Abstract
Dye contaminants in industrial effluents contribute significantly to environmental pollution. Conventional wastewater treatment methods are expensive and energy-consuming. These limitations could be overcome by the use of nanobioremediation processes. The present work was an effort to study decolorization of azo dyes by ZnO nanoparticles (NPs). Rubine GDB (R-GDB) and Congo Red (CR) were the azo dyes selected for the study. The ZnO NPs were successfully synthesized by chemical and biological methods. Chemical synthesis of ZnO NPs was carried out by co-precipitation method; biosynthesis was done using bacteria Bacillus subtilis. The synthesized nanoparticles were characterized by UV–Vis Spectroscopy, SEM, and XRD. The UV spectrophotometer showed peaks in the range of 300–400 nm. SEM pictures indicated the presence of NPs in the size of 100–200 nm. XRD results were analyzed based on the peaks that were seen. EDX analysis showed the presence of Zn particles and oxygen particles majorly. Decolorization efficiency was evaluated by calculating % decolorization by Meyer’s method. Chemically synthesized NPs showed maximum % decolorization of the R-GDB (89.55 ± 0.44%) and CR (88.52 ± 0.90%). The biosynthesized NPs showed the least decolorization (R-GDB, 18.46 ± 0.45% and CR, 21.41 ± 1.02%). However, moderate percentages of decolorization of both the azo dyes were observed when a combination of the NPs was used (R-GDB, 36.25 ± 0.22% and CR, 39.47 ± 0.94%). Nanoparticles showed good potential for the decolorization of the azo dyes. With further optimization of the parameters, the present findings show that dye decolorization by chemically synthesized ZnO NPs could be used as a probable nanobioremediation approach to treat wastewaters.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Discharges of colored wastewater consisting of a mixture of dyes polluting the environment are a matter of great concern and challenge. This issue is being faced throughout the world. India, along with other developing countries, faces a huge part of this worry as hardly any treatment is being carried out before the discharge. The cause of all of these issues is textile industries [1,2,3]. The disposal of wastewater generated from these industries is an alarming threat to nature. It accounts for 15–20% of the total wastewater in the country [4]. Azo dyes are a large group that is most commonly used in dyeing textile fibers, i.e., cotton, wool, silk, and synthetic fibers. They are the most common as they are affordable and easy to use and provide strong colors that are easily distinguished. There are approximately 2000 dyes available.
Around 4–5% azo dyes cleave to form aromatic amines [5]. Cancer and aromatic amines are found to be in close relation. Azo dyes owe their carcinogenic and detrimental properties to the presence of aromatic amines and their water-soluble nature [5]. Rubine-GDB (R-GDB) is an azo dye that is used to impart a red color to fabrics. It is also used in paints, printing inks, textile printing, and plastics, and it is carcinogenic in nature. Congo Red (CR) is an azo dye that is water soluble. In earlier times, it was used to impart a lighter shade of red color to cotton which then became more popular as a staining agent in histology and as an acid–base indicator. The most effective and substantially efficient methods to date have been biological techniques [6]. Consistent research has been taking place for developing newer and better techniques for the degradation of dyes.
Nanoparticles (NPs) can be used for the treatment of wastewater released from industries. The synthesis of NPs can be carried out by various physical, chemical, and biological methods. The biosynthesized NPs have drawn enormous focus due to their non-toxic and eco-friendly nature. Consequently, nanobiotechnology is now regarded as the epitome of producing eco-friendly and cost-effective nanoparticles for the treatment of wastewater and dye degradation [7]. NPs have a structural dimension of less than 100 nm which makes them comparable to sub-cellular structures thereby enabling them to get incorporated into biological systems [8]. Zinc oxide NPs have a large surface area and high catalytic activity. These nanoparticles have different physical and chemical properties by the source of their synthesis. Recently, these ZnO NPs have gained much acclamation for dye degradation procedures [9,10,11].
