Abstract
Pigs are valuable animal models in pre-clinical research due to their anatomical and similarity to human-beings. Little is known about porcine embryonic development and porcine pluripotent stem cells. Recently, porcine-induced pluripotent stem cells (piPSCs) have been generated with Oct4 (Pou5f1), Sox2, Klf4 and c-Myc (termed OSKM, 4 F). Here, we found two other factors (Tbx3 and Nr5α2, termed TN), with important roles in piPSCs induction. They could improve the generation of piPSCs by supplementing these two factors on the basis of OSKM (OSKMTN, 6 F) orientated to mouse ESCs-like. Surprisingly, Nr5α2 alone could induce piPSCs formation in the presence or absence of c-Myc. These results suggested that Tbx3 and Nr5α2 may have vital roles in Sus scrofa and proposed new insights into pig pluripotent stem cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pigs are ideal animal models for a variety of diseases. However, porcine embryonic stem cells (pESCs) have not so far been successfully established, thus impeding the gene target research. Induced pluripotent stem cells were established by infecting mouse embryonic fibroblasts with retroviral vectors carrying four pluripotent genes Oct4, Sox2, c-Myc, and Klf4 [1]. Subsequently, a number of groups confirmed the reliability of this finding, and improved the quality of the mouse-induced pluripotent stem cells (miPSCs) [2, 3]. In 2009, pig-induced pluripotent stem cells (piPSCs) were established simultaneously by three groups with Yamanaka’s classic four-factor OSKM approach, and these cells have been proved as having pluripotency [4–6]. As far as we know, there exist many defects in piPSCs. For example, the pluripotency maintenance of piPSCs needs the expression of exogenous genes. Some groups claimed that piPSCs-derived chimeric offspring have been obtained; however, this still needs to be further confirmed [7, 8]. The developmental efficiency of piPSCs nuclear transfer (NT)-derived embryos was lower than that of porcine embryonic fibroblasts (PEFs) NT-derived ones both in vitro and in vivo, but this was not the case in mouse-induced pluripotent stem cells (miPSCs) [9, 10]. So, this suggested that the existed piPSCs may not be the true pluripotent stem cells. Therefore, factors and the medium which is used in piPSCs need to be further optimized.
The liver receptor homolog-1(Lrh-1, Nr5α2) is an orphan nuclear receptor family member [11] which could replace Oct4 in the process of miPSCs induction when using the classic OSKM approach [12], and mESCs like human-induced pluripotent stem cells (hiPSCs) could be induced by the addition of Nr5α2 and retinoic acid receptor gamma (RAR gamma) to OSKM [13]. Mouse epiblast stem cells (mEpiSCs) could be reset to the naïve state by ectopic expression of Nr5α2 [14], while T-box transcription factor, Tbx3, could improve the germline transmission of miPSC chimeras [15]. Therefore, in this study, Nr5α2 and Tbx3 have been chosen for induction of piPSCs and to determine whether these two transcription factors play similar roles in porcine stem cells.
We optimized the medium by modified the LBX medium which had roles in pluripotent states conversion and the improvements of hiPSCs [16]. This modified medium was named MX. In our study, TN was supplemented in the piPSCs induction system on the basis of OSKM. The results showed that TN increased the number of alkaline phosphatase-positive colonies. In addition, we observed the morphology of piPSCs, and some conventional pluripotency characteristics in 6 F group were very similar to mESCs.
Materials and Methods
Reagents and Media
The reagents and media were purchased from Life Technologies, R&D, Millipore and BD (all USA). The porcine embryonic fibroblast cells (PEF) were derived from Duroc and Landrace pigs which were provided by Harbin Sanyuan pig farm. ICR mice for mouse embryonic stem cells (MEF) were purchased from Beijing Vital River.
