Abstract
The Siah ubiquitin ligases are members of the RING finger E3 ligases. The Siah E3s are conserved from fly to mammals. Primarily implicated in cellular stress responses, Siah ligases play a key role in hypoxia, through the regulation of HIF-1α transcription stability and activity. Concomitantly, physiological conditions associated with varying oxygen tension often highlight the importance of Siah, as seen in cancer and neurodegenerative disorders. Notably, recent studies also point to the role of these ligases in fundamental processes including DNA damage response, cellular organization and polarity. This review summarizes the current understanding of upstream regulators and downstream effectors of Siah.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ubiquitin is a small 76 amino acid polypeptide, which is conjugated to protein substrates through a multistep process involving the sequential action of three classes of enzymes: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 substrate-specific ubiquitin ligases [1]. Ubiquitin conjugation can result in a single molecule per substrate (mono-ubiquitination), multiple molecules conjugated as single ubiquitin along the substrate (multi-ubiquitination), or multiple ubiquitin that are conjugated to each other thereby generating a ubiquitin chain (poly-ubiquitination). Notably, ubiquitin chains assume different topologies depending on the lysines utilized for conjugation of ubiquitin molecules to each other. Conjugation through lysine 48 or 11 results in topology of ubiquitin chains that are primarily recognized by the 26S proteasomes and implicated in protein degradation. Conjugation through lysine 63 has been implicated in signaling complexes whereby ubiquitin chains play a role in assembly of select protein complexes. On the other hand, mono or multi-mono-ubiquitination of substrate has been associated with subcellular localization of the marked substrate within the cell [2]. E2 conjugating enzymes play a primary role in determining the type and topology of the ubiquitin conjugation. Among the E3, ubiquitin ligases is the class of RING finger ubiquitin ligases, which consists of over 600 members. The activity of most E3s in this family is specified by a RING domain, which binds to an E2-ubiquitin thioester and activates the discharge of its ubiquitin cargo. Common to the RING finger E3 ligases is the conserved organization of histidine and cysteine residues, which help maintain the overall structure through binding two atoms of zinc, resulting in a globular platform for interactions with E2s.
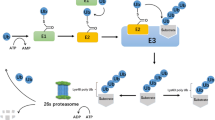
The Siah family of E3 ubiquitin ligases, mammalian homologs of the Drosophila sina protein, are RING finger ubiquitin ligases composed of a catalytic RING domain, two zinc finger domains, and a substrate binding domain [3–5]. Mice express three members of this gene family; Siah1a, Siah1b, and Siah2, whereas humans express Siah1 and Siah2 [6]. A number of Siah substrates have been identified (Table 1). Some substrates interact directly with Siah, whereas others require adaptor proteins, such as phyllopod (PHYL) and Siah-interacting protein (SIP) [7, 8]. Knockout of both Siah1a and Siah2 genes is embryonically lethal in mice, indicating an essential function for the enzymes in early development [9]. Under normal physiological conditions, Siah2 −/− mice exhibit mild phenotypes, such as a small increase in the number of hematopoietic progenitor cells [9], but marked phenotypes are observed when the mice are subjected to stress, suggesting a central role for these enzymes in maintaining normal cellular homeostasis and in the response to stress (Fig. 1). The importance of Siah2 in fundamental cellular processes such as hypoxia, mitochondrial dynamics and the implication of such regulation for diverse pathological conditions, including cancer, point to the importance of understanding how this ubiquitin ligase is being regulated as well as the conditions required for its association with- and ubiquitination of- downstream substrates.
Outline of Siah regulation and function. Factors involved in the regulation of Siah ligases are associated with stress response, including hypoxia, ER stress and genotoxic stress, which induce respective transcription factors, microRNA to induce Siah2 transcription, as well as post-translational modifications that determine Siah subcellular localization and activity. Consequently, Siah activities as an E3 ubiquitin ligase affects growing number of substrates associated with fundamental cellular processes including hypoxia, DNA damage response, neurodegeneration and cancer
Upstream: Transcriptional and Post-Translational Regulation of Siah Ligases
The levels and activity of Siah1 and Siah2 are increased in response to various forms of cellular stress, including oxygen deprivation (hypoxia) [35], glucose deprivation [33], glucose elevation [36], DNA damage, and apoptosis [16, 37], supporting their possible roles in normal homeostasis and the stress response. Siah2 expression is also elevated in several types of cancers including melanoma [38], prostate [39], breast [40], lung [41], pancreas [42], and liver cancers [43, 44], suggesting that Siah2 may contribute to tumor development and progression.
Despite these clear indications that the expression of Siah proteins can be modulated, little is known of their regulation at the transcriptional level. In breast cancer, Siah2 expression is found to be amplified at the genomic level [40] and is transcriptionally regulated by estrogen [45]. Characterization of the human Siah1 promoter has shown that the transcription factor Sp1 regulates the basal promoter activity [46]. Siah1b is a direct transcriptional target of p53 and undergoes p53-induced upregulation during apoptosis [37]. Interestingly, such regulation has not been reported for Siah2, suggesting Siah ligases may have distinct functions in stress and DNA damage responses. Additional pathways implicated in the induction of Siah ligases include the transcription factor E2F1 and WNT5a, which induce Siah1 and Siah2 expression, respectively [47–49].
