Abstract
The lipids produced by oleaginous microbes are considered sustainable resources for biofuels. To facilitate controlled lipid production and lipid analysis, more efficient lipid extraction methods are required. This study describes the automated pressurized liquid extraction (APLE) method for lipid extraction from dried cells of the oleaginous yeast species Rhodosporidium toruloides and Cryptococcus curvatus. Cells were mixed with diatomite in a mortar, added to the sample chamber, and treated with a mixture of chloroform and methanol at 105 °C. More than 95% lipids were extracted. Analysis by using high-performance thin-layer chromatography showed that the neutral lipid contents in the obtained samples by APLE method were similar to those by the ball milling–assisted extraction method. The lipids had an essentially identical fatty acid composition compared with lipids extracted with the acid-heating extraction (AHE) method. This demonstrated that lipids can be efficiently extracted from oleaginous yeasts in less time and without harsh pretreatment procedures.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Along with vegetable oils, animal fats, and other waste oils, microbial lipids could also be used for biodiesel production. Oleaginous yeasts are excellent candidates for the production of microbial lipids [1]. Rhodosporidium toruloides and Cryptococcus curvatus in particular are two yeasts that can accumulate more than 60% of lipids in their dry cell mass on corn stover hydrolysate [2]. Lipid extraction is a significant part of whole-microbial lipid technology and is especially important for lipid component analysis and process tracking. Reliable and simple methods are required for the qualitative and quantitative analysis of the lipid components extracted from oleaginous yeasts.
Acid-heating extraction (AHE) is a conventional lipid extraction method, which requires complex procedures that include four main steps: drying, cell disruption, extraction of lipids, and removal of the solvents [3,4,5,6]. The AHE method can extract the total lipids of cells but requires a long extraction time and harsh pretreatment [4]. A further conventional method is Soxhlet extraction, which requires at least 4 h to reach 90% of lipid [7]. Several methods have been developed to overcome the problems of these conventional methods [3, 5, 6]. The use of ultrasound for cell disruption and lipid extraction has been