1-3) and showed no mRNA expression of differentiation markers such as SOX17 and CXCR4 (Fig. 2B3,4). Immunocytochemical stainings for the stem cell marker OCT4 (POU5F1) demonstrated a very high percentage of OCT4 immuno-positive nuclei ranging from 95 to 99 % which was supported by a homogenous staining for the stem cell surface marker SSEA-4 (representative pictures in Fig. 1b). In order to assess the robustness of the DEF-CS across multiple hPSC lines, we calculated the generation time, i.e., the time needed for a doubling of the cell population, for eleven hPSC lines by evaluating between seven and 95 passages per line (Fig. 1C2). Interestingly, very similar generation times with an average of 32.6 h (Fig. 1C1) were observed for these lines, and each line showed only small variation of the generation time between passages (error bars in Fig. 1C1). In addition, karyotyping of hPSC lines maintained in the DEF-CS for up to 29 passages showed no genetic aberrations (representative pictures in Fig. 1D1-6). Taken together, the DEF-CS is a highly robust culture system across multiple hPSC lines reproducibly generating highly homogenous undifferentiated hPSC cultures.
The DEF-CS is a robust culturing system for multiple hiPSC and hESC lines. a Representative phase contrast pictures of undifferentiated hPSC cultured in the DEF-CS: hiPSC lines ChiPSC4 (A1) and ChiPSC18 (A2), and the hESC line Val9 (A3). The scale bar represents 100 μM. b Immunocytochemical stainings of undifferentiated hPSC from the lines ChiPSC4 (B1-4), ChiPSC18 (B5-8), and Val9 (B9-12) for the nuclear stem cell marker OCT4 (B1,5,9; nuclear staining DAPI in B2,6,10) and the membrane-associated stem cell marker SSEA-4 (B3,7,11; nuclear staining DAPI in B4,8,12). The scale bar represents 50 μM. A high percentage of nuclei is OCT4 immuno-positive ranging from 95.1 % (ChiPSC4; Fig. B1,2), 95.9 % (Val9; Fig. B9,10) to 98.5 % (ChiPSC18; Fig. B5,6). c (C1) The generation time of 6 hiPSC lines (ChiPSC4, ChiPSC7, ChiPSC9, ChiPSC18, ChiPSC22, and P11012) and 5 hESC lines (SA121, SA167, SA181, SA461, and Val9) in the DEF-CS system is between 30 to 40 h with an average of 32.6 h (indicated by black line). Data is presented as mean ± SEM. (C2) The generation time was calculated based on 7 to 95 evaluated passages per line including both low and high passages (5 to 31). d Karyotype of undifferentiated hPSC from ChiPSC4 (D1, p14), ChiPSC18 (D2, p20), Val9 (D3, p29), SA167 (D4, p26), SA181 (D5, p22), and SA461 (D6, p28) after between 14 and 29 enzymatic passages in the DEF culture system. Abbreviations: DAPI 4′,6-diamidino-2-phenylindol, hPSC human pluripotent stem cells, p passage number
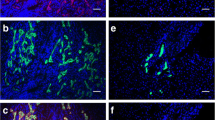
Derivation of homogenous definitive endoderm (DE) from 25 hPSC lines. a Representative phase contrast pictures of DE after 7 days of differentiation derived from the hiPSC lines ChiPSC4 (A1), ChiPSC6b (A2), ChiPSC7 (A3), ChiPSC8 (A4), ChiPSC9 (A5), ChiPSC10 (A6), ChiPSC11 (A7), ChiPSC13 (A8), ChiPSC15 (A9), ChiPSC17 (A10), ChiPSC18 (A11), ChiPSC19 (A12), ChiPSC20 (A13), ChiPSC21 (A14), ChiPSC22 (A15), ChiPSC23 (A16), ChiPSC24 (A17), P11012 (A18), P11021 (A19), P11025 (A20), and the hESC lines SA121 (A21), SA167 (A22), SA181 (A23), SA461 (A24), and Val9 (A25). All lines differentiated into a morphologically homogenous DE population. Occasionally, small bright clusters of dead cells were observed (e.g., A13, A17) that were eliminated in the subsequent enzymatic dissociation and re-seeding of the DE. The scale bar represents 100 μM. b QPCR analysis of DE derived from 24 hiPSC and hESC lines. Expression of the stem cell markers OCT4 (B1) and NANOG (B2) is strongly down-regulated in DE compared to undifferentiated hPSC (d0). In contrast, mRNA expression of DE markers CXCR4 (B3) and SOX17 (B4) is strongly up-regulated in DE compared to levels in undifferentiated hPSC. Low expression of the extraembryonic marker SOX7 (B5) indicates a low contamination of extraembryonic endoderm. c Immunocytochemical stainings for the DE marker SOX17 (C2,5,8) and the stem cell marker OCT4 (C3,6,9) reveal only very few OCT4 immuno-positive nuclei and a majority of SOX17 immuno-positive nuclei in DE derived from the hESC line SA121 and the hiPSC lines ChiPSC6b and P11025 on day 7 in the differentiation protocol. Nuclear counter-staining with DAPI (C1,4,7). The scale bar represents 50 μM. Abbreviations: CXCR4 C-X-C chemokine receptor type 4, DAPI 4′,6-diamidino-2-phenylindol, DE definitive endoderm