The abundance and activity of Siah1 and Siah2 is also regulated through post-translational modification. Like other RING finger ubiquitin ligases, Siah ligases self-ubiquitinate under normal physiological conditions to limit their availability and activity. The degree of Siah2 autoubiquitination is determined by the post-translational modification of Siah2, as well as by select deubiquitinating (DUB) enzymes. The DUB USP13 regulates Siah2 availability and activity as interaction of Siah2 with USP13 results in a more stable, albeit less active, ubiquitin ligase [27]. Notably, hypoxia reduces the expression of USP13, thus contributing to the hypoxia-induced increase in Siah2 activity [27]. The effect of USP13 on Siah2 highlights the fact that the activity of Siah2, and possibly other E3 ubiquitin ligases, is not directly proportional to its protein level. Siah2 level is elevated under stress conditions also due to its phosphorylation by stress activated protein kinases. For example, Siah1 protein levels are stabilized by JNK-dependent phosphorylation of tyrosines 100 and 126 [50] whereas ATM/ATR-dependent phosphorylation of Siah1 at serine 19 prevents its degradation during the DNA damage response [16].
During hypoxia, several enzymes regulate the subcellular localization and the activity of Siah2 through phosphorylation. p38 MAPK phosphorylates serine 29, and threonines 24 and 29 of Siah2, which increases its activity towards selected substrates, including prolyl hydroxylase 3 (PHD3) [51]. Similarly, hypoxia induces phosphorylation of serines 16, 28, and 69 and threonines 26 and 119 of Siah2 by DYRK2 (dual specificity tyrosine-phosphorylation-regulated kinase 2), which results in increased Siah2-dependent ubiquitination and degradation of PHD3 [52]. Under normoxic conditions, homeodomain-interacting protein kinase 2 (HIPK2) phosphorylates Siah2 at positions 26, 28, and 68, destabilizing Siah2 and weakening its interactions with substrates. In contrast, HIPK2 phosphorylation of Siah2 is reduced during hypoxia, which results in increased Siah2 activity and subsequent degradation of HIPK2 [15]. In human breast cancer cell lines, the protein kinase Src has been shown to activate Siah2 by phosphorylation of tyrosines 86, 140, and 263 [24].
Additional regulators of Siah protein include vitamin K3, which was identified in a small molecule screen to affect Siah2 levels and activity [53], largely through a redox-independent mechanism. Lastly, hypoxia induction of p75 neurotrophin receptor was associated with stabilization of Siah2, by decreasing its self-ubiquitination [54].
Siah expression is also regulated at the mRNA level by small noncoding microRNAs. MiR-146b targets Siah2 mRNA for destruction in response to TGFβ signaling. Expression of a miR-146b inhibitor prior to TGFβ treatment prevented the reduction in Siah2 [55]. Infection of HEK-293 cells by the Epstein-Barr virus induces the expression of miR-424, which reduces Siah1 expression and attenuates the level of Siah1-induced apoptosis [56]. Regulation of Siah1a by miR-135a has been implicated in preimplantation embryo development [57].
Downstream: Emerging Roles for Siah Ligases in Neuronal Function and in the Hypoxia and DNA Damage Responses
Over the past decade, a number of proteins have been identified that interact with Siah. Identification of these binding partners has shed light not only on the individual pathways associated with Siah1 and Siah2 but also on their diverse physiological and pathophysiological functions.
Siah Functions in Neurons
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by the progressive destruction of dopaminergic neurons. A hallmark of PD is the accumulation of cytoplasmic inclusion bodies called Lewy bodies. The mechanisms by which Lewy bodies form and their contribution to the pathology of PD remain unclear. However, Siah has been detected in Lewy bodies in the brains of PD patients, and Siah is known to ubiquitinate synphilin-1 and α-synuclein, which are two major components of Lewy bodies.
Siah1 has been reported to mono-ubiquitinate α-synuclein, consistent with previous observations that α-synuclein is ubiquitinated [58, 59]. The lysine residues modified by Siah1 show partial overlap with the ubiquitinated residues identified in α-synuclein purified from Lewy bodies, suggesting a possible pathological role for Siah1 in PD [60, 61]. Consistent with this, synphilin-1 undergoes Siah-dependent ubiquitination and proteasomal degradation. The interaction between synphilin-1 and α-synuclein is thought to facilitate the formation of Lewy bodies [62]. Thus, Siah may limit the availability of synphilin-1 for binding to α-synuclein and formation of inclusion bodies [59, 63]. An aggregation-prone isoform of synphilin-1, termed synphilin-1A, was recently shown to interact with Siah, but was not as effectively destabilized as synphilin-1. Synphilin-1A may thus sequester and limit the activity of Siah, consistent with reduction in Siah self-ubiquitination, synphilin-1A degradation, and α-synuclein mono-ubiquitination. The role of Siah in Lewy body formation may thus be dictated by the relative expression of synphilin-1 and synphilin-1A in the neurons [64, 65].
Recently, Siah has been implicated in the control of cell adhesion during neuronal cell exit from the germinal zone and migration in the developing brain [21]. The PAR complex is involved in the formation and regulation of cell polarity and is composed of PARD3A (partitioning defective-3), PARD6 (partitioning defective-6), and PKCζ. Siah interacts with and regulates the stability of PARD3A. Consistent with this, expression of PARD3A protein is low in the external granular layer of the cerebellum, where Siah expression is high. Siah-dependent PARD3A destabilization limits the interaction between PARD3A and junctional adhesion molecule C and reduces cell adhesion, which prevents germinal zone exit by the neurons. As expected, depletion of Siah activity or increased expression of PARD3A accelerates neuronal exit into the external granular layer. Thus, it will be interesting to determine if Siah plays a role in regulating cell–cell adhesion and cell polarity in other tissues, both in physiological and pathological conditions.