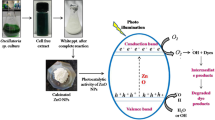
Biosynthesis of NPs using bacteria has been of special interest as it involves easy handling procedures and eco-friendly disposal methods and is very easy for downstream processing. Recently, eco-friendly synthesis of metal NPs using various strains of bacteria such as Bacillus species and their use in dye decolorization and degradation has been reported [12]. The present investigation was an effort to study the efficiency of azo dye decolorization of R-GDB and CR by ZnO NPs synthesized following chemical and biological routes.
Materials and Methods
Chemical synthesis was carried out by co-precipitation method and biosynthesis was done using Bacillus subtilis.
Synthesis of ZnO NPs by Chemical Route
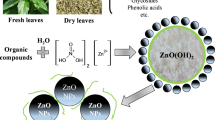
Zinc nitrate hexahydrate (ZnNO3.6H2O; 0.1 M) and sodium hydroxide (NaOH; 0.8 M) solutions were prepared using distilled water. After complete dissolution of zinc nitrate, NaOH was added drop-wise with continuous stirring for 45 min. A white colloid was formed which was allowed to settle overnight. The supernatant was removed and the remaining solution was centrifuged at 5000 rpm for 10 min. The precipitate was collected and washed with de-ionized water and ethanol to remove the by-products. It was later subjected to drying at 60 °C for 2–3 days [13]. The absorption spectrum for the synthesized nanoparticles was analyzed by UV–Visible spectroscopy (Shimadzu, UV 1700 PharmaSpec, scanning range: 200–400 nm). The size and morphology of the powder were studied by scanning electron microscope (SEM) and X-ray diffraction (XRD) was analyzed [13].
Synthesis of ZnO NPs by Biological Route
Zinc acetate dihydrate (Zn(CH3COO)2 · 2H2O; 0.001 M) was added into B. subtilis inoculum prepared in nutrient broth and was incubated at 37 °C overnight. The culture was centrifuged at 5000 rpm for 15 min. Pellet was collected, washed, and was dried completely in a hot air oven at 60 °C until a fine powder was obtained. The size and morphology of the powder were studied by SEM and XRD was recorded [14].
Characterization of Nanoparticles
UV–Vis Spectroscopy
ZnO nanoparticles obtained by the chemical synthesis and biosynthesis were characterized using UV–Vis spectroscopy. A suspension was prepared by adding NPs in milli-q water, it was transferred to the cuvette, and analysis was carried out using UV–Visible spectrophotometer (Shimadzu, UV-1700 Pharma Spec).
XRD Analysis
The crystallite structure of NPs was analyzed using XRD (Rigaku X-ray diffractometer at CeNSE laboratory, IISc, Bengaluru). The size was determined with the use of Scherrer’s equation,
where is D is the average crystallite size, K is the Scherrer constant, θ is the Bragg angle, β is the line broadening in radians, and λ is the X-ray wavelength [15].
Scanning Electron Microscope Analysis
The SEM analysis was done to determine the surface structure, shape, and size of the NPs. This was performed using Zeiss, Ultra 55 Field Emission Scanning Electron Microscope (CeNSE laboratory, Indian Institute of Science (IISc), Bengaluru) [16].
Energy-Dispersive X-ray Spectroscopy (EDX)
The elemental composition, chemical purity, and stoichiometry of the synthesized ZnO nanoparticles were determined by the EDX analysis [16]. It is performed by using Zeiss, Ultra 55 Field Emission Scanning Electron Microscope (CeNSE laboratory, IISc., Bengaluru).
Decolorization of Azo Dyes Using NPs
Stock solutions of the azo dyes at a concentration of 1 mg mL−1 were prepared in milli-q water. Decolorization studies were carried out in three sets in a closed batch culture system at room temperature (28 ± 2 °C). NPs were added to the dye solutions taken in glass screw-capped tubes at a concentration of 250 ppm with continuous stirring at 75 rpm on a rotary shaker. The first set had chemically synthesized ZnO NPs, the second set had NPs biosynthesized, and the third set had both chemically and biosynthesized NPs. Samples were withdrawn every 24 h and centrifuged at 3000 rpm for 10 min. The clear supernatant was pipetted and absorbance was read at λmax of 580 nm. Dye decolorization percentage was calculated by the following formula [17]:
Results and Discussion
NPs were successfully synthesized following chemical and biological routes. During biosynthesis, reduction of zinc ions into zinc NPs occurred on addition of bacterial cell extracts; as a result, a color change was observed. Generally, the metal NPs have free electrons, which help in the formation of the Surface Plasmon Resonance absorption band. It happens due to the united vibration of the electrons of metal nanoparticles in resonance with light wave [18].