Derivation of Definitive Endoderm and Hepatocyte Cultures from 25 hPSC Lines
Next, we evaluated if we could derive homogenous hepatocyte cultures from a panel of 25 hPSC lines using one standardized procedure. First, we investigated the DE derived from all lines. After 7 days of differentiation, highly homogenous cultures displaying typical DE morphology could be obtained from all lines (Fig. 2A1-25). QPCR analyses showed that mRNA expression of the stem cell markers OCT4 and NANOG was strongly down-regulated (Fig. 2B1 2) in DE (NB: in DE derived from ChiPSC9, ChiPSC19 and SA461, low residual expression of OCT4 and NANOG is present), whereas expression of the DE markers SOX17 and CXCR4 was up-regulated in DE compared to hPSC (Fig. 2B3,4). Low SOX7 mRNA expression across the panel of hPSC lines indicated low contamination with extra-embryonic endoderm (Fig. 2B5). OCT4 and SOX17 immunostainings revealed that 97.4 % ± 0.002 (mean ± SEM) of the DAPI-stained nuclei were immuno-positive for the DE-marker SOX17 and only 2.6 % ± 0.002 for the stem cell marker OCT4 (representative pictures in Fig. 2c). Taken together, these data indicate a stringent first stage of the protocol which is crucial for obtaining homogenous hepatocyte cultures subsequently.
Next, hepatocyte cultures were derived from the panel of hPSC lines and investigated regarding their homogeneity. To this end, we investigated if we could detect other, non-hepatocyte, cell types in the cultures after 3 to 4 weeks of differentiation. In all lines tested, we detected a very low degree of contaminating cell types based on morphological inspections (Fig. 3A1-25). This was confirmed by the observation that on average 93.8 % ± 0.7 (mean ± SEM) of DAPI-stained nuclei were immuno-positive for the hepatic marker HNF4α (Fig. 3b; Suppl. Fig. 1) and that no OCT4 immuno-positive nuclei could be detected (data not shown). In accordance with this, the majority of cells were also immuno-positive for the hepatic markers Cytokeratin 18 and α1-Antitrypsin, and this again was very similar for different lines (Fig. 4A1,3; B1,3; Suppl. Fig. 2). The consistent α1a-Antitrypsin expression across different lines was confirmed by qPCR analysis: α1-Antitrypsin (SERPINA1) mRNA expression was similar across the 25 lines and varied only by about five fold (Fig. 4c). Compared to cryo hphep (directly after thawing), α1-Antitrypsin mRNA levels in the hPSC-hep were on average about seven times lower (Fig. 4c). Cryo and fresh hphep from different donors also displayed similar α1-Antitrypsin mRNA levels (Fig. 4c; Suppl. Fig. 3A). In contrast to the high percentage of HNF4α, CK18, and α1-Antitrypsin immuno-positive cells in the hPSC-hep cultures, we observed that only small groups or scattered cells were strongly immuno-positive for Albumin (Fig. 4A5, B5; Suppl. Fig. 2). In addition, hPSC-hep derived from some lines, e.g., ChiPSC18, (Fig. 4A5), appeared to have more Albumin-positive cells than others, e.g., P11012 (Suppl. Fig. 2B5). This observation was supported by the analysis of the Albumin mRNA levels in hPSC-hep which revealed that Albumin mRNA levels varied by about 30 fold between lines and that e.g., ChiPSC18-hep had higher Albumin mRNA levels than P11012-hep (Fig. 4d). Compared to cryo hphep, Albumin mRNA levels were on average about 400 times lower in hPSC-hep (Fig. 4d). Interestingly, donor variation of Albumin expression was also higher for hphep than variation of α1-Antitrypsin expression (10 to 15 times; cryo hphep in Fig. 4d; fresh hphep in Suppl. Fig. 3B).