Hypoxia-Associated Functions of Siah
Hypoxic conditions trigger major adaptive changes in cell physiology. Expression of hypoxia-responsive genes is regulated in large part by the transcription factor hypoxia-inducible factor (HIF). HIF is regulated in response to reduced oxygen tension by two hydroxylation-dependent mechanisms: proteasomal degradation and transcriptional inactivation. The three oxygen sensing proline hydroxylases (PHD1–3) hydroxylate the α subunit of HIF, which results in its ubiquitination by von Hippel Lindau E3 ligase and subsequent proteasomal degradation. In addition, HIF-1α transcriptional activity is also inhibited by hydroxylation on asparagine residues by factor inhibiting HIF (FIH), which blocks the interaction between HIF-1α and p300/CBP, a transcriptional activator.
Notably, the stability of PHD1/3 and FIH is regulated by Siah-dependent ubiquitination. Thus, increased expression of Siah leads to stabilization of HIF-1α and increased expression of hypoxia-responsive genes. These findings implicate Siah as a critical regulator of HIF-1α activity and the cellular response to hypoxia [35, 66]. As mentioned previously, HIPK2 is not only a substrate of Siah2 but also phosphorylates and negatively regulates Siah2 activity under normoxic conditions [15]. Under oxygen deprivation conditions, however, HIPK2 phosphorylation of Siah2 is reduced, which increases Siah2 activity and results in the increased transcription of hypoxia-responsive genes [18, 67]. Thus, the hypoxia-dependent ubiquitination and phosphorylation of Siah and HIPK2 constitutes a novel layer of regulation in the expression of hypoxia-responsive genes.
Because mitochondria are the main site of oxygen consumption in mammalian cells, fluctuations in oxygen levels affect many aspects of mitochondrial biology. Recent findings have suggested a novel role for Siah in regulating mitochondrial function during hypoxia [33]. AKAP1 (A kinase anchoring protein 1; also known as AKAP121 in the mouse and AKAP149 in humans) is a Siah substrate located at the outer mitochondrial membrane. AKAP121 is ubiquitinated and degraded in a Siah2-dependent manner in cells cultured under conditions of oxygen or glucose deprivation, and in vivo in cerebral and cardiac ischemia models [32, 33]. Interestingly, abrogation of AKAP121 expression impairs not only mitochondrial activity but also mitochondrial morphology [32, 33, 68]. In normoxia, AKAP121 recruits the cAMP-regulated protein kinase A (PKA) to the mitochondrial membrane where PKA phosphorylates Drp1 and inhibits the association of Drp1 with Fis1, which is required for mitochondrial fission. Upon oxygen deprivation, Siah-dependent degradation of AKAP121 abrogates the inhibitory functions of AKAP121, which allows association of active (dephosphorylated) Drp1 at the mitochondria and ultimately leads to mitochondria fission [32]. The significance of Siah2 regulation of mitochondrial fission is demonstrated in myocardial infarction, where impaired expression of Siah or inhibition of mitochondrial fission by Drp1 inhibitor, mdivi-1, were shown to protect heart muscle from necrosis [32, 69]. A full understanding of this novel function of Siah in the regulation of mitochondrial activity and dynamics is still emerging, and it will be interesting to determine how Siah activity at the mitochondria is itself regulated by changing oxygen conditions.
The role of Siah in the DNA Damage Response and Cellular Senescence
As noted above HIPK2 and ATM/ATR both regulate the activity of Siah in a phosphorylation-dependent manner. HIPK2 is a DNA-damage-activated kinase and plays an important role in DNA-damage-induced apoptosis by phosphorylating Ser46 of p53 and phosphorylation or degradation of CtBP, a corepressor to growth inhibitory and apoptotic gene expression [70–72]. HIPK2 is stabilized following DNA damage signals. Upon sublethal DNA damage, Siah1-dependent HIPK2 degradation occurs during recovering phase. However, under lethal DNA damage, activated ATM/ATR phosphorylates Ser19 of Siah1 that abrogates its interaction with and degradation of HIPK2. Ultimately, accumulated HIPK2 increases p53 phosphorylation at Ser46 and decreases expression of CtBP [16]. These observations suggest that both hypoxia and DNA damage signaling regulate HIPK2, through diverse mechanisms involving Siah ubiquitin ligases [16, 67].
Mammalian telomeres, composed of unique DNA components, recruit a specific protein complex termed shelterin complex to protect DNA ends from unwanted DNA damage responses [73]. Two components of shelterin complex, TRF2 and TIN2, were reported as Siah-interacting proteins [23, 74]. Cellular senescence is associated with reduced expression of TRF2 and increased expression of Siah1. Notably, depletion of Siah1 expression increases replicative lifespan of cells, which is consistent with previous observations seen with overexpression of TRF2 [74, 75]. Given the intrinsic function of TRF2 in inhibition of DNA damage response including ATM activation, p53-Siah1-TRF2 pathway offers an integral feed-forward loop in telomere-initiated DNA damage response. How the ATM/ATR-mediated modification of Siah-1 affects TRF2 regulation remains to be determined [16, 74].
In an independent study, TIN2, a component of shelterin complex, is also found as a Siah-interacting protein and a substrate for the ubiquitin proteasome-dependent degradation [23]. Depletion of Siah2 significantly increases TIN2 expression as well as slightly increasing TRF1 and TRF2. Interestingly, overexpression of Siah2 leads to the loss of TIN2 expression and inhibition of TPP1 (a known interacting component of shelterin complex) localization to shelterin complex in telomere [76]. Thus, regulation of TIN2 availability by Siah may be integral for remodeling of shelterin complex under conditions requiring dynamic telomere function.
Downstream: Siah Ligases in Cancer
Accumulating functional evidence support a tumor-promoting role of Siah2 in multiple cancers, including breast [77], lung [41], pancreatic [42], prostate [39], liver [44] and melanoma [38]. In support of its functional roles, Siah2 protein levels are upregulated in high-grade breast cancer [40], lung cancer [41], and prostate cancer [39], compared with its expression in the corresponding low-grade cancers or in normal tissues.
