Abstract
5-Fluorouracil (5-FU) is one of the most commonly used anticancer drugs in the treatment of colon cancer. However, acquired chemoresistance is becoming one of the major challenges for patients with advanced stages of colon cancer. Currently, the mechanisms underlying cancer cell resistance to 5-FU are not fully understood. MicroRNAs (miRNA) have been suggested to play important roles in tumorigenesis and drug resistance in colon cancer. In this study, we generated 5-FU-resistant colon cancer cell lines from which we found that miR-122 was downregulated in 5-FU-resistant cells compared with sensitive cells. Meanwhile, the glucose metabolism is significantly upregulated in 5-FU-resistant cells. We report that PKM2 is a direct target of miR-122 in colon cancer cell. Importantly, overexpression of miR-122 in 5-FU-resistant cells resensitizes 5-FU resistance through the inhibition of PKM2 both in vitro and in vivo. In summary, these findings reveal that the dysregulated glucose metabolism contributes to 5-FU resistance, and glycolysis inhibition by miR-122 might be a promising therapeutic strategy to overcome 5-FU resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
5-Fluorouracil (5-FU) is the most commonly used drug in colon cancer therapy with the capability to induce apoptosis in normal and tumor intestinal cells [1]. The main mechanism of 5-FU activation is through its conversion to fluorouridine monophosphate (FUMP) either directly by orotate phosphoribosyltransferase (OPRT) with phosphoribosyl pyrophosphate (PRPP) as the cofactor, or indirectly via fluorouridine (FUR) through the sequential actions of uridine phosphorylase (UP) and uridine kinase (UK) [1–3]. However, despite its initial efficacy, acquired resistance to 5-FU develops in the majority of patients who have advanced colon tumors [4, 5]. Possible mechanisms of 5-FU resistance, such as amplification or mutation of drug target genes and the modification in the expression levels of p53, bax, Bcl-2, and Bcl-xL, have been identified and studied [6]. Currently, the detailed mechanism for 5-FU resistance in colon cancer remains elusive.
Pyruvate kinase type M2 (PKM2) is an isoenzyme of the glycolytic enzyme, pyruvate kinase, which catalyzes the last step of glycolysis, the dephosphorylation of phosphoenolpyruvate to pyruvate, and is responsible for net ATP production during the glycolytic sequence [7]. PKM2 has been reported to be involved in aerobic glycolysis and cell growth in various tumors. A recent study reported that the expression of PKM2 was increased in colon cancer cells, and the increased PKM2 expression was associated with later stage and lymph metastasis of the tumors [8]. In addition, switching pyruvate kinase expression to the M1 (adult) isoform leads to reversal of the Warburg effect by PKM2, leading to increased lactate production and reduced oxygen consumption that are the typical features of the metabolism for cancer cells [9].
In this study, we explored the mechanisms for the microRNA-mediated 5-FU resistance in colon cancer cells. By generating 5-FU-resistant colon cancer cell lines, we identified that miR-122 was downregulated in 5-FU-resistant cells. Meanwhile, we reported that PKM2 was a direct target of miR-122. Importantly, overexpression of miR-122 in 5-FU-resistant cells resensitized 5-FU resistance through inhibition of PKM2 both in vitro and in vivo. These findings have important implications for the future development of therapeutics for 5-FU-resistant colon cancers.
Methods
Cell Lines and Reagents
HCT-116 and HT-29 human colon cancer cell lines were purchased from ATCC. Cells were cultured in RPMI-1640 supplemented with 10 % FBS and 1× penicillin–streptomycin–glutamine (10378-016; Life Technology) at 37 °C in a humidified incubator with 95 % air and 5 % CO2. The reagents used in this project were LDHA (Cell signaling #2012), β-actin (Cell Signaling #4967), GLUT1 (Santa Cruz: sc-7903), PKM2 (Cell Signaling#3198), and 5-FU, purchased from Sigma-Aldrich (Hong Kong, China).
Generation of 5-FU-Resistant Cell Line
The 5-FU-resistant cell lines were generated from HCT-116 and HT-29 cells. Briefly, parental cells were treated with gradually increasing concentrations of 5-FU in regular cell culture conditions for the selection of resistant cells. After successive treatments for up to 3 months, resistant cell clones were pooled and used for all the subsequent experiments in this study. The resistant cells were treated by 5-FU each month for repeating the selection.