Homogenous hepatocyte cultures derived from 25 hPSC lines. a Representative phase contrast pictures of near-homogenous hepatocyte populations obtained from the hiPSC lines ChiPSC4 (A1), ChiPSC6b (A2), ChiPSC7 (A3), ChiPSC8 (A4), ChiPSC9 (A5), ChiPSC10 (A6), ChiPSC11 (A7), ChiPSC13 (A8), ChiPSC15 (A9), ChiPSC17 (A10), ChiPSC18 (A11), ChiPSC19 (A12), ChiPSC20 (A13), ChiPSC21 (A14), ChiPSC22 (A15), ChiPSC23 (A16), ChiPSC24 (A17), P11012 (A18), P11021 (A19), P11025 (A20), and the hESC lines SA121 (A21), SA167 (A22), SA181 (A23), SA461 (A24), and Val9 (A25) on day 25 (A4, A13, A14, A22), 23 (A1, A10, A12, A20, A24) or 21 (all others) after start of differentiation. NB: hepatocytes derived from ChiPSC8 tend to grow in a slightly wavy manner so that some cells are out of focus in the phase contrast picture (A4). The scale bar represents 100 μm. Occasionally large droplets inside the hepatocytes were observed (see e.g., arrows in A6 and A25). Those were negative in an Oil Red O staining indicating that they do not contain neutral lipids or triglycerides (data not shown). Time lapse recordings revealed that these droplets were released by the hepatocytes into the medium leaving behind healthy looking hepatocytes. b Percentage of HNF4α immuno-positive nuclei in hepatocyte cultures derived from 16 hPSC lines (12 hiPSC and 4 hESC lines) on day 28 in the differentiation protocol. On average 93.8 % of nuclei are HNF4α-immuno-positive which is indicated by the black line. Abbreviations: HNF4α hepatocyte nuclear factor 4 alpha
Expression of hepatic markers in hPSC-derived hepatocytes after 28 days of differentiation. a, b Representative pictures of immunocytochemical stainings of hepatocytes derived from ChiPSC18 (A1-A6) and Val9 (B1-B6) for CK18 (A1, B1), α1-Antitrypsin (A3, B3), and Albumin (A5, B5), respectively. Nuclear counterstaining with DAPI is presented in A2, 4, and 6 for ChiPSC18, and in B2, 4, and 6 for Val9. Scale bars represent 50 μM. c, d Levels of mRNA expression of the liver-enriched markers α1-Antitrypsin (c), and Albumin (d) in hepatocytes derived from 25 hPSC lines and cryo hphep from four different donors (named 1–4) directly after thawing. Expression levels are normalized to CEBPα serving as a house-keeping gene and a calibrator mix (set as 1) and presented as relative quantification. Abbreviations: α1-AT α1-Antitrypsin, Alb Albumin, CEBPα CCAAT/enhancer binding protein α, CK18 Cytokeratin 18, cryo hphep cryoplateable human primary hepatocytes, DAPI 4′,6-diamidino-2-phenylindol
In order to investigate the reproducibility of the differentiation protocol, we repeatedly performed experiments for nine hPSC lines (ChiPSC17, ChiPSC22, P11012, P11032, SA121, SA461, Val9: all n = 2; ChiPSC18: n = 34; ChiPSC4: n = 102) and obtained near-homogenous hepatocyte cultures in all experiments performed during a 3 year period (Suppl. Fig. 4). This is in line with our previous studies in which we repeatedly could derive near-homogenous hepatocyte cultures from ChiPSC4 and SA181 [29, 42]. Taken together, all 25 hPSC lines evaluated in this study could be differentiated into near-homogenous DE and hepatocyte cultures.
Hepatocyte Cultures Derived from Multiple hPSC Lines Display Inter-individual Functional Variation
Next, we investigated if the hPSC-hep expressed drug-metabolizing enzymes and transporters, and started by testing for CYP activities. We observed that CYP1A and 3A activities in hPSC-hep, on average, approached similar levels as hphep cultured for 68 h (Fig. 5a, d), whereas CYP2C9 and 2D6 activities were, on average, about 100 times lower in hPSC-hep compared to hphep cultured for 68 h (Fig. 5b, c; see also [29]). Importantly, CYP activities varied substantially in hPSC-hep derived from different lines, similar to the variability observed in hphep from different donors (see error bars for hphep; Fig. 5). The largest variation was observed for CYP2C9 that varied by about 70 fold between the highest and lowest hPSC line, whereas CYP2D6 and CYP3A activities varied by about 15 fold, and CYP1A activity only by about 5 fold. In order to assess if this diversity was due to variations between experiments or between hPSC lines, we repeatedly derived hepatocytes from ChiPSC4 (n = 9) and ChiPSC18 (n = 4) and performed CYP activity assays on the different batches. Importantly, only small batch-to-batch variations were detected (see error bars in Fig. 5a–d and mean values ± SEM in Table 1), indicating a reproducible functionality of hPSC-hep derived in separate experiments (see also [29]) and that the diversity in CYP activities in hPSC-hep derived from different lines is due to inherent line variation and not inter-experiment variation.