Cell Culture
The KOSR medium consisted of 76 % knockout Dulbecco’s modified eagle medium (DMEM), 20 % knockout serum replacement, 8 ng/ml bFGF, 2 mmol/L L-glutamine, 0.1 mmol /L β-mercaptoethanol, 1 % MEM nonessential amino acids, and 1 % penicillin-streptomycin. MX medium is mixture of 1:1 MX-1 and MX-2 medium, but the concentrations of bFGF and LIF were not changed. The MX-1 medium consisted of 76 % knockout DMEM, 20 % knockout serum replacement, 8 ng/ml bFGF, 2 mmol/L L-glutamine, 0.1 mmol /L β-mercaptoethanol, 1 % MEM nonessential amino acids, and 1 % penicillin-streptomycin. MX-2 medium consisted of 48 % DMEM/F12, 48 % Neurobasal, 1,000U/ml LIF (human), 0.5 % N2, 1 % B27, 0.5 mg/mL BSA and 1 % penicillin-streptomycin. FBS medium consisted of 86 % high glucose DMEM, 10 % FBS, 1 % NEAA, 2 mmol/L L- glutamine, and 1 % penicillin-streptomycin. Freezing medium consisted of 90 % FBS and 10 % dimethyl Sulfoxide (DMSO). Pig embryonic fibroblasts (PEF) were obtained from 33.5-day-old pig fetus via collagenase IV and DnaseIdigestion followed by culturing in FBS medium and then passaged by 0.25 % Trypsin-EDTA enzyme. Mouse embryonic fibroblasts (MEFs) were acquired from 13.5-day-old mouse embryo and were treated with 10 μg/mL mitomycin C as feeder layer. MEF and feeder layer were cultured in FBS medium. piPSCs were cultured in MX medium and dissociated with Tryple™. All the porcine and mouse cells were cultured in an incubator under the conditions of 5 % CO2 and 37 °C. These cells were frozen according to a program freezing method. In brief, after digestion, the cells were suspended with 500 μl freezing medium and subsequently transferred into a −80 °C freezer for storage.
Retroviral Transduction
pMX plasmids containing mouse Oct4, Sox2, Klf4, c-Myc, Nr5α2 and Tbx3, and VSV-G and Gag-Pol were purchased from Addgene. A total of 293 T cells were transfected with 42 μl LTX and 21 μl of PLUS regent. The ratio of VSV-G, Gag-Pol, and pMX was 5:10:16. Supernatants were collected at 24 and 48 h after transfecting, and filtered through a 0.45-μm filter. Finally, 104 PEF per well were infected with concentrated virus.
Immunofluorescence Analysis and Alkaline Phosphatase
piPSCs were fixed with 4 % paraformaldehyde for 30 min at room temperature. After three times washing in PBS, they were permeabilized with 0.5 % triton for 1 h at 37 °C, followed by blocking with 2 % bovine serum albumin (BSA). Subsequently, these cells were incubated with primary antibodies to Oct4 (Santa Cruz, Sc-8628, 1:200), Sox2 (Santa Cruz, sc-17320, 1:200) and Nanog (Abcam) overnight at 4 °C. The following day, the cells were washed three times with PBS and incubated with secondary antibodies for 1 h at 37 °C. Finally, nuclei were stained with Hochest for 8 min. A Nikon 80i was used for the analysis of these cells. Alkaline phosphatase (AP) staining was performed with BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime) following the manufacturer’s instructions.
Karyotype Analysis
When piPSCs reached 70∼80 % confluence, these cells were treated with 0.4 μg/ml colchicine for 2.5 h. Subsequently, these cells were harvested, plated into a gelatin-coated dish for 10 min, and further cultured in a hypotonic solution at 37 °C for 15 min. Later, piPSCs were fixed in fixative (3:1 methanol and glacial acetic acid) at room temperature for 5 min. The cells were collected by centrifugation at 800 rpm for 10 min, followed by cold fixative with 6 ml at 4 °C for 20 min, and the process repeated once again. Then, 2 ml of cold fixative was added and centrifuged. Finally, spreads were made by “huffing” on slides, holding at 45 °C, and dropping one drop of cells onto the top of the slide. After drying for 5 min, the sides were observed under a Nikon 80i microscope. Thirty samples were selected for chromosome number analysis.