Characterization of ZnO NPs
The synthesized NPs were characterized using UV–Visible spectroscopy, XRD, SEM, energy-dispersive X-ray analysis (EDX).
UV–Visible Absorption Analysis of ZnO NPs
Using UV–Vis spectroscopy, the absorption spectra of ZnO NPs were obtained. The maximum absorption (λmax) was obtained in the range of 380 to 390 nm for chemical ZnO NPs and 370 to 380 nm for biosynthesized NPs. The results obtained in the present study show similarities with the findings of previous literature where they reported absorption maxima in a range of 320–390 nm which is a characteristic band for the ZnO [19].
X-ray Diffraction Analysis
An XRD pattern obtained for the NPs is indicated in Figs. 1 and 2. The crystallite size of chemically synthesized NPs determined by Scherrer’s equation was 24.419 nm. A sharp peak was obtained at 101 (Fig. 1). This confirms the sample to be ZnO NPs in polycrystalline form. However, biosynthesized NPs were amorphous and no crystals were formed. Hence, the size of crystallite size could not be estimated since a clear graph was not obtained. This could be due to the presence of impurities in the sample (Fig. 2). The broadening line of the diffraction peaks indicates that the synthesized particles are in the nm range [20]. Peaks obtained other than the characteristic peaks are also due to impurities.
SEM Analysis of ZnO NPs
The SEM images of chemically synthesized ZnO NPs showed spherical non-agglomerated, uniform-shaped, and evenly distributed nanoflakes (Fig. 3). The average diameter of the structures ranged from 100 to 200 nm. Results are similar to previous literature having dimension around 200 nm × 200 nm [21].
SEM images of biosynthesized NPs were nanorod shape embedded nanoparticle structures with hints of agglomeration in the bacterial biomass. The average diameters of the structures were in the range of 100–200 nm and the length in 1–2 µm range. The structures are seen to be distributed evenly and are of similar shapes. SEM images indicated the length (nm) and shape of the synthesized NPs (Table 1). These images can be correlated with the previous literature having average diameter of 100–200 nm and length to be in micrometers [22]. Synthesis of NPs using bacteria has been studied by several researchers but the exact mechanism is still not clear; however, cells of bacteria play role in the formation of NPs [23].
Energy-Dispersive X-ray Analysis
The results indicate the sample is made of ZnO, with major amounts of Zn (Zinc) and O (Oxygen) is present in both chemically synthesized and biosynthesized NPS. Carbon present in the sample is due to the carbon tape which was used as a sample holder. Hence, it can be said that the purification has been carried out well. The weight percentage is 65.95, and the sample had fairly 65–66% of Zn (Table 2). Previous literatures indicate that the obtained characteristic peaks of Zn and O confirm the presence of ZnO NPs with no impurities [21]. Trace amounts of impurities were also detected in the analysis of biosynthesized NPs, which indicates that the samples were impure. Since the weight percentage is 65.97, the sample had fairly 65–66% of Zn (Table 2). The impurities were detected because biological molecules combine with the NPs in the bacteria [24].
Decolorization of Dyes using NPs
Azo dye decolorization was observed in all three sets. Decolorization percentages of R-GDB by the NPs at 48 h, 72 h, and 96 h of incubation are indicated in Fig. 4. For the Rubine dye, as is evident from Table 3 and Fig. 4, the maximum decolorization was observed at 96 h of incubation. Chemically synthesized NPs (89.55 ± 0.44%) showed the maximum decolorization percentage followed by a combination of both NPs (36.25 ± 0.22%) and then the biosynthesized NPs (18.46 ± 0.45%).