CYP1A, 2C9, 2D6, and 3A activities in hepatocytes derived from 25 hPSC lines and four donors of cryo hphep. CYP1A (a), 2C9 (b), 2D6 (c), and 3A (d) activities in hepatocyte cultures after 29 days of differentiation were determined by LC/MS, normalized to the protein content per well and the duration of CYP activity assay and are presented as pmol metabolite per mg protein per minute. For two hPSC lines, analysis of several individual experiments were performed (ChiPSC4 n = 9; ChiPSC18 n = 4) and for these hPSC lines and for four donors of cryo hphep mean values ± SEM are presented. Abbreviations: CYP Cytochrome P450, cryo hphep cryoplateable human primary hepatocytes, LC/MS liquid chromatography / mass spectrometry
Since the CYP activity assay does not allow distinction of different CYP1A and CYP3A iso-enzymes, we performed qPCR for six CYP enzymes. Regarding the CYP1A family, we found that CYP1A1 mRNA levels were substantially higher (about 200 fold; Fig. 6a) in hPSC-hep than in cryo hphep (RNA prepared directly post-thawing), while the opposite was observed for CYP1A2 mRNA levels (about 1000 fold lower in hPSC-hep; Fig. 6b). This indicates that the CYP1A activity detected in hPSC-hep is probably largely due to CYP1A1 expression. Regarding the CYP3A family, we found that CYP3A5 mRNA expression was about 10 fold lower (Fig. 6e), CYP3A7 about 40 fold lower (Fig. 6f), and CYP3A4 about 3000 fold lower (Fig. 6d) in hPSC-hep compared to cryo hphep directly after thawing. This suggests that CYP3A activity measured in the hPSC-hep is probably mainly due to CYP3A5 expression which is supported by the observation that the lines with the highest CYP3A5 expression also displayed the highest CYP3A activities (ChiPSC4, CHiPSC10, CHiPSC18, P11021; Figs. 5d, 6e). In line with the lower CYP1A2 and CYP3A4 mRNA levels in hPSC-hep, also the CYP2C9 mRNA levels were about 100 fold lower in hPSC-hep (Fig. 6c), which is in agreement with about 100 fold lower CYP2C9 activity levels in hPSC-hep compared to cryo hphep (see Fig. 5b).
Inter-individual variation of mRNA expression in hepatocytes derived from 25 hPSC lines and four donors of cryoplateable human primary hepatocytes. QPCR analysis of mRNA expression of the drug-metabolizing enzymes CYP1A1, 1A2, 2C9, 3A4, 3A5, and 3A7 (a–f), the phase II enzymes GSTA1-1 and UGT2B7 (g, h), and the transporters NTCP and OATP1B1 (i, j) in hepatocytes derived from 25 hPSC lines after 28 days of differentiation and cryoplateable human primary hepatocytes from 4 different donors (RNA collected directly after thawing). Expression levels are normalized to CEBPα serving as a house-keeping gene and a calibrator mix (set as 1) and presented as relative quantification. For ChiPSC4 and SA181, analysis of CYP expression in 4 individual experiments was performed and mean values ± SEM are presented. Abbreviations: CEBPα CCAAT/enhancer binding protein α, cryo hphep cryoplateable human primary hepatocytes, CYP Cytochrome P450 enzyme, GSTA1-1 glutathione-S-transferase A1-1, NTCP Sodium taurocholate co-transporting polypeptide, OATP1B1 organic anion transporter family, member 1B1, SEM standard error of the mean, UGT2B7 UDP-glucuronosyltransferase 2B7
In contrast to the lower CYP expression, mRNA expression of the phase II enzymes GSTA1-1 and UGT2B7 was, on average, higher or similar in hPSC-hep compared to hphep (Fig. 6g, h). This is in accordance with a previous study reporting substantial GSTA1-1 expression and activity in hESC-derived hepatocytes comparable to levels in hphep [14]. Regarding transporters, we found that NTCP mRNA levels were, on average, higher in hPSC-hep than in hphep, whereas OATP1B1 mRNA levels were, on average, 200 fold lower in hPSC-hep (Fig. 6i, j; see [29] for similar results on ChiPSC4- and SA181-hep). As expected, substantial variations (between 10 and 50 fold) in CYPs, phase II enzymes, and transporters mRNA levels were observed in both cryo and fresh hphep from different donors (Fig. 6; Suppl. Fig. 3) and, importantly, the same was true for hPSC-hep from different lines (Fig. 6). The largest variations (>50 fold) were found for CYP3A4 and CYP3A7 mRNA levels both in cryo and fresh hphep as well as in hPSC-hep. In order to assess the inter-experiment variability of mRNA expression levels, we repeatedly differentiated one hiPSC and one hESC line (ChiPSC4 and SA181; 4 experiments per line) and analyzed the mRNA expression of 6 CYPs which varied substantially across the hPSC lines and hphep batches (Fig. 6). Importantly, only little variation in CYP mRNA levels was observed between separate experiments (error bars in Fig. 6; mean values ± SEM in Table 2) which is in agreement with the small inter-experiment variation observed for CYP activities (Fig. 5; Table 1). These results suggest that the differentiation protocol is robust and generates hPSC-hep cultures with low batch-to-batch variation. In conclusion, hPSC-hep derived from different lines show substantial metabolic diversity, well comparable to the variability observed in hphep from different donors.