Western Blot Analysis
The whole cells were lysed in 1× SDS sample buffer and resolved by electrophoresis using SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed with primary antibodies overnight, and then incubated with appropriate horseradish peroxidase-conjugated secondary antibodies for 3 h followed by detection with a SuperSignal enhanced chemiluminescence kit (Pierce, Rockford, IL). For sequential blotting, the membranes were stripped with Stripping Buffer (Pierce) and re-probed with proper antibodies.
Cell Viability Assay
A colorimetric assay using the tetrazolium salt, MTT, was used to assess the cytotoxicity of the anticancer agent—5-FU. Single-cell suspensions were prepared, and the cell density was measured. MTT assays were performed according to the manufacturer’s instructions.
Pre-miRNA and Plasmid DNA Transfection
miRNA precursors (pre-miRNAs) and Pre-miR negative control were purchased from Applied Biosystems. Lipofectamine 2000 (Life Technology) was used for the transfection of pre-miRNAs. 48 h after transfection, the expression of miR-122 was detected by Real-time PCR, and the expression of PKM2, a target of miR-122, was measured by Western blotting. Plasmid DNA transfection was performed using the Lipofectamine 2000 Transfection reagent (Life Technology) according to the manufacturer’s protocol. The overexpression vectors containing wild-type PKM2 (Myc-DDK-tagged) were purchased from Origene. 48 h after transfection, cells were collected or the whole cell lysates were prepared for further analysis.
miRNA Real-time PCR
For miRNA expression analysis, qRT-PCR was done using the TaqMan microRNA reverse transcription kit (Applied Biosystems) and TaqMan microRNA assays kit (Applied Biosystems) by following the manufacturer’s protocols. All the reactions were performed in triplicate. Human U6 served as an internal control. The relative amounts of mRNA were calculated by the comparative CT method. Experiments were triplicated.
Glucose Uptake and Lactate Product Assay
Glucose uptake was measured using an Amplex Red Glucose/Glucose Oxidase assay kit (Molecular Probes). Absorbance was measured at 563 nm using a SpectraMax M5 plate reader (Molecular Devices), and the results were normalized to the amount of total protein. Lactate production in the medium was detected using a Lactate assay kit (BioVision). Results were normalized to the amount of total protein compared with the control cells.
Animal Experiments
The athymic BALB/c nude mice (5–8 week old) were housed in the Biological Resource Centre of Southwest Cancer Center, Third Military Medical University, Chongqing, China. Mice were implanted subcutaneously into a mouse mammary fat pad mfp with 1 × 107 HCT-116 5-FU-resistant cells. Tumor progress was monitored by tumor size measurements on every other day. When the tumor reached a size of >150 mm3, the mice were randomly divided into 4 groups (8 mice per group) as follows: HCT-116 cells with control miRNA were treated with PBS-treated control or 5-FU, and HCT-116 cells with overexpression of miR-112 were treated with PBS-treated control or 5-FU. All the experimental procedures involving mouse models were approved by the Ethics Committee of the Third Military Medical University.
Statistical Analysis
The unpaired Student’s t test was used for the data analysis. All data were shown as mean ± standard error (SE). A statistical difference of P < 0.05 was considered significant.
Results
Establishment of 5-FU-Resistant Cell Lines from Human Colon Cancer Cells
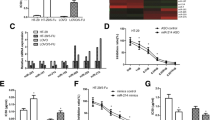
Since multiple microRNAs have been reported to play an essential role in cancer chemosensitivity and correlate with 5-FU resistance [10], we started to establish 5-FU-resistant cell lines from human colon cancer cell lines HCT-116 and HCT-29. After successive treatments for 3 months, several 5-FU-resistant cell clones were established and pooled for the following experiments. To verify the resistance, parental cells and resistant pooled cells were treated with 5-FU at indicated concentrations for 72 h. As we expected, cell viability assays showed that 5-FU-resistant cells could tolerate much higher concentrations of 5-FU compared with sensitive cells. The IC50 of HCT-116 cells for 5-FU was eightfold higher in the resistant cells (26.83 µM) compared with the parent cells (3.25 µM) (Fig. 1a top). The IC50 of HCT-29 cells was sixfold higher in the resistant cells (4.86 µM) than in the parent cells (0.78 µM) (Fig. 1a bottom).