Decolorization of CR by NPs
As it is evident from Table 4 and Fig. 5, the maximum decolorization of CR dye was obtained at 96 h of incubation. Chemically synthesized NPs showed maximum decolorization (88.52 ± 0.90%) followed by a combination of both NPs (39.47 ± 0.94%) and then biosynthesized NPs (21.41 ± 1.02%).
Present findings of decolorization studies of azo dye by NPs showed that chemically synthesized ZnO NPs could decolorize azo dyes which are similar to earlier findings [25]. Percent decolorization of the dye increased with an increase in the treatment duration, there was maximum at 96 h (R-GDB, 89.55 ± 0.44; CR, 88.52 ± 0.90%), and the least was at 48 h (R-GDB, 4.34 ± 0.08; CR, 15.55 ± 0.22%). With the same concentration of NPs taken, decolorization percentage increases with the treatment time; also, the more the number of NPs, the more is it likely for the catalyst (NPs) to attack the dye’s chromopheric system [26]. Previous studies indicate that the decolorization potential of immobilized cells was also compared with the free cells. At optimum experimental conditions, the complete dye removal was observed at 18 h by free cells whereas nanoparticle coated cells showed complete decolorization at 24 h for 100 ppm of RB5 dye concentration, which is in accordance to the current study. In the current work, the biosynthesized NPs showed maximum decolorization at 96 h [27].
The enzymatic degradation of azo dyes by the bacteria occurs either by extracellular or intracellular enzymes. The enzymes that are commonly involved in azo dye degradation are azoreductase, laccase, peroxidases, etc. These enzymes require appropriate environmental conditions to maintain their stability and activity. Laccase is copper-containing oxidases that do not require redox mediators for degrading azo dyes [28]. In the current study, biosynthesized NPs showed the least dye decolorization when compared to chemically synthesized and combined NPs. The probable reason could be due to the lesser quantity of intercellular enzymes which aid in decolorization.
ZnO is known to have antimicrobial properties; hence, this property of ZnO can inhibit the growth of Bacillus subtilis [29]. ZnO NPs have anti-bacterial properties and they tend to disrupt the cell membranes of bacteria [30]. The dye degrading efficiency depends on the size, shape, and efficiency of the NPs. The size of the nanoparticles has a vital role, i.e., the smaller the size of the nanoparticles, the greater is its efficiency [31, 32]. Nanoparticles having smaller size get easily adhered to the reactants and thus, the rate of dye degradation also increases [33]. These facts can be correlated with the results obtained in the current study, as the percentage degradation with chemically synthesized NPs was seen to be higher as compared to the biosynthesized NPs due to the difference in size and shape of the NPs obtained (Table 1).
NPs synthesized via biological method have limitations as its presence in effluent water is seen even after degradation. Thus, recovery and reusability are needed [34]. Hence, this might be the reason that both the dyes were not efficiently decolorized with biosynthesized NPs.
Conclusion
Industrial wastewater has very high quantity of pollutants that could be removed by chemical and biological methods. In the current study, NPs were synthesized using chemical and biological reduction systems. This study reports the novel approach to decolorize Rubine GDB and Congo Red dyes commonly found textile effluents. Chemically synthesized NPs showed high percentage of decolorization for Rubine GDB (89.55 ± 0.44%) and Congo Red (88.52 ± 0.90%). Optimization of the process parameters to get uniform shaped and sized NPs which could further enhance the decolorization efficiency. Understanding the mechanisms that are involved in the biosynthesis of ZnO NPs using Bacillus subtilis needs further investigation. The process could be scaled up at commercial level. In the coming years, NPs would hold prominent role in wastewater treatment and could become an effective approach for dye removal from textile wastewaters. Nanomaterials are believed to completely replace conventional methods in degradation of dye [31].
Code Availability
Not applicable.
References
Govindarajalu, K., & Govindarajalu, K. (2003, December). Industrial effluent and health status: A case study of Noyyal river basin. In Proceedings of the third international conference on environment and health, Chennai, India (Vol. 15, No. 17, pp. 150–157).