Embryoid Body Formation
piPSCs were digested into single cells, transferred into gelatin-coated plates, and incubated in FBS medium for 10 min. Subsequently, the suspensions were harvested and transferred to a dish with FBS medium. The medium was changed every other day and, 6 days later, the EBs formed.
Teratoma Formation
Before harvesting, the piPSCs were transferred to gelatin-coated plates. Then, piPSCs were transferred into 1.5-ml eppendorf tube with 600 μl DPBS at 4 °C. Next, a 300 μl suspension containing 1 × 107 piPSCs was injected under the skin of non-obese diabetic/severe combined immune-deficient (NOD/SCID) mice. The majority of piPSCs lines at 15–20 passages can form teratomas. These teratomas were harvested for histological analysis at 4 weeks.
RT-PCR and Quantitative PCR
Total RNA was extracted using TRIzol reagent. RT-PCR was performed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, 4368814) according to the manufacturer’s instructions. Electrophoresis was performed on a 2 % agarose gel. QPCR was performed using SYBRPremix Ex TaqTM (TaKaRa) and the 7300 Real-Time PCR System (Applied Biosystems). Primers are listed in Table 1.
Results
Tbx3 and Nr5α2 Could Improve pig iPSCs Generation
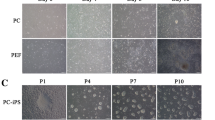
Porcine embryonic fibroblast cells (PEF_N and PEF-9) were infected by the retroviral factors OSKM (4 F), OSKMN, OSKMT, and OSKMNT (6 F). The infected cells were cultured in two kinds of culture media, KOSR medium (hESCs culturing) and MX medium. AP staining was carried out in these groups and positive colonies were counted (Fig. 1b). The number of AP positive colonies in the 6 F group were significantly increased compared with the other groups (Fig. 1c). On day 7 after infection, the majority of the colonies induced by 4 F were flattened similar to the hESCs colonies. However, 6 F-induced colonies were domed, similar to mESCs colonies. The colonies were dissociated mechanically and were then passaged using type III (6 F groups)or type IV collagenase (4 F groups)(Fig. 1a). Two and 12 pig iPSCs cell lines were established from 4 F and 6 F induction separately. The 6 F-induced pig iPSCs lines were morphologically similar to mESCs and could be digested into single cells, while the 4 F-induced cell lines were like hESCs whose single cell colony formation ability was poor (Fig. 1a). Attempts to culture piPSCs have been made in different feeder-free systems, in which the plates were treated with matrigel (BD), FBS, or gelatin, or in a matrix-free culture. The results showed that none of these systems is suitable for stable culture of 4 F-induced piPSCs, while the matrigel and matrix-free culturing system could produce 6 F-induced piPSCs which survived. In the latter subculturing, piPSCs could be cultured for more than 30 passages in the matrix-free systemwhereas, when cultured on matrigel, these cells readily formed EBs (Fig. 3e).