MiR-122 is downregulated in 5-FU-resistant colon cancer cells, and PKM2 is a direct target of miR-122. a Generation of 5-FU-resistant cells from HCT-116 and CT-29 colon cancer cells. Parental cells were treated with gradually increasing concentrations of 5-FU in regular cell culture conditions for the selection of resistant cells. 5-FU-resistant clones were pooled and analyzed by the treatments of 5-FU at indicated concentrations for 72 h. b The expression of miR-122 was downregulated in 5-FU-resistant cell lines compared with the sensitive cell lines. c Target prediction from Targetscan.org, the position 520–527 of PKM2 3′ UTR contains putative binding sites for miR-122. d HCT-116 cells were transfected with 100 nM pre-miR negative (Ctr), pre-miR-122, or anti-miR-122 for 48 h. Cell lysates were prepared for Western blotting analysis. β-actin was used as a loading control. e HCT-116 cells were co-transfected with luciferase reporter plasmids with wild-type 3′-UTR of PKM2 or mutant 3′-UTR of PKM2, and pre-miR-122 or pre-miR negative (Control miR) using Lipofectamine 2000 reagent. 48 h post transfection, cells were harvested and lysed with passive lysis buffer. Luciferase activities were measured by a dual luciferase reporter assay. The pRL-TK vector was used as an internal control. The results were expressed as relative luciferase activity (firefly LUC/Renilla LUC). Columns, mean of three independent experiments; bars, SE. **P < 0.01
Downregulation of miR-122 in 5-FU-Resistant Colon Cancer Cells
To investigate the role of microRNAs in 5-FU resistance in human colon cells, we measured the expression of multiple microRNAs in 5-FU-resistant colon cancer cell lines compared with the parental cells. Among them, the expression of miR-122 was significantly downregulated in both the 5-FU-resistant cell lines (Fig. 1b), suggesting that miR-122 might be a tumor suppressor in human colon cancers and play an essential role in 5-FU resistance.
PKM2 is a Direct Target of miR-122 in Colon Cancer Cells
We next investigated the mechanism of miR-122 in 5-FU resistance. By searching miRNA databases for the prediction of miR-122 targets that may possibly contribute to 5-FU resistance, we found that the public miRNA databases (TargetScan) predicted that PKM2 might be a target for miR-122, and the 3′-UTR of PKM2 contains a highly conserved binding site for miR-122 (Fig. 1c). To determine whether miR-122 could target PKM2 in colon cancer cells, we transfected the pre-miR-122 into HCT-116 cells. The overexpression of miR-122 significantly downregulated PKM2 protein expression. Furthermore, the knockdown of endogenous miR-122 recovered the PKM2 protein expression (Fig. 1d). We next investigated whether miR-122 could directly target the 3′-UTR of PKM2 mRNA. We performed Luciferase reporter analysis by co-transfecting a vector containing pMIR-REPORTER luciferase fused with the original sequence or predicted binding site mutant sequence of the 3′-UTR of PKM2 mRNA with pre-miR-122 or with control microRNA. The overexpression of miR-122 decreased the luciferase activity of the reporter with wild-type 3′-UTR of PKM2 by about 60 % in HCT-116 cells (Fig. 1e). However, no inhibitory effects of miR-122 on the activity of the reporter with binding site mutant of 3′-UTR of 122 were detected (Fig. 1e). Taken together, our results demonstrated that PKM2 is a direct target of miR-122 in colon cancer cells.