Inhibition of Siah2 in cancer cell lines (pancreatic, lung, liver, breast, and melanoma) attenuates tumor cell proliferation in vitro and/or tumor formation or metastasis in mouse xenograft models [38, 41, 42, 44, 78]. Importantly, inhibition of Siah2 in fibroblasts and in the non-transformed mammary epithelial MCF10A cells confers resistance to the transforming activity of Ras and Src oncogenes, respectively [24, 42].
Genetic evidence for the role of Siah2 in tumor development comes from mouse models in which Siah2−/− mice were crossed with the TRAMP or MMTV mouse models of prostate and breast cancers, respectively. Siah2 deficiency abolished the formation of malignant prostate tumors [39], and delayed the initiation and progression of mammary tumors [77], thereby establishing a clear tumor-promoting role for Siah2. Moreover, Siah2 deficiency in the MMTV breast cancer model increased perfusion and pericyte-associated vasculature, similar to that occurring with antiangiogenic therapy, resulting in an increased response to chemotherapy and prolonged survival [77]. Inhibition of Siah2 expression also directly sensitizes human hepatocellular carcinoma (HCC) cells to treatment with cytostatic drugs [44]. Collectively, these observations provide a rationale for the development of Siah2-targeted therapeutic agents for cancer therapy.
As HIF proteins have been implicated in cancer, an important focus of recent studies has been the link between Siah and HIF in tumor development and progression. As described above, increased expression of Siah2 in hypoxia increases HIF activity and expression of hypoxia-responsive genes. As expected, inhibition of Siah2 in melanoma [38], breast cancer [78], and HCC [44] cells reduces HIF levels under hypoxic conditions. Consistent with this, HIF-1α levels are also reduced in tissues of Siah2−/− TRAMP or MMTV mice [39, 78], which phenocopies the HIF-1α−/+ mouse mammary tumor model [79]. Moreover, re-expression of HIF-1α or specific HIF-1α target genes partially restored tumorigenesis and metastasis in mouse melanoma cells or TRAMP prostate tumor cells expressing a Siah-inhibitory peptide [38, 39]. These studies established a causal role for Siah2 in tumorigenesis and metastasis through its regulation of HIF-1α and the hypoxia response.
Siah2 enhances the MAPK-ERK signaling pathway, primarily through direct ubiquitination-dependent modulation of Sprouty2, a Ras inhibitory protein that negatively regulates MAPK-ERK signaling. Accordingly, inhibition of Siah2 reduces phospho-ERK levels in cultured melanoma and pancreatic, lung, and liver cancer cell lines and inhibits growth of tumor xenografts [38, 41, 42]. Moreover, attenuation of Sprouty2 expression effectively restores phospho-ERK levels in Siah2-inhibited melanoma cells and partially rescues tumor formation in vivo [38]. Consistent with the important role of Siah2 in HIF and ERK signaling, vitamin D3, a Siah2 inhibitor, reduces HIF and phospho-ERK levels in a human melanoma cell line and inhibits tumor formation in the mouse xenograft model [53].
Siah2 was recently shown to facilitate the ubiquitination-dependent degradation of the transcription factor CCAAT/enhancer-binding protein delta (C/EBPδ), a tumor suppressor that is downregulated during breast cancer progression. This function is promoted by Src phosphorylation of Siah2, which increases C/EBPδ binding, ubiquitination, and degradation. Src/Siah2-induced degradation of C/EBPδ is required for cyclin D1 expression in breast tumor cell lines, and has been associated with enhanced cell motility and invasion. Moreover, ectopic expression of degradation-resistant C/EBPδ mutants blocks the full transformation of MCF-10A cells by activated Src in vitro [24]. Thus, Siah2 activity reduces the expression of C/EBPδ, a tumor suppressor implicated in the progression of breast cancer.
In contrast to Siah2, the role of Siah1 in cancer development and/or progression is poorly understood. Siah1 expression is frequently reduced in HCCs [43, 80]. Inactivating mutations of Siah1 have been reported in malignant gastric cancers [81]. These findings point to a putative tumor suppressor role for Siah1, which is distinct from the oncogenic, tumor-promoting functions implied for Siah2. Paradoxically, inhibition of Siah1 expression reduces proliferation and migration of Hep3B or HuH-7 HCC-derived cells [43]. Interestingly, Siah1 is primarily located in the cytoplasm of normal hepatocytes but is mainly in the nucleus of HCC cells. Siah1 has been shown to direct the ubiquitination-dependent proteasomal degradation of the hepatitis B viral X protein (HBx), a multifunctional trans-activator that has been implicated in hepatitis B virus replication and hepatocarcinogenesis. Thus, Siah1 may play a role in suppressing the progression of HCC [25]. The possibility that the function of Siah1 in promoting or suppressing HCC may be dictated by its subcellular localization deserves careful examination.
The expression of Siah1 is downregulated in human breast cancer tissues, consistent with the likely opposing functions of Siah1 and Siah2 in cancer [82]. In addition, inhibition of Siah1 expression increases phospho-ERK levels in breast cancer cells and enhances their invasive activity [83], suggesting that Siah1 acts in a Sprouty2-independent manner. Inhibition of Siah1 also reduces apoptosis and promotes survival of irradiated breast cancer cells [84], suggesting that Siah1 may contribute to the resistance of breast cancer to radiotherapy.
Overexpression of Siah1 induces ubiquitination-dependent degradation of several key oncogenic proteins associated with leukemia pathogenesis, including AML1-ETO, PML-RARα [85, 86], AF4, AF4-MLL [87], and the constitutively active mutant FLT3 (FLT3-ITD) [88]. It remains to be determined whether Siah1 regulates the expression of the endogenous oncogenic proteins in the same manner and whether Siah1 acts as a suppressor of leukemogenesis in vivo.