Tbx3 and Nr5α2 can improve piPSCs generation. a The PEF was infected by 6 F and 4 F, respectively. D1 shows infected PEF 1 day later. After 7 days, some colonies came out and picked for establishing cell lines. 6 F piPSCs were passaged routinely using Tryple™ and appeared domed. 4 F piPSCs were passaged routinely using 1 mg/mL type IV collagenase and appeared flattened. b AP staining for counting the positive colonies of diverse factors groups in MX medium. OSKM represents Oct4, Sox2, Klf4 and c-Myc; OSKMN represents Oct4, Sox2, Klf4, c-Myc and Nr5α2; OSKMT represents Oct4, Sox2, Klf4, c-Myc and Tbx3; OSKM represents Oct4, Sox2, Klf4, c-Myc, Nr5α2 and Tbx3. Each had three replicates. c Number of AP-positive colonies of the four groups in KOSR and MX medium. The numbers of each group were counted from 3 wells, repectively, and each well was counted for three times. Values with different superscripts are significantly different on the same histogram figure by one-way ANOVA. P < 0.05
Characterization of piPSCs
The piPSCs had a normal karyotype of 38 chromosomes both in 4 F and 6 F groups (Fig. 2b and Supplementary Fig. 1B). We characterized the pluripotency of these pig iPSCs by using immunofluorescence staining and RT-PCR. The results showed that the classic pluripotent markers (OCT4, SOX2 and NANOG) were expressed in both 4 F and 6 F groups (Fig. 2a and Supplementary Fig. 1A). RT-PCR showed that the exogenous genes were not silenced and the endogenous pluripotent genes including Oct4, Sox2 and Nanog were expressed in both the 4 F and 6 F groups. However, rex1 was only expressed in the 6 F group (Fig. 2c). Both the 4 F and 6 F-induced piPSCs could form embryoid bodies (EBs) which indicated the differentiation ability of piPSCs in vitro (Fig. 2d and Supplementary Fig. 1C). The in vivo differentiation ability of 6 F-induced piPSCs was checked by injection of these cells into severe combined immune-deficient (SCID) mice and detecting teratoma formation. Teratomas were observed in the 6 F group by 3 weeks, while in the 4 F group, teratomas appeared a little later. Histological examination showed that the teratomas contained three germ layers including hepatocyte (endoderm), cartilage (mesoderm), and nervous tissue (ectoderm) as shown by the arrows in Fig. 2e.
Characterization of piPSCs. a Immunostaining for pluripotency markers of 6 F piPSCs. Positive OCT4 (green), SOX2 (green) and NANOG (purple) were observed. DNA was stained with Hoechst 33342 (blue) and propidium iodide (PI, red). b Karyotype analyses of 6 F piPSCs. More than 80 % of the cells showed normal pig karyotype of 38 chromosomes. c RT-PCR for transgenes (mOct4, mSox2, mKlf4, mc-Myc, mNr5α2 and mTbx3) and endogenous (pOct4, pSox2, pNanog and pRex1) expression. The first column is the 6 F piPSCs and the second and third columns are 4 F piPSCs and H2O, respectively. d Embryoid body (EB) formation of 6 F piPSCs. Scale bars 200 μm. e Histological examination of stained teratomas derived from 6 F piPSCs. Nervous tissues represent the ectoderm; cartilage tissue represents the mesoderm; hepatocyte represents the endoderm (arrows)
Tbx3 and Nr5α2 Generated piPSCs That are Morphologically Similar to Mouse ESCs
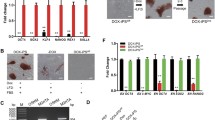
As the 6 F-induced piPSCs satisfied the conventional pluripotency, their characteristics were further detected. Morphologically, 6 F-induced piPSCs colonies were similar to the mESCs, while the 4 F-induced piPSCs colonies were similar to hESCs (Fig. 3a). Then, we assayed the growth of piPSCs by measuring the cell cumulative increase. The 6 F-induced piPSCs grew faster whose doubling time was 18.5 h similar to mESCs (Fig. 3b). To test the piPSCs viability, single-cell colony efficiency was performed and the results showed that the colony efficiency was decreased in the 4 F-induced piPSCs compared with the 6 F-induced piPSCs. Meanwhile, we also found that the single-cell cloning efficiency was decreased in piPSCs when cultured in KOSR medium compared with that in MX medium in both the 4 F and 6 F groups (Fig. 3c). To determine which signaling pathway involved in the regulation of single-cell colony efficiency of piPSCs, key downstream effector molecules of LIF and bFGF pathways were examined by using real-time PCR. The results showed that the expression of Lifr, Smad4, and Bmp4 were higher in the 6 F than in the 4 F group, and, reversely, the expression of Fgfr1, Fgfr2, and bFGF were higher in the 4 F than that in the 6 F piPSCs (Fig. 3d).