5-FU-Resistant Colon Cancer Cells Exhibit Increased Glucose Metabolism
Many tumor cells exhibit altered metabolism such as higher rate of glycolysis and reduced mitochondrial oxidative phosphorylation. In addition, previous studies revealed that dysregulated cellular metabolism is linked to 5-FU resistance in cancer therapy [11]. To test whether the 5-FU-resistant colon cancer cells have altered aerobic glycolysis, we measured several key glycolytic parameters, including glucose consumption, lactate production, and expression levels of glycolytic enzymes in the resistant cells. As shown in Fig. 2a, both HCT-116 and HT-29 5-FU-resistant cells consumed more glucose than their parental cells. In addition, the resistant cells released more lactate into the media (Fig. 2b). Consistently, the protein expression levels of glucose transporter (Glut-1), pyruvate kinase M2 (PKM2), and lactate dehydrogenase-A (LDHA) were all significantly upregulated in 5-FU-resistant cells (Fig. 2c). Taken together, our above data revealed that dysregulated glucose metabolism was correlated with 5-FU resistance in colon cancer cells.
Glycolysis pathway is upregulated in 5-FU-resistant colon cancer cells. a Glucose uptake of HCT-116 and HT-29 5-FU-resistant cells were increased. b Lactate products of HCT-116 and HT-29 5-FU-resistant cells were increased. c Protein expression levels of GLUT1, PKM2, and LDHA in HCT-116 and HT-29 5-FU-resistant cells were upregulated by Western blotting analysis. Columns, mean of three independent experiments; bars, SE. *P < 0.05; **P < 0.01; ***P < 0.001
Overexpression of miR-122 Sensitizes 5-FU-Resistant Cells Through the Inhibition of PKM2
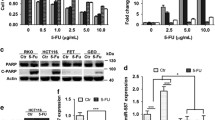
Next, we examined whether the overexpression of miR-122 was capable of sensitizing 5-FU-resistant colon cancer cells to 5-FU. HCT-116 5-FU-sensitive and -resistant cells were transfected with pre-miR-negative or pre-miR-122 (Fig. 3a); then, the cells were treated with increasing concentrations of 5-FU. Compared with negative control, the overexpression of miR-122 markedly enhanced 5-FU-induced cell cytotoxicity in both 5-FU-sensitive and -resistant cells (Fig. 3b). The IC50 of miR-122 overexpressing HCT-116 5-FU-sensitive cells was decreased to 1.02 µM compared with the negative control cells (3.23 µM). Meanwhile, the IC50 of miR-122 overexpressing HCT-116 5-FU-resistant cells was decreased to 10.7 µM compared with the negative control cells (26.83 µM).
Overexpression of miR-122 resensitizes 5-FU-resistant cells through the inhibition of PKM2. a Overexpression of miR-122 in HCT-116 5-FU-sensitive (left) and -resistant cells (right). b HCT-116 5-FU-sensitive cells (left) and -resistant cells (right) were transfected with miR-122 for 48 h followed by the treatments of 5-FU at indicated concentrations for 72 h followed by cell viability analysis. c Western blotting experiments showed the overexpression of PKM2 in miR-122-pre-transfected HCT-116 cells. d Exogenous overexpression of PKM2 into miR-122-pre-transfected HCT-116 parental cells restored resistance to 5-FU at the indicated treatments for 72 h. Columns, mean of three independent experiments; bars, SE. *P < 0.05; **P < 0.01; ***P < 0.001
We next investigated whether miR-122 affects 5-FU resistance through the suppression of PKM2 expression. We, therefore, transfected pre-miR-122 or pre-miR-122 in combination with overexpression vector containing wild-type PKM2 into HCT-116 cells (Fig. 3c); then, we treated the cells with the indicated concentrations of 5-FU for 72 h. Transfection with the pre-miR-122 alone decreased the expression of PKM2, which renders HCT-116 cells sensitive to 5-FU, and restoration of PKM2 by transfection with vector contacting PKM2 recovered 5-FU resistance (Fig. 3d). Taken together, these results clearly demonstrated that PKM2 plays an important role in the regulation of 5-FU sensitivity, and the overexpression of miR-122 overcomes 5-FU resistance through the direct suppression of PKM2 expression.