Appasamy, P. P., & Nelliyat, P. (2007). Compensating the loss of ecosystem services due to pollution in Noyyal River basin, Tamil Nadu. Madras School of Economics.
Sivakumar, K. K., Balamurugan, C., Ramakrishnan, D., & Bhai, L. H. (2011). Assessment studies on wastewater pollution by textile dyeing and bleaching industries at Karur Tamil Nadu. Rasayan Journal of Chemistry, 4(2), 264–269.
Gopal, B. (1999). Natural and constructed wetlands for wastewater treatment: Potentials and problems. Water Science and Technology, 40(3), 27–35.
Nijkamp, M. M., Maslankiewics, L., Delmaar, J. E., & Muller, J. J. A. (2015). Hazardous substances in textile products.
Sarkar, S., Banerjee, A., Halder, U., Biswas, R., & Bandopadhyay, R. (2017). Degradation of synthetic azo dyes of textile industry: A sustainable approach using microbial enzymes. Water Conservation Science and Engineering, 2(4), 121–131.
Nomoto, K. I., Tominaga, N., Umeda, H., Kobayashi, C., & Maeda, K. (2006). Nucleosynthesis yields of core-collapse supernovae and hypernovae, and galactic chemical evolution. Nuclear Physics A, 777, 424–458.
Orgeig, S., Morrison, J. L., Sullivan, L. C., & Daniels, C. B. (2014). The development of the pulmonary surfactant system. In The Lung (pp. 183–209). Academic Press.
Kuo, C. L., Kuo, T. J., & Huang, M. H. (2005). Hydrothermal synthesis of ZnO microspheres and hexagonal microrods with sheetlike and platelike nanostructures. The Journal of Physical Chemistry B, 109(43), 20115–20121.
El-Rafie, H. M., El-Rafie, M., & Zahran, M. K. (2013). Green synthesis of silver nanoparticles using polysaccharides extracted from marine macro algae. Carbohydrate Polymers, 96(2), 403–410.
Wei, M., Casey Boutwell, R., Faleev, N., Osinsky, A., & Schoenfeld, W. V. (2013). Growth of high quality ZnO thin films with a homonucleation on sapphire. Journal of Vacuum Science & Technology B, Nanotechnology and Microelectronics: Materials, Processing, Measurement, and Phenomena, 31(4), 041206.
Velusamy, P., Kumar, G. V., Jeyanthi, V., Das, J., & Pachaiappan, R. (2016). Bio-inspired green nanoparticles: Synthesis, mechanism, and antibacterial application. Toxicological Research, 32(2), 95–102.
Talam, S., Karumuri, S. R., & Gunnam, N. (2012). Synthesis, characterization, and spectroscopic properties of ZnO nanoparticles. International Scholarly Research Notices, 2012, 2.
Willner, I., Basnar, B., & Willner, B. (2007). Nanoparticle–enzyme hybrid systems for nanobiotechnology. The FEBS Journal, 274(2), 302–309.
Becheri, A., Dürr, M., Nostro, P. L., & Baglioni, P. (2008). Synthesis and characterization of zinc oxide nanoparticles: Application to textiles as UV-absorbers. Journal of Nanoparticle Research, 10(4), 679–689.
Chaudhuri, S. K., & Malodia, L. (2017). Biosynthesis of zinc oxide nanoparticles using leaf extract of Calotropis gigantea: Characterization and its evaluation on tree seedling growth in nursery stage. Applied Nanoscience, 7(8), 501–512.
Nekouei, S., & Nekouei, F. (2018). Comparative procedure of photodegradation of methylene blue using N doped activated carbon loaded with hollow 3D flower like ZnS in two synergic phases of adsorption and catalytic. Journal of Photochemistry and Photobiology A: Chemistry, 364, 262–273.
Haes, A. J., & Van Duyne, R. P. (2002). A nanoscale optical biosensor: Sensitivity and selectivity of an approach based on the localized surface plasmon resonance spectroscopy of triangular silver nanoparticles. Journal of the American Chemical Society, 124(35), 10596–10604.