6 F piPSCs were similar to mESCs. a The morphology of 6 F and 4 F piPSCs. b Proliferation detection of the 6 F piPSCs. Double time: TD=txlog2/(logNtlogN0 ) = 18.5 h. c Single-cell cloning efficiency of piPSCs. 6 F and 4 F piPSCs were cultured in MX medium. d Quantitative RT-PCR for Fgfr1, Fgfr2, Lifr, Lif, Bmp4, and Smad4. The expressions of these genes were relative to the expression of β-actin. e 6 F piPSCs could be cultured in feeder-free system. The 4 F and 6 F piPSCs were cultured in three matrix (matrigel, FBS, gelatin) and without matrix. Scale bars 200 μm
PiPSCs Could be Induced by two Factors (Nr5α2 and c-Myc, MN) or a Single Factor (Nr5α2, N)
So far, we could generate piPSCs by 6 F efficiently. In order to use fewer transcription factors to induce piPSCs formation, we found that two factors, Nr5α2 together with c-Myc, could generate piPSCs in place of Oct4. MN-induced piPSCs had normal proliferative ability and could be passaged stably (Fig. 4a). Surprisingly, we found Nr5α2 alone could induce piPSCs generation but Oct4 alone never got piPSCs. Single-factor Nr5α2-induced piPSCs could be cultured routinely and AP staining was positive (Fig. 4b). The expression of Nr5α2 and c-Myc were detected in MN and N-induced piPSC by RT-PCR, and no other transgenes were found to be involved in this process (Fig. 4b). Further detection will be carried out in the future experiments
piPSCs generated with MN or N. a piPSCs generated with MN. D10 the morphology of the colonies on the tenth day after PEFs were infected. P5 piPSCs morphology at the fifth passage. AP piPSCs were positive for AP activity. b piPSCs generated with N. D10 the morphology of the colonies on the tenth day after PEFs were infected. P5 piPSCs morphology at the fifth passage. AP piPSCs were positive for AP activity. c RT-PCR for detection of transgenes
Discussion
Morphologically, 6 F-derived piPSCs were similar to mESCs but different from those derived by the 4 classical factors. The 6 F-induced piPSCs colonies were domed. The cells in these colonies were closely connected with each other. 6 F-induced piPSCs can be digested into single cells and cultured in a feeder-free system. The differences between 6 F-induced piPSCs and 4 F-induced piPSCs indicated that Nr5a2 and Tbx3 played an important role in establishing mESCs-like piPSCs. The 6 F-induced piPSCs possess better viability and more rapid proliferative ability than the 4 F-induced iPSCs. Nr5α2 can bind both the proximal enhancer and proximal promoter regions of Oct4 and regulate Oct4 expression in the epiblast stage of mouse embryonic development [17]. And it can replace Oct4 in miPSCs induction [12]. The combination of Nr5α2 and RAR gamma can increase the miPSCs generation due to the activation of endogenous Oct4 and can generate mESCs-like hiPSCs by maintaining Oct4 expression [13]. Therefore, in the induction of piPSCs, Nr5α2 may also play a similar role. It can enhance the expression of endogenous Oct4, and activate the LIF signal pathway (Fig. 3d). Tbx3 can maintain ESC self-renewal in an undifferentiated state by its persistent expression in the absence of LIF [18, 19] and can improve the germ-line competency of miPSCs [12]. Practically, 6 F piPSCs in this study had a dominant position in culture. Therefore, in this study, the existence of Nr5α2 and Tbx3 has promoted piPSCs to grow more like mESCs which were distinct from the 4 F piPSCs.
6 F-induced piPSCs’ mESCs-like pluripotent state can be proved through the signal genes analysis. Lifr and Smad4 expressions were higher in 6 F piPSCs compared to 4 F piPSCs (Fig. 3d). In mESCs, Lifr had a higher expression and Smad4 can maintain the pluripotent state through taking part in inhibiting ERK activity [20, 21]. These findings suggested that Nr5a2 and Tbx3 may have some functional roles in pig. Single cell colony formation ability can be used for transgenic porcine production. 6 F piPSCs can be cultured and passaged in a feeder-free system and this can accelerate the piPSCs application for transgenic research and screening for some drugs.