Discussion
Here we present results from an evaluation of 25 hPSC lines which all, independent of donor background, reprogramming technique or sourced tissue, could be differentiated into near-homogenous hepatocyte cultures using one standardized protocol without adaptations for individual lines. Nine lines were repeatedly subjected to the protocol and generated homogenous hepatocyte cultures each time (Suppl. Fig. 4), indicating a high reproducibility and robustness of the protocol. This observation appears to be in contrast to earlier reports which describe a marked differentiation propensity of hPSC lines resulting in the inability of some lines to differentiate into homogenous DE or hepatocytes [30–36]. In our study, we did not observe such variability, suggesting that our procedure is generally applicable across hPSC lines originating from different donors. Earlier studies evaluated up to 35 or even 48 hPSC lines [30, 31], but used only up to 16 different donors. Thus, many lines in these studies were “sibling lines” (i.e., derived from the same donor) which have been reported to display similar differentiation behavior suggesting that the observed differentiation propensity is due to donor specificity [30, 36]. Therefore, in order to increase the diversity of the genetic background in our study material, we evaluated 25 hPSC lines derived from 24 different donors.
An important corner stone for the robust derivation of near-homogenous hepatocyte cultures in this study is the DEF-CS that reproducibly maintains homogenous hPSC cultures (Fig. 1a, b) which is crucial for successful and reproducible differentiation into various cell types [38]. All lines used in this study, except for two, were derived in other culture systems and subsequently transferred into the DEF-CS. Despite that, all lines could be cultured using standard conditions and had very similar generation times in the DEF-CS (Fig. 1c). Taken together, the DEF-CS robustly delivers high quality undifferentiated hPSC, supports easy adaptation of lines from other culture systems, and allows user-friendly scale-up of multiple lines. This, together with the serum- and feeder-free differentiation protocol, enables large-scale production of hepatocytes and opens up the possibility to develop cGMP protocols for hepatocyte derivation which both are essential for applications within the regenerative medicine field. However, for such applications a better understanding of the about 6 % of HNF4α-immuno-negative cells is required (Fig. 3b, Suppl. Fig. 1).
Starting with a stable hPSC culture system, we could successfully derive near-homogenous DE (Fig. 2) and hepatocyte cultures (Figs. 3 and 4; Suppl. Figs. 1, 2) from all hPSC lines tested. Several hepatic markers, e.g., HNF4α, Cytokeratin 18, and α1-Antitrypsin, were expressed at considerable levels across the panel of lines (Figs. 3 and 4; Suppl. Figs. 1, 2). Regarding genes associated with drug metabolism, we observed some to be expressed in a similar range in hPSC-hep as in hphep, e.g., CYP1A and 3A activities (Fig. 5a, d), and mRNA levels of CYP3A5, GSTA1-1, UGT2B7, and NTCP (Fig. 6e, g, h, i). Other functions were lower than in hphep, e.g., CYP2C9 and 2D6 activities (Fig. 5b, c), and CYP1A2, CYP3A4, and OATP1B1 mRNA levels (Fig. 6b, d, j). Also Albumin was expressed at much lower levels in hPSC-hep compared to hphep (Fig. 4a, b, d; Suppl. Fig. 2). A possible explanation for the lower functionality of hPSC-hep could be an immature fetal-like phenotype which is however contradicted by low mRNA levels of the fetal gene CYP3A7 in hPSC-hep (Fig. 6f). Another potential reason for the lower functionality is likely to be the 2D culture system that also causes hphep to rapidly lose functionality (e.g., compare CYP activities in hphep cultured for 20 and 68 h in Fig. 5). However, despite the lower functionality, hPSC-hep derived with the present protocol have proven to be useful for some applications such as chronic toxicity studies [42]. Nevertheless, for applications requiring high expression levels of multiple drug metabolism genes, it is clear that improvements are necessary and further studies are underway to address this issue [43].
In the present study, two hiPSC lines were included which were derived from different tissues of the same donor: ChiPSC20 from dermal fibroblasts and P11021 from blood cells. Interestingly, P11021- and ChiPSC20-derived hepatocytes displayed differences in CYP activities (Fig. 5) and gene expression (Fig. 6). Similar differences in hPSC derived from different tissues of the same donor and the potentially underlying mechanism, namely retention of epigenetic memory, have been described in earlier studies [44, 45]. Interestingly, the P11021-hPSC used in this screen were in a low passage number (p.6) and might retain some epigenetic memory. However, all other hPSC lines were in passage 10 or higher and therefore it is likely that their epigenetic memory has largely faded as reported in previous studies [46, 47]. Supporting this assumption, Kajiwara et al. reported no substantial differences between fibroblast- and blood-derived hiPSC lines originating from the same donor [30]. It is clear that additional studies focused on investigating the underlying mechanism are warranted.