Resensitization of 5-FU-Resistant Cells by miR-122 In Vivo
The above results showed that the overexpression of miR-122 in colon cancer cells increased the sensitivity of both 5-FU-sensitive and -resistant cells to 5-FU treatments in vitro. To further support our conclusions, we performed in vivo experiment in a xenograft nude mice model. 5-FU-resistant cells were transfected with pre-miR-122 or negative control followed by the injection into the mammary fat pads of 6-week-old nude mice. After tumors were formed, we treated the tumor xenograft with 5-FU or PBS control for 6 weeks. As expected, compared with the control group, the overexpression of miR-122 into 5-FU-resistant colon cancer cells inhibits tumor growth under the 5-FU treatments (Fig. 4a, b). To test whether the overexpression of miR-122 sensitizes 5-FU-resistant cells through the inhibition of glucose metabolism, we evaluated the rates of glucose metabolism of the 5-FU-treated or -untreated HCT-116 5-FU-resistant cell-derived tumors. Consistently, the glucose uptake (Fig. 4c) and lactate product (Fig. 4d) were down-regulated in miR-122 overexpressing cells by 5-FU treatments. Taken together, our results demonstrated that glycolysis inhibition by miR-122 contributes to 5-FU-inhibited tumor growth in vivo.
In vivo experiments show that the overexpression of miR-122 resensitizes 5-FU-resistant colon cancers. a Pre-established 5-FU-resistant HCT-116 cells with or without the overexpression of miR-122 were treated with control (PBS) or 5-FU (500 mg/kg, i.p., 2 times/week × 6 weeks), and the tumor size was measured with caliper twice a week over a period of 40 days. b Tumorigenicity of transfection with control miR or miR-122 in HCT-116 cells (1 × 107) after subcutaneous injection in the flanks of nude mice (n = 8). c Measurements of glucose uptake and d lactate product from HCT-116 5-FU-resistant cells with or without the overexpression of miR-122-derived tumors. Columns, mean of three independent experiments; bars, SE
Discussion
The “Warburg effect” described that tumor cells had high glycolytic rate with the production of lactate even in the presence of oxygen [12]. Furthermore, cancer cells were found to be addicted to glucose and very sensitive to glucose concentration changes [13]. It has become clear that the altered metabolism gives several advantages to cancer cells [14]. Moreover, the high glycolytic flux ensures that there are sufficient intermediates for keeping the pentose phosphate pathway active to generate NADPH and ribose-5-phosphate for the synthesis of nucleotides [15]. Therefore, the altered metabolism in cancer cells is contributing to chemoresistance, but the precise mechanisms are unclear [16, 17]. In this study, we reported that the dysregulated glucose metabolism of colon cancer cells was correlated in 5-FU-resistant cells. The glucose uptake and lactate product were upregulated in 5-FU-resistant colon cancer cells. In addition, the glucose metabolic key enzymes were upregulated. Our data suggested that increased glucose metabolism might be a mechanism underlying the 5-FU resistance in human colon cancer cells.
Many miRNAs have been identified to have an oncogenic or tumor suppressor-like function that is shown to be involved in cell proliferation, differentiation, apoptosis, and drug resistance [18]. Multiple microRNAs were found to be involved in the development of colon cancer, and potential miRNAs have been investigated as biomarkers in diagnosis and prognosis of colon cancer [19]. It has been reported that the transient introduction of miR-34a into two human colon cancer cell lines, HCT-116 and RKO, completely suppressed the cell proliferation and induced senescence-like phenotypes through downregulation of the E2F pathway and upregulation of the p53 pathway [20]. Moreover, the overexpression of miR-451 in gastric cancer and colon cancer cells resulted in the reduction of cell proliferation and increased their susceptibility to radiotherapy [21]. Our study illustrated a novel tumor suppressor function of miR-122 in colon cancer cells. The expression of miR-122 was significantly downregulated in 5-FU-resistant cells, indicating that it might contribute to chemoresistance through the activation of anti-apoptosis pathway. Among the targets of miR-122, we identified PKM2 as its direct target. PKM2 protein expression level was downregulated by overexpression of miR-122. In addition, luciferase reporter assay demonstrated that the 3′UTR region of PKM2 can be the targeting site of miR-122. Importantly, the overexpression of miR-122 in 5-FU-resistant colon cancer cells rendered the resistant cells sensitive to 5-FU through the downregulation of PKM2 both in vitro and in vivo. These findings presented the essential role of miR-122 in chemoresistance and will provide important implications in the development of targeted therapeutics for overcoming 5-FU resistance in human colon cancers.
References
Longley, D. B., Harkin, D. P., & Johnston, P. G. (2003). 5-Fluorouracil: Mechanisms of action and clinical strategies. Nature Reviews Cancer, 3, 330–338.