In this study, piPSCs have been successfully induced by two factors (MN) or a single factor (N). It has been reported that piPSCs have been obtained by other two factors (Oct4 and Klf4) [22, 23] and miPSCs can be obtained by Oct4 alone [24]. In murine, Nr5α2 can mediate pluripotent genes expression [25]. Nanog was able to rescue Nr5α2 knockdown but not Oct4 knockdown in ESCs [12]. So far, there has been no report on the generation of miPSCs by a single factor or two factors induction without Oct4 being involved. The MN and N piPSCs have not been detected for more pluripotent markers, but the proliferation ability of these cells indicated that Nr5α2 may had an important function in porcine stem cell which is similar to the role of Oct4 in mESCs.
Researchers have tried to establish porcine embryo stem cells (pESCs) for about 20 years, but it has proved to be very difficult [26, 27]. It seems that these pluripotent genes which are important in mouse and human ESCs, such as Oct4 and Nanog, do not have crucial roles in porcine ESCs, but some other pluripotent genes may affect the establishment of pESCs. In this study, we proved that Nr5α2 and Tbx3 play important roles in piPSCs induction. Tracking the expression of Nr5α2 and Tbx3 in the early porcine embryo and the analysis of Nr5a2- and Tbx3-related signaling pathways may contribute to the future establishment of pESCs.
In summary, we found that the genes Nr5α2 and Tbx3 improved the generation of piPSCs, and the generated cells exhibited outstanding cell viability. The features of the 6 F piPSCs imply that they could be a good source for future domestic applications as well as for the basic research that utilizes porcine pluripotent cells.
References
Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663–676.
Wernig, M., Meissner, A., Foreman, R., Brambrink, T., Ku, M., Hochedlinger, K., et al. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature, 448, 318–324.
Maherali, N., Sridharan, R., Xie, W., Utikal, J., Eminli, S., Arnold, K., et al. (2007). Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell, 1, 55–70.
Esteban, M. A, Xu, J., Yang, J., Peng, M., Qin, D., Li, W., et al. (2009). Generation of induced pluripotent stem cell lines from tibetan miniature pig. Journal of Biological Chemistry, 284, 17634–17640.
Ezashi, T., Telugu, B. P., Alexenko, A. P., Sachdev, S., Sinha, S., & Roberts, R. M. (2009). Derivation of induced pluripotent stem cells from pig somatic cells. Proceedings of the National Academy of Sciences of the United States of America, 106, 10993–10998.
Wu, Z., Chen, J., Ren, J., Bao, L., Liao, J., Cui, C., et al. (2009). Generation of pig induced pluripotent stem cells with a drug-inducible system. Journal of Molecular Cell Biology, 1, 46–54.
West, F. D., Terlouw, S. L., Kwon, D. J., Mumaw, J. L., Dhara, S. K., Hasneen, K., et al. (2010). Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells and Development, 19, 1211–1220.
West, F. D., Uhl, E. W., Liu, Y., Stowe, H., Lu, Y., Yu, P., et al. (2011). Chimeric pigs produced from induced pluripotent stem cells demonstrate germline transmission and no evidence of tumor formation in young pigs, Stem Cells, 29, 1640–1643.
Fan, N., Chen, J., Shang, Z., Dou, H., Ji, G., Zou, Q., et al. (2013). Piglets cloned from induced pluripotent stem cells. Cell Research, 23, 162–166.
Zhou, S., Ding, C., Zhao, X., Wang, E., Dai, X., Liu, L., et al. (2010). Successful generation of cloned mice using nuclear transfer from induced pluripotent stem cells. Cell Research, 20, 850–853.
Fayard, E., Auwerx, J., & Schoonjans, K. (2004). LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends in Cell Biology, 14, 250–260.
Heng, J. C., Feng, B., Han, J., Jiang, J., Kraus, P., Ng, J. H., et al. (2010). The nuclear receptor Nr5α2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell, 6, 167–174.