Jamdagni, P., Khatri, P., & Rana, J. S. (2018). Green synthesis of zinc oxide nanoparticles using flower extract of Nyctanthes arbor-tristis and their antifungal activity. Journal of King Saud University-Science, 30(2), 168–175.
Selvarajan, E., & Mohanasrinivasan, V. (2013). Biosynthesis and characterization of ZnO nanoparticles using Lactobacillus plantarum VITES07. Materials Letters, 112, 180–182.
Samanta, P. K., & Saha, A. (2015). Wet chemical synthesis of ZnO nanoflakes and photoluminescence. Optik, 126(23), 3786–3788.
Bhakat, C., & Singh, P. P. (2012). Zinc oxide nanorods: Synthesis and its applications in solar cell. International Journal of Modern Engineering Research, 2, 2452–2454.
Mahdi, Z. S., Talebnia Roshan, F., Nikzad, M., & Ezoji, H. (2020). Biosynthesis of zinc oxide nanoparticles using bacteria: A study on the characterization and application for electrochemical determination of bisphenol A. Inorganic and Nano-Metal Chemistry, 1–9.
Arshad, A. (2017). Bacterial synthesis and applications of nanoparticles. Nanoscience and Nanotechnology, 11(2), 119.
Saleh, S. M. (2019). ZnO nanospheres based simple hydrothermal route for photocatalytic degradation of azo dye. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 211, 141–147.
Kale, R. D., & Kane, P. B. (2017). Colour removal using nanoparticles. Textiles and Clothing Sustainability, 2(1), 1–7.
Vinayak, A., & Singh, G. B. (2021). Biodecolorization of reactive black 5 using magnetite nanoparticles coated Bacillus sp. RA5. Materials Today: Proceedings.
Sridharan, R., Krishnaswamy, V. G., Archana, K. M., Rajagopal, R., Kumar, D. T., & Doss, C. G. P. (2021). Integrated approach on azo dyes degradation using laccase enzyme and Cul nanoparticle. SN Applied Sciences, 3(3), 1–12.
Padmavathy, N., & Vijayaraghavan, R. (2008). Enhanced bioactivity of ZnO nanoparticles—An antimicrobial study. Science and technology of advanced materials.
Xie, Y., He, Y., Irwin, P. L., Jin, T., & Shi, X. (2011). Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Applied and Environmental Microbiology, 77(7), 2325–2331.
Sha, Y., Mathew, I., Cui, Q., Clay, M., Gao, F., Jackie, X., & Gu, Z. (2016). Rapid degradation of azo dye methyl orange using hollow cobalt nanoparticles. Chemosphere, 144, 1530–1535. https://doi.org/10.1016/j.chemosphere.2015.10.04027
Nandhini, N. T., Rajeshkumar, S., & Mythili, S. (2019). The possible mechanism of eco-friendly synthesized nanoparticles on hazardous dyes degradation. Biocatalysis and Agricultural Biotechnology, 19, 101138.
Suvith, V. S., & Philip, D. (2014). Catalytic degradation of methylene blue using biosynthesized gold and silver nanoparticles. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 118, 526–532.
Yi, T. F., Jiang, L. J., Shu, J., Yue, C. B., Zhu, R. S., & Qiao, H. B. (2010). Recent development and application of Li4Ti5O12 as anode material of lithium ion battery. Journal of Physics and Chemistry of Solids, 71(9), 1236–1242.
Acknowledgements
We gratefully acknowledge the CeNSE lab members (Indian Institute of Science, Bengaluru) for helping us with XRD and SEM facility. Thanks to Mr. Pradeep, Mr. Amit, Mr. Abhishek, Ms. Vanitha, Mr. Arun Babu, and Ms. Suma for the guidance to conduct and analyze the XRD and SEM results.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
The participants gave their consent for publication.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Swain, J., Kulkarni, P. & Manuel, S. Decolorization of Azo Dyes by Chemically and Biosynthesized ZnO Nanoparticles. Appl Biochem Biotechnol 194, 1027–1038 (2022). https://doi.org/10.1007/s12010-021-03789-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03789-1