Epilogue
More than 30 substrates of the Siah ubiquitin ligases have been identified to date. Siah2 is clearly a key player in a number of fundamental cellular processes, including mitochondrial dynamics, the hypoxia response, and the DNA damage response (Fig. 1). Notably, these processes are often deregulated in pathological conditions, including neurodegenerative disorders and cancer. Although our understanding of the importance of Siah ligases in normal and pathological conditions is increasing, we do not yet have a full understanding of how these ligases are normally regulated, or how such regulation changes in individual pathological conditions. The possible tumor suppressor role of Siah1 and oncogenic activity of Siah2 are of particular interest. Lastly, the wealth of evidence supporting the importance of Siah enzymes in key biological processes suggests that therapeutics designed to control their ligase activity, individually and combined, will be beneficial for numerous diseases, including cancer. For example, once could envision inhibitors of Siah2 would be beneficial in cancer, as they would reduce the level of HIF1α, and attenuate tumorigenesis and metastasis that are HIF-1α-dependent, as demonstrated in previous reports [38, 39, 53, 78]. Targeting a non-catalytic substrate has been deemed among the more difficult tasks, although more advanced structure-based design holds promise [89].
References
Hershko, A., & Ciechanover, A. (1998). The ubiquitin system. Annual Review of Biochemistry, 67, 425–479.
Schnell, J. D., & Hicke, L. (2003). Non-traditional functions of ubiquitin and ubiquitin-binding proteins. Journal of Biological Chemistry, 278, 35857–35860.
Reed, J. C., & Ely, K. R. (2002). Degrading liaisons: Siah structure revealed. Natural Structural Biology, 9, 8–10.
Genbacev, O., Zhou, Y., Ludlow, J. W., & Fisher, S. J. (1997). Regulation of human placental development by oxygen tension. Science, 277, 1669–1672.
Tang, A. H., Neufeld, T. P., Kwan, E., & Rubin, G. M. (1997). PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell, 90, 459–467.
Della, N. G., Senior, P. V., & Bowtell, D. D. (1993). Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development, 117, 1333–1343.
Matsuzawa, S. I., & Reed, J. C. (2001). Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Molecular Cell, 7, 915–926.
Li, S., Xu, C., & Carthew, R. W. (2002). Phyllopod acts as an adaptor protein to link the sina ubiquitin ligase to the substrate protein tramtrack. Molecular and Cellular Biology, 22, 6854–6865.
Frew, I. J., Hammond, V. E., Dickins, R. A., Quinn, J. M., Walkley, C. R., Sims, N. A., et al. (2003). Generation and analysis of Siah2 mutant mice. Molecular and Cellular Biology, 23, 9150–9161.
Liu, M., Hsu, J., Chan, C., Li, Z., & Zhou, Q. (2012). The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Molecular Cell, 46, 325–334.
Grishina, I., Debus, K., Garcia-Limones, C., Schneider, C., Shresta, A., Garcia, C., et al. (2012). SIAH-mediated ubiquitination and degradation of acetyl-transferases regulate the p53 response and protein acetylation. Biochimica et Biophysica Acta, 1823, 2287–2296.
Kilroy, G., Kirk-Ballard, H., Carter, L. E., & Floyd, Z. E. (2012). The ubiquitin ligase Siah2 regulates PPARgamma activity in adipocytes. Endocrinology, 153, 1206–1218.
Zhao, H. L., Ueki, N., & Hayman, M. J. (2010). The Ski protein negatively regulates Siah2-mediated HDAC3 degradation. Biochemical and Biophysical Research Communications, 399, 623–628.
Wu, H., Lin, Y., Shi, Y., Qian, W., Tian, Z., Yu, Y., et al. (2010). SIAH-1 interacts with mammalian polyhomeotic homologues HPH2 and affects its stability via the ubiquitin-proteasome pathway. Biochemical and Biophysical Research Communications, 397, 391–396.
Calzado, M. A., de la Vega, L., Moller, A., Bowtell, D. D., & Schmitz, M. L. (2009). An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nature Cell Biology, 11, 85–91.
Winter, M., Sombroek, D., Dauth, I., Moehlenbrink, J., Scheuermann, K., Crone, J., et al. (2008). Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nature Cell Biology, 10, 812–824.
Buchwald, M., Pietschmann, K., Brand, P., Gunther, A., Mahajan, NP., Heinze,l T., and Kramer OH (2012) SIAH ubiquitin ligases target the nonreceptor tyrosine kinase ACK1 for ubiquitinylation and proteasomal degradation. Oncogene. 2012.
Zhou, Y., Li, L., Liu, Q., Xing, G., Kuai, X., Sun, J., et al. (2008). E3 ubiquitin ligase SIAH1 mediates ubiquitination and degradation of TRB3. Cellular Signalling, 20, 942–948.
Yun, S., Moller, A., Chae, S. K., Hong, W. P., Bae, Y. J., Bowtell, D. D., et al. (2008). Siah proteins induce the epidermal growth factor-dependent degradation of phospholipase Cepsilon. Journal of Biological Chemistry, 283, 1034–1042.
Nadeau, R. J., Toher, J. L., Kovalenko, D., & Friesel, R. (2007). Regulation of Sprouty2 stability by mammalian Seven-in-Absentia homolog 2. Journal of Cellular Biochemistry, 100, 151–160.
Famulski, J. K., Trivedi, N., Howell, D., Yang, Y., Tong, Y., Gilbertson, R., et al. (2010). Siah regulation of Pard3A controls neuronal cell adhesion during germinal zone exit. Science, 330, 1834–1838.