Wang, W., Yang, J., Liu, H., Lu, D., Chen, X., Zenonos, Z., et al. (2011). Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proceedings of the National Academy of Sciences of the United States of America, 108, 18283–18288.
Guo, G., & Smith, A. (2010). A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development, 137, 3185–3192.
Han, J., Yuan, P., Yang, H., Zhang, J., Soh, B. S., Li, P., et al. (2010). Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature, 463, 1096–1100.
Gu, Q., Hao, J., Zhao, X. Y., Li, W., Liu, L., Wang, L., et al. (2012). Rapid conversion of human ESCs into mouse ESC-like pluripotent state by optimizing culture conditions. Protein & Cell, 3, 71–79.
Gu, P., Goodwin, B., Chung, A. C., Xu, X., Wheeler, D. A., Price, R. R., et al. (2005). Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Molecular and Cellular Biology, 25, 3492–3505.
Ivanova, N., Dobrin, R., Lu, R., Kotenko, I., Levorse, J., DeCoste, C., et al. (2006). Dissecting self-renewal in stem cells with RNA interference. Nature, 442, 533–538.
Niwa, H., Ogawa, K., Shimosato, D., & Adachi, K. (2009). A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature, 460, 118–122.
Cartwright, P., McLean, C., Sheppard, A., Rivett, D., Jones, K., & Dalton, S. (2005). LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development, 132, 885–896.
Li, Z., Fei, T., Zhang, J., Zhu, G., Wang, L., Lu, D., et al. (2012). BMP4 Signaling Acts via dual-specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell, 10, 171–182.
Telugu, B. P., Ezashi, T., Sinha, S., Alexenko, A. P., Spate, L., Prather, R. S., et al. (2011). Leukemia inhibitory factor (LIF)-dependent, pluripotent stem cells established from inner cell mass of porcine embryos. Journal of Biological Chemistry, 286, 28948–28953.
Liu, K., Ji, G., Mao, J., Liu, M., Wang, L., Chen, C., et al. (2012). Generation of porcine-induced pluripotent stem cells by using OCT4 and KLF4 porcine factors. Cellular Reprogramming, 14, 505–513.
Li, Y., Zhang, Q., Yin, X., Yang, W., Du, Y., Hou, P., et al. (2011). Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Research, 21, 196–204.
Wagner, R. T., Xu, X., Yi, F., Merrill, B. J., & Cooney, A. J. (2010). Canonical Wnt/beta-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells, 28, 1794–1804.
Flechon, J. E., Degrouard, J., & Flechon, B. (2004). Gastrulation events in the prestreak pig embryo: ultrastructure and cell markers. Genesis, 38, 13–25.
Gandolfi, F., Pennarossa, G., Maffei, S., & Brevini, T. (2012). Why is it so difficult to derive pluripotent stem cells in domestic ungulates? Reproduction in Domestic Animals, 47(Suppl 5), 11–17.
Acknowledgments
This study was supported in part by grants from China National Basic Research Program of China 2009 CB941000, 2012CBA01300, 2011CB944100 and 2011CB965300.
Conflict of Interest
The authors declare no potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Jianyu Wang, Qi Gu and Jie Hao contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Figure 1
Characterization of 4 F piPSCs. (A) Immunostaining for pluripotency markers of 4 F piPSCs. Positive OCT4 (green) and SOX2 (green) were observed. DNA was stained with Hoechst 33342 (blue). (B) Karyotype of 4 F piPSCs. More than 80 % of the cells showed normal pig karyotype of 38 chromosomes. (C) Embryoid body formation detection. Bars scale, 200 μm. (PPTX 573 kb)
Rights and permissions
About this article
Cite this article
Wang, J., Gu, Q., Hao, J. et al. Tbx3 and Nr5α2 Play Important Roles in Pig Pluripotent Stem Cells. Stem Cell Rev and Rep 9, 700–708 (2013). https://doi.org/10.1007/s12015-013-9439-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-013-9439-2