Curtin, N. J., Harris, A. L., & Aherne, G. W. (1991). Mechanism of cell death following thymidylate synthase inhibition: 2′-Deoxyuridine-5′-triphosphate accumulation, DNA damage, and growth inhibition following exposure to CB3717 and dipyridamole. Cancer Research, 51, 2346–2352.
Peters, G. J., van Triest, B., Backus, H. H., Kuiper, C. M., van der Wilt, C. L., & Pinedo, H. M. (2000). Molecular downstream events and induction of thymidylate synthase in mutant and wild-type p53 colon cancer cell lines after treatment with 5-fluorouracil and the thymidylate synthase inhibitor raltitrexed. European Journal of Cancer, 36, 916–924.
Zhang, N., Yin, Y., Xu, S. J., & Chen, W. S. (2008). 5-Fluorouracil: Mechanisms of resistance and reversal strategies. Molecules, 13, 1551–1569.
De Angelis, P. M., Svendsrud, D. H., Kravik, K. L., & Stokke, T. (2006). Cellular response to 5-fluorouracil (5-FU) in 5-FU-resistant colon cancer cell lines during treatment and recovery. Molecular Cancer, 5, 20.
Violette, S., Poulain, L., Dussaulx, E., Pepin, D., Faussat, A. M., Chambaz, J., et al. (2002). Resistance of colon cancer cells to long-term 5-fluorouracil exposure is correlated to the relative level of Bcl-2 and Bcl-X(L) in addition to Bax and p53 status. International Journal of Cancer, 98, 498–504.
Wu, S., & Le, H. (2013). Dual roles of PKM2 in cancer metabolism. Acta Biochimica et Biophysica Sinica (Shanghai), 45, 27–35.
Israelsen, W. J., Dayton, T. L., Davidson, S. M., Fiske, B. P., Hosios, A. M., Bellinger, G., et al. (2013). PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell, 155, 397–409.
Christofk, H. R., Vander Heiden, M. G., Harris, M. H., Ramanathan, A., Gerszten, R. E., Wei, R., et al. (2008). The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature, 452, 230–233.
Schetter, A. J., Okayama, H., & Harris, C. C. (2012). The role of microRNAs in colorectal cancer. Cancer Journal, 18, 244–252.
Zhou, Y., Tozzi, F., Chen, J., Fan, F., Xia, L., Wang, J., et al. (2012). Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Research, 72, 304–314.
Warburg, O. (1956). On respiratory impairment in cancer cells. Science, 124, 269–270.
Kroemer, G., & Pouyssegur, J. (2008). Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell, 13, 472–482.
Vander Heiden, M. G., Cantley, L. C., & Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science, 324, 1029–1033.
Dang, C. V. (2010). Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Research, 70, 859–862.
Zhao, Y., Butler, E. B., & Tan, M. (2013). Targeting cellular metabolism to improve cancer therapeutics. Cell Death and Disease, 4, e532.
Vander Heiden, M. G. (2011). Targeting cancer metabolism: A therapeutic window opens. Nature Reviews Drug Discovery, 10, 671–684.
Ameres, S. L., & Zamore, P. D. (2013). Diversifying microRNA sequence and function. Nature Reviews Molecular Cell Biology, 14, 475–488.
Yang, L., & Belaguli, N. (2009). Berger DH MicroRNA and colorectal cancer. World Journal of Surgery, 33, 446–638.
Tazawa, H., Tsuchiya, N., Izumiya, M., & Nakagama, H. (2007). Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proceedings of the National Academy of Sciences of the United States of America, 104, 15472–15477.
Bandres, E., Bitarte, N., Arias, F., Agorreta, J., Fortes, P., Agirre, X., et al. (2009). microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clinical Cancer Research, 15, 2281–2290.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, J., Xie, G., Tong, J. et al. Overexpression of MicroRNA-122 Re-sensitizes 5-FU-Resistant Colon Cancer Cells to 5-FU Through the Inhibition of PKM2 In Vitro and In Vivo. Cell Biochem Biophys 70, 1343–1350 (2014). https://doi.org/10.1007/s12013-014-0062-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-0062-x