Nagano, Y., Fukushima, T., Okemoto, K., Tanaka, K., Bowtell, D. D., Ronai, Z., et al. (2011). Siah1/SIP regulates p27(kip1) stability and cell migration under metabolic stress. Cell Cycle, 10, 2592–2602.
Bhanot, M., & Smith, S. (2012). TIN2 stability is regulated by the E3 ligase Siah2. Molecular and Cellular Biology, 32, 376–384.
Sarkar, T. R., Sharan, S., Wang, J., Pawar, S. A., Cantwell, C. A., Johnson, P. F., et al. (2012). Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPdelta expression and contributes to transformation of breast tumor cells. Molecular and Cellular Biology, 32, 320–332.
Zhao, J., Wang, C., Wang, J., Yang, X., Diao, N., Li, Q., et al. (2011). E3 ubiquitin ligase Siah-1 facilitates poly-ubiquitylation and proteasomal degradation of the hepatitis B viral X protein. FEBS Letters, 585, 2943–2950.
Wu, H., Shi, Y., Lin, Y., Qian, W., Yu, Y., & Huo, K. (2011). Eukaryotic translation elongation factor 1 delta inhibits the ubiquitin ligase activity of SIAH-1. Molecular and Cellular Biochemistry, 357, 209–215.
Scortegagna, M., Subtil, T., Qi, J., Kim, H., Zhao, W., Gu, W., et al. (2011). USP13 enzyme regulates Siah2 ligase stability and activity via noncatalytic ubiquitin-binding domains. Journal of Biological Chemistry, 286, 27333–27341.
Nagel, C. H., Albrecht, N., Milovic-Holm, K., Mariyanna, L., Keyser, B., Abel, B., et al. (2011). Herpes simplex virus immediate-early protein ICP0 is targeted by SIAH-1 for proteasomal degradation. Journal of Virology, 85, 7644–7657.
Garrison, J. B., Correa, R. G., Gerlic, M., Yip, K. W., Krieg, A., Tamble, C. M., et al. (2011). ARTS and Siah collaborate in a pathway for XIAP degradation. Molecular Cell, 41, 107–116.
Twomey, E., Li, Y., Lei, J., Sodja, C., Ribecco-Lutkiewicz, M., Smith, B., et al. (2010). Regulation of MYPT1 stability by the E3 ubiquitin ligase SIAH2. Experimental Cell Research, 316, 68–77.
Ban, R., Matsuzaki, H., Akashi, T., Sakashita, G., Taniguchi, H., Park, S. Y., et al. (2009). Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. Journal of Biological Chemistry, 284, 28367–28381.
Kim, H., Scimia, M. C., Wilkinson, D., Trelles, R. D., Wood, M. R., Bowtell, D., et al. (2011). Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Molecular Cell, 44, 532–544.
Carlucci, A., Adornetto, A., Scorziello, A., Viggiano, D., Foca, M., Cuomo, O., et al. (2008). Proteolysis of AKAP121 regulates mitochondrial activity during cellular hypoxia and brain ischaemia. EMBO Journal, 27, 1073–1084.
Szczepanowski, M., Adam-Klages, S., Kruse, M. L., Pollmann, M., Klapper, W., Parwaresch, R., et al. (2007). Regulation of repp 86 stability by human Siah2. Biochemical and Biophysical Research Communications, 362, 485–490.
Nakayama, K., Frew, I. J., Hagensen, M., Skals, M., Habelhah, H., Bhoumik, A., et al. (2004). Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell, 117, 941–952.
Yego, E. C., & Mohr, S. (2010). Siah-1 protein is necessary for high glucose-induced glyceraldehyde-3-phosphate dehydrogenase nuclear accumulation and cell death in Muller cells. Journal of Biological Chemistry, 2010(285), 3181–3190.
Fiucci, G., Beaucourt, S., Duflaut, D., Lespagnol, A., Stumptner-Cuvelette, P., Geant, A., et al. (2004). Siah-1b is a direct transcriptional target of p53: identification of the functional p53 responsive element in the siah-1b promoter. Proc Natl Acad Sci U S A., 101, 3510–3515.
Qi, J., Nakayama, K., Gaitonde, S., Goydos, J. S., Krajewski, S., Eroshkin, A., et al. (2008). The ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by HIF-dependent and -independent pathways. Proc Natl Acad Sci USA, 105, 16713–16718.
Qi, J., Nakayama, K., Cardiff, R. D., Borowsky, A. D., Kaul, K., Williams, R., et al. (2010). Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell, 18, 23–38.
Chan, P., Moller, A., Liu, M. C., Sceneay, J. E., Wong, C. S., Waddell, N., et al. (2011). The expression of the ubiquitin ligase SIAH2 (seven in absentia homolog 2) is mediated through gene copy number in breast cancer and is associated with a basal-like phenotype and p53 expression. Breast Cancer Research, 13, R19.
Ahmed, A. U., Schmidt, R. L., Park, C. H., Reed, N. R., Hesse, S. E., Thomas, C. F., et al. (2008). Effect of disrupting seven-in-absentia homolog 2 function on lung cancer cell growth. Journal of the National Cancer Institute, 100, 1606–1629.
Schmidt, R. L., Park, C. H., Ahmed, A. U., Gundelach, J. H., Reed, N. R., Cheng, S., et al. (2007). Inhibition of RAS-mediated transformation and tumorigenesis by targeting the downstream E3 ubiquitin ligase seven in absentia homologue. Cancer Research, 67, 11798–11810.
Brauckhoff, A., Malz, M., Tschaharganeh, D., Malek, N., Weber, A., Riener, M. O., et al. (2011). Nuclear expression of the ubiquitin ligase seven in absentia homolog (SIAH)-1 induces proliferation and migration of liver cancer cells. Journal of Hepatology, 55, 1049–1057.