Although some functions need to be improved for certain applications, considerable metabolic diversity could be observed in hPSC-hep from different lines (Figs. 5 and 6) which is reminiscent of the genotype-dependent metabolic diversity in humans (Figs. 5 and 6; Suppl. Fig. 3; see also e.g., [48]). For example, we observed that hPSC-hep from four lines had high CYP3A5 mRNA levels while most had lower CYP3A5 levels (Fig. 6e), which is in accordance with the reported CYP3A5 variations in humans [49]. Importantly, the hPSC-hep show low batch-to-batch variation (Figs. 5 and 6; Tables 1 and 2), suggesting that the observed variability is not due to inter-experiment variation but instead due to genotypic diversity of the donors. A similar finding was described in a recent study: 12 hiPSC lines were generated from hphep and subsequently differentiated into hepatocytes that reflected the donor-specific CYP metabolism capacity [50]. The observation that different genotypes translate into diverse hPSC-hep phenotypes opens up the possibility to specifically select hPSC lines with appropriate genotypes and compile panels of functionally diverse hPSC-hep representing different drug metabolizing phenotypes.
Conclusion
Here we report the derivation of near-homogenous DE and hepatocyte cultures from a large panel of hPSC lines, using a standardized protocol without any adaptation to individual lines. To our knowledge, this is the first time a virtually universal differentiation procedure towards DE and hepatocytes has been reported. In addition, hPSC-hep derived from different cell lines mirror the metabolic diversity typically observed in hphep from different donors. Based on advances in the hiPSC technology, hiPSC lines can today easily be derived from individuals with specific genotypes or phenotypes. This progress, in combination with the robust DEF-CS and hepatic differentiation protocols, allows the generation of homogenous hepatocyte cultures from customized panels of hiPSC lines at large scale and opens up new possibilities for drug discovery, toxicology studies, and regenerative medicine.
References
Kola, I., & Landis, J. (2004). Can the pharmaceutical industry reduce attrition rates? Nature Reviews Drug Discovery, 3, 711–715.
Lee, W. M. (2003). Acute liver failure in the United States. Seminars in Liver Disease, 23, 217–226.
Hewitt, N. J., Gomez-Lechon, M. J., Houston, J. B., et al. (2007). Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metabolism Reviews, 39, 159–234.
Gomez-Lechon, M. J., Donato, M. T., Castell, J. V., et al. (2004). Human hepatocytes in primary culture, the choice to investigate drug metabolism in man. Current Drug Metabolism, 5, 443–462.
Li, A. P., Lu, C., Brent, J. A., et al. (1999). Cryopreserved human hepatocytes: characterization of drug-metabolizing enzyme activities and applications in higher throughput screening assays for hepatotoxicity, metabolic stability, and drug-drug interaction potential. Chemico-Biological Interactions, 121, 17–35.
Richert, L., Liguori, M. J., Abadie, C., et al. (2006). Gene expression in human hepatocytes in suspension after isolation is similar to the liver of origin, is not affected by hepatocyte cold storage and cryopreservation, but is strongly changed after hepatocyte plating. Drug Metabolism and Disposition, 34, 870–879.
Rodriguez-Antona, C., Donato, M. T., Boobis, A., et al. (2002). Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica, 32, 505–520.
Gerets, H. H. J., Tilmant, K., Gerin, B., et al. (2012). Characterization of primary human hepatocytes, HepG2 cells and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biology and Toxicology, 28, 69–87.
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science, 282, 1145–1147.
Takahashi, K., Tanabe, K., Ohnuki, M., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872.
Baxter, M. A., Rowe, C., Alder, J., et al. (2010). Generating hepatic cell lineages from pluripotent stem cells for drug toxicity screening. Stem Cell Research, 5, 4–22.
Sartipy, P., & Björquist, P. (2011). Concise review: human pluripotent stem cell-based models for cardiac and hepatic toxicity assessment. Stem Cells, 29, 744–748.
Li, A. P. (2008). Human hepatocytes as an effective alternative experimental system for the evaluation of human drug properties: general concepts and assay procedures. ALTEX, 25, 33–42.
Ek, M., Söderdahl, T., Küppers-Munther, B., et al. (2007). Expression of drug metabolizing enzymes in hepatocyte-like cells derived from human embryonic stem cells. Biochemical Pharmacology, 74, 496–503.
Söderdahl, T., Küppers-Munther, B., Heins, N., et al. (2007). Glutathione transferases in hepatocyte-like cells derived from human embryonic stem cells. Toxicology In Vitro, 21, 929–937.
Hay, D. C., Fletcher, J., Payne, C., et al. (2008). Highly efficient differentiation of human embryonic stem cells to functional hepatic endoderm requires Activin A and Wnt3a signaling. Proceedings of the National Academy of Sciences of the United States of America, 105, 2301–2306.
Hay, D. C., Zhao, D., Fletcher, J., et al. (2008). Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells, 26, 894–902.
Song, Z., Cai, J., Liu, Y., et al. (2009). Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Research, 19, 1233–1242.
Basma, H., Soto-Gutiérrez, A., Yannam, G. R., et al. (2009). Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology, 136, 990–999.
Brolen, G., Sivertsson, L., Björquist, P., et al. (2010). Hepatocyte-like cells derived from human embryonic stem cells specifically via definitive endoderm and a progenitor stage. Journal of Biotechnology, 145, 84–94.