Malz, M., Aulmann, A., Samarin, J., Bissinger, M., Longerich, T., Schmitt, S., et al. (2012). Nuclear accumulation of seven in absentia homologue-2 supports motility and proliferation of liver cancer cells. International Journal of Cancer, 131, 2016–2026.
Frasor, J., Danes, J. M., Funk, C. C., & Katzenellenbogen, B. S. (2005). Estrogen down-regulation of the corepressor N-CoR: mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc Natl Acad Sci USA, 102, 13153–13157.
Maeda, A., Yoshida, T., Kusuzaki, K., & Sakai, T. (2002). The characterization of the human Siah-1 promoter(1). FEBS Letters, 512, 223–226.
Topol, L., Jiang, X., Choi, H., Garrett-Beal, L., Carolan, P. J., & Yang, Y. (2003). Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. Journal of Cell Biology, 162, 899–908.
Xie, W., Mei, Y., & Wu, M. (2009). E2F1 represses beta-catenin/TCF activity by direct up-regulation of Siah1. Journal of Cellular and Molecular Medicine, 13, 1719–1727.
MacLeod, R. J., Hayes, M., & Pacheco, I. (2007). Wnt5a secretion stimulated by the extracellular calcium-sensing receptor inhibits defective Wnt signaling in colon cancer cells. American Journal of Gastrointestinal and Liver Physiology, 293, G403–G411.
Xu, Z., Sproul, A., Wang, W., Kukekov, N., & Greene, L. A. (2006). Siah1 interacts with the scaffold protein POSH to promote JNK activation and apoptosis. Journal of Biological Chemistry, 281, 303–312.
Khurana, A., Nakayama, K., Williams, S., Davis, R. J., Mustelin, T., & Ronai, Z. (2006). Regulation of the ring finger E3 ligase Siah2 by p38 MAPK. Journal of Biological Chemistry, 281, 35316–35326.
Perez, M., Garcia-Limones, C., Zapico, I., Marina, A., Schmitz, M. L., Munoz, E., et al. (2012). Mutual regulation between SIAH2 and DYRK2 controls hypoxic and genotoxic signaling pathways. J Mol Cell Biol, 4, 316–330.
Shah, M., Stebbins, J. L., Dewing, A., Qi, J., Pellecchia, M., & Ronai, Z. A. (2009). Inhibition of Siah2 ubiquitin ligase by vitamin K3 (menadione) attenuates hypoxia and MAPK signaling and blocks melanoma tumorigenesis. Pigment Cell Melanoma Res, 22, 799–808.
Le Moan, N., Houslay, D. M., Christian, F., Houslay, M. D., & Akassoglou, K. (2011). Oxygen-dependent cleavage of the p75 neurotrophin receptor triggers stabilization of HIF-1alpha. Molecular Cell, 44, 476–490.
Liao Y, Zhang M and Lonnerdal B (2012) Growth factor TGF-beta induces intestinal epithelial cell (IEC-6) differentiation: miR-146b as a regulatory component in the negative feedback loop. Genes Nutr. 2012.
Imig, J., Motsch, N., Zhu, J. Y., Barth, S., Okoniewski, M., Reineke, T., et al. (2011). microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Research, 39, 1880–1893.
Pang, R. T., Liu, W. M., Leung, C. O., Ye, T. M., Kwan, P. C., Lee, K. F., et al. (2011). miR-135A regulates preimplantation embryo development through down-regulation of E3 Ubiquitin Ligase Seven In Absentia Homolog 1A (SIAH1A) expression. PLoS ONE, 6, e27878.
Tofaris, G. K., Razzaq, A., Ghetti, B., Lilley, K. S., & Spillantini, M. G. (2003). Ubiquitination of alpha-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. Journal of Biological Chemistry, 278, 44405–44411.
Liani, E., Eyal, A., Avraham, E., Shemer, R., Szargel, R., Berg, D., et al. (2004). Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson’s disease. Proc Natl Acad Sci USA, 101, 5500–5505.
Anderson, J. P., Walker, D. E., Goldstein, J. M., de Laat, R., Banducci, K., Caccavello, R. J., et al. (2006). Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. Journal of Biological Chemistry, 281, 29739–29752.
Lee, J. T., Wheeler, T. C., Li, L., & Chin, L. S. (2008). Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Human Molecular Genetics, 17, 906–917.
Engelender, S., Kaminsky, Z., Guo, X., Sharp, A. H., Amaravi, R. K., Kleiderlein, J. J., et al. (1999). Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nature Genetics, 22, 110–114.
Nagano, Y., Yamashita, H., Takahashi, T., Kishida, S., Nakamura, T., Iseki, E., et al. (2003). Siah-1 facilitates ubiquitination and degradation of synphilin-1. Journal of Biological Chemistry, 278, 51504–51514.
Eyal, A., Szargel, R., Avraham, E., Liani, E., Haskin, J., Rott, R., et al. (2006). Synphilin-1A: an aggregation-prone isoform of synphilin-1 that causes neuronal death and is present in aggregates from alpha-synucleinopathy patients. Proc Natl Acad Sci USA, 103, 5917–5922.
Szargel, R., Rott, R., Eyal, A., Haskin, J., Shani, V., Balan, L., et al. (2009). Synphilin-1A inhibits seven in absentia homolog (SIAH) and modulates alpha-synuclein monoubiquitylation and inclusion formation. Journal of Biological Chemistry, 284, 11706–11716.
Fukuba, H., Takahashi, T., Jin, H. G., Kohriyama, T., & Matsumoto, M. (2008). Abundance of aspargynyl-hydroxylase FIH is regulated by Siah-1 under normoxic conditions. Neuroscience Letters, 433, 209–214.