Duan, Y., Ma, X., Zou, W., et al. (2010). Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells, 28, 674–686.
Si-Tayeb, K., Noto, F. K., Nagaoka, M., et al. (2010). Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology, 51, 297–305.
Sullivan, G. J., Hay, D. C., Park, I. H., et al. (2010). Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology, 51, 329–335.
Touboul, T., Hannan, N. R., Corbineau, S., et al. (2010). Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology, 51, 1754–1765.
Rashid, S. T., Corbineau, S., Hannan, N., et al. (2010). Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. Journal of Clinical Investigation, 120, 3127–3136.
Ghodsizadeh, A., Taei, A., Totonchi, M., et al. (2010). Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Reviews and Reports, 6, 622–632.
Yildirimman, R., Brolén, G., Vilardell, M., et al. (2011). Human embryonic stem cell derived hepatocyte-like cells as a tool for in vitro hazard assessment of chemical carcinogenicity. Toxicological Sciences, 124, 278–290.
Funakoshi, N., Duret, C., Pascussi, J. M., et al. (2011). Comparison of hepatic-like cell production from human embryonic stem cells and adult liver progenitor cells: CAR transduction activates a battery of detoxification genes. Stem Cell Reviews, 7, 518–531.
Ulvestad, M., Nordell, P., Asplund, A., et al. (2013). Drug metabolizing enzyme and transporter protein profiles of hepatocytes derived from human embryonic and induced pluripotent stem cells. Biochemical Pharmacology, 86, 691–702.
Kajiwara, M., Aoi, T., Okita, K., et al. (2012). Donor-dependent variations in hepatic differentiation from human induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America, 109, 12538–12543.
Hannan, N. R., Segeritz, C. P., Touboul, T., et al. (2013). Production of hepatocyte-like cells from human pluripotent stem cells. Nature Protocols, 8, 430–437.
Hannan, N. R., Fordham, R. P., Syed, Y. A., et al. (2013). Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports, 1, 293–306.
Mikkola, M., Olsson, C., Palgi, J., et al. (2006). Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Developmental Biology, 6, 40.
Osafune, K., Caron, L., Borowiak, M., et al. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nature Biotechnology, 3, 313–315.
Itaba, N., Wairagu, P. M., Aramaki, N., et al. (2014). Nuclear receptor gene alteration in human induced pluripotent stem cells with hepatic differentiation propensity. Hepatology Research, 44(14), 408–439.
Bock, C., Kiskinis, E., Verstappen, G., et al. (2011). Reference maps of human embryonic and induced pluripotent stem cell variation enable high-throughput characterization of pluripotent cell lines. Cell, 144, 439–452.
Heins, N., Englund, M. C., Sjöblom, C., et al. (2004). Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells, 22, 367–370.
Sjögren-Jansson, E., Zetterström, M., Moya, K., et al. (2005). Large-scale propagation of four undifferentiated human embryonic stem cell lines in a feeder-free culture system. Developmental Dynamics, 233, 1304–1314.
Caisander, G., Park, H., Frej, K., et al. (2006). Chromosomal integrity maintained in five human embryonic stem cell lines after prolonged in vitro culture. Chromosome Research, 14, 131–137.
Aguilar-Gallardo, C., Poo, M., Gomez, E., et al. (2010). Derivation, characterization, differentiation, and registration of seven human embryonic stem cell lines (VAL-3, -4, -5, -6M, -7, -8, and -9) on human feeder. In Vitro Cellular and Developmental Biology - Animal, 46, 317–326.
Funa, N. S., Schachter, K. A., Lerdrup, M., et al. (2015). Beta-Catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell, 16(6), 639–652.
Holmgren, G., Sjögren, A. K., Barragan, I., et al. (2014). Long-term chronic toxicity testing using human induced pluripotent stem cell-derived hepatocytes. Drug Metabolism and Disposition, 42, 1401–1406.
Sartipy, P. (2013). Advancing pluripotent stem cell culture: it is a matter of setting the standard. Stem Cells and Development, 22, 1159–1161.
Kim, K., Doi, A., Wen, B., et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature, 467, 285–290.
Ohi, Y., Qin, H., Hong, C., et al. (2011). Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human induced pluripotent stem cells. Nature Cell Biology, 13, 541–549.
Bar-Nur, O., Russ, H. A., Efrat, S., et al. (2011). Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cell-derived from human beta-cells. Cell Stem Cell, 9, 17–23.
Lee, S. B., Seo, D., Choi, D., et al. (2012). Contribution of hepatic lineage stage-specific donor memory to the differential potential of induced mouse pluripotent stem cells. Stem Cells, 30, 997–1007.
Ponsoda, X., Pareja, E., Gomez-Lechon, M. J., et al. (2001). Drug biotransformation by human hepatocytes. In vitro/in vivo metabolism by cells from the same donor. Journal of Hepatology, 34, 19–25.
Kuehl, P., Zhang, J., Lin, Y., et al. (2001). Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature Genetics, 27, 383–391.
Takayama, K., Morisaki, Y., Kuno, S., et al. (2014). Prediction of interindividual differences in hepatic functions and drug sensitivity by using human induced pluripotent stem cell-derived hepatocytes. Proceedings of the National Academy of Sciences of the United States of America, 111, 16772–16777.
Acknowledgments
We thank other members of Takara Bio Europe AB and Skövde University who have provided assistance and technical help through-out this project.
Supported by the IMI-JU project MIP-DILI (grant 115336), the Seventh Framework Program project InnovaLiv (grant 278152), and the Knowledge Foundation (grants 2010/0069, 2012/0310, 2013/89).
Disclosure of Potential Conflicts of Interest
All authors are or have been employed by Takara Bio Europe AB (former Cellartis AB) when this study was performed.
Author Contributions
Annika Asplund: Collection, assembly, analysis, and interpretation of data, manuscript writing.
Anders Aspegren: Conception and design, data analysis and interpretation.
Arvind Pradip, Mariska van Giezen, Marie Rehnström, Susanna Jacobsson, Nidal Ghosheh, Dorra El Hajjam, Sandra Holmgren, Susanna Larsson, Jörg Benecke, Mariela Butron, Annelie Wigander: Collection, assembly and analysis of data.
Helena Choukair, Karin Noaksson: Analysis and interpretation of data.
Peter Sartipy, Josefina Edsbagge: Interpretation of data, manuscript writing.
Petter Björquist: Conception and design, manuscript writing.
Barbara Küppers-Munther: Conception and design, assembly, analysis, and interpretation of data, manuscript writing.
All: Final approval of manuscript
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Figure 1
Homogenous expression of the hepatic marker HNF4α in hepatocyte cultures derived from 16 hPSC lines. Immunocytochemical staining of hepatocyte cultures derived from 12 hiPSC and 4 hESC lines after 28 days of differentiation for the hepatic marker HNF4α. Nuclear counterstaining with DAPI. On average 93.8 % ± 0.7 (SEM) of DAPI-stained nuclei are immuno-positive for HNF4α. Percentages for the individual hPSC lines are given in the respective HNF4α-picture. The scale bars represent 100 μM. Abbreviations: DAPI = 4′,6-diamidino-2-phenylindol; HNF4α = hepatocyte nuclear factor 4α. (GIF 415 kb)
Suppl. Figure 2
Expression of hepatic markers in hPSC-derived hepatocytes after 28 days of differentiation. Immunocytochemical stainings of hepatocyte cultures derived from the hiPSC lines ChiPSC4 and P11012, and the hESC lines SA121 and SA461 for the hepatic markers Cytokeratin 18 (CK18), α1-Antitrypsin (a1-AT), and Albumin (Alb). Scale bars represent 100 μM. Abbreviations: α1-AT = α1-Antitrypsin; Alb = Albumin; CK18 = Cytokeratin 18. (GIF 106 kb)
Suppl. Figure 3
Inter-individual variation of mRNA expression of hepatic genes in freshly isolated primary human hepatocytes. QPCR analysis of mRNA expression of the hepatic markers α1-Antitrypsin and Albumin (A,B), the drug-metabolizing enzymes CYP1A1, 1A2, 2C9, 3A4, 3A5, and 3A7 (C-H), the phase II enzymes GSTA1-1 and UGT2B7 (I,J), and the transporters NTCP and OATP1B1 (K,L) in freshly isolated human primary hepatocytes from 7 different donors. Expression levels are normalized to CEBPα serving as a house-keeping gene and a calibrator mix (set as 1) and presented as relative quantification. Abbreviations: α1-AT = α1-Antitrypsin; CEBPα = CCAAT/enhancer binding protein α; CYP = Cytochrome P450 enzyme; GSTA1-1 = glutathione-S-transferase A1-1; hphep = human primary hepatocytes; NTCP = Sodium taurocholate co-transporting polypeptide; OATP1B1 = organic anion transporter family, member 1B1; UGT2B7 = UDP-glucuronosyltransferase 2B7. (GIF 97 kb)
Suppl. Figure 4
Homogenous hepatocyte cultures repeatedly derived from 9 hPSC lines. A Representative phase contrast pictures of homogenous hepatocyte populations obtained in repeated experiments from the hiPSC lines ChiPSC4, ChiPSC17, ChiPSC22, ChiPSC18, P11012, P11032, and the hESC lines SA121, SA461, Val9 on between day 21 and 28 after start of differentiation. Per repeated differentiation experiment one picture is shown. The scale bar represents 100 μm. (GIF 1444 kb)
Rights and permissions
About this article
Cite this article
Asplund, A., Pradip, A., van Giezen, M. et al. One Standardized Differentiation Procedure Robustly Generates Homogenous Hepatocyte Cultures Displaying Metabolic Diversity from a Large Panel of Human Pluripotent Stem Cells. Stem Cell Rev and Rep 12, 90–104 (2016). https://doi.org/10.1007/s12015-015-9621-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-015-9621-9