Calzado, M. A., de la Vega, L., Munoz, E., & Schmitz, M. L. (2009). Autoregulatory control of the p53 response by Siah-1L-mediated HIPK2 degradation. Biological Chemistry, 390, 1079–1083.
Merrill, R. A., Dagda, R. K., Dickey, A. S., Cribbs, J. T., Green, S. H., Usachev, Y. M., et al. (2011). Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biology, 9, e1000612.
Ong, S. B., Subrayan, S., Lim, S. Y., Yellon, D. M., Davidson, S. M., & Hausenloy, D. J. (2010). Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation, 121, 2012–2022.
D’Orazi, G., Cecchinelli, B., Bruno, T., Manni, I., Higashimoto, Y., Saito, S., et al. (2002). Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nature Cell Biology, 4, 11–19.
Hofmann, T. G., Moller, A., Sirma, H., Zentgraf, H., Taya, Y., Droge, W., et al. (2002). Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nature Cell Biology, 4, 1–10.
Zhang, Q., Yoshimatsu, Y., Hildebrand, J., Frisch, S. M., & Goodman, R. H. (2003). Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell, 115, 177–186.
Longhese, M. P., Anbalagan, S., Martina, M., & Bonetti, D. (2012). The role of shelterin in maintaining telomere integrity. Frontiers in Bioscience, 17, 1715–1728.
Fujita, K., Horikawa, I., Mondal, A. M., Jenkins, L. M., Appella, E., Vojtesek, B., et al. (2010). Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nature Cell Biology, 12, 1205–1212.
Karlseder, J., Smogorzewska, A., & de Lange, T. (2002). Senescence induced by altered telomere state, not telomere loss. Science, 295, 2446–2449.
Liu, D., Safari, A., O’Connor, M. S., Chan, D. W., Laegeler, A., Qin, J., et al. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nature Cell Biology, 6, 673–680.
Wong, C. S., Sceneay, J., House, C. M., Halse, H. M., Liu, M. C., George, J., et al. (2012). Vascular normalization by loss of Siah2 results in increased chemotherapeutic efficacy. Cancer Research, 72, 1694–1704.
Moller, A., House, C. M., Wong, C. S., Scanlon, D. B., Liu, M. C., Ronai, Z., et al. (2009). Inhibition of Siah ubiquitin ligase function. Oncogene, 28, 289–296.
Liao, D., Corle, C., Seagroves, T. N., & Johnson, R. S. (2007). Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Research, 67, 563–572.
Matsuo, K., Satoh, S., Okabe, H., Nomura, A., Maeda, T., Yamaoka, Y., et al. (2003). SIAH1 inactivation correlates with tumor progression in hepatocellular carcinomas. Genes, Chromosomes & Cancer, 36, 283–291.
Kim, C. J., Cho, Y. G., Park, C. H., Jeong, S. W., Nam, S. W., Kim, S. Y., et al. (2004). Inactivating mutations of the Siah-1 gene in gastric cancer. Oncogene, 23, 8591–8596.
Wen, Y. Y., Yang, Z. Q., Song, M., Li, B. L., Yao, X. H., Chen, X. L., et al. (2010). The expression of SIAH1 is downregulated and associated with Bim and apoptosis in human breast cancer tissues and cells. Molecular Carcinogenesis, 49, 440–449.
Wen, Y. Y., Yang, Z. Q., Song, M., Li, B. L., Zhu, J. J., & Wang, E. H. (2010). SIAH1 induced apoptosis by activation of the JNK pathway and inhibited invasion by inactivation of the ERK pathway in breast cancer cells. Cancer Science, 101, 73–79.
He, H. T., Fokas, E., You, A., Engenhart-Cabillic, R., & An, H. X. (2010). Siah1 proteins enhance radiosensitivity of human breast cancer cells. BMC Cancer, 10, 403.
Kramer, O. H., Muller, S., Buchwald, M., Reichardt, S., & Heinzel, T. (2008). Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. FASEB J., 22, 1369–1379.
Fanelli, M., Fantozzi, A., De Luca, P., Caprodossi, S., Matsuzawa, S., Lazar, M. A., et al. (2004). The coiled-coil domain is the structural determinant for mammalian homologues of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. Journal of Biological Chemistry, 279, 5374–5379.
Bursen, A., Moritz, S., Gaussmann, A., Moritz, S., Dingermann, T., & Marschalek, R. (2004). Interaction of AF4 wild-type and AF4.MLL fusion protein with SIAH proteins: indication for t(4;11) pathobiology? Oncogene, 23, 6237–6249.
Buchwald, M., Pietschmann, K., Muller, J. P., Bohmer, F. D., Heinzel, T., & Kramer, O. H. (2010). Ubiquitin conjugase UBCH8 targets active FMS-like tyrosine kinase 3 for proteasomal degradation. Leukemia, 24, 1412–1421.
Qi, J., Pellecchia, M., & Ronai, Z. A. (2010). The Siah2-HIF-FoxA2 axis in prostate cancer—new markers and therapeutic opportunities. Oncotarget, 5, 379–385.
Acknowledgments
Support by NCI Grants CA099961 and CA128814 (to ZR) and CA154888 (to JQ) is gratefully acknowledged. We thank Allan Weisman and Serge Fuchs for the critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jianfei Qi and Hyungsoo Kim contributed equally to this study.
Rights and permissions
About this article
Cite this article
Qi, J., Kim, H., Scortegagna, M. et al. Regulators and Effectors of Siah Ubiquitin Ligases. Cell Biochem Biophys 67, 15–24 (2013). https://doi.org/10.1007/s12013-013-9636-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-013-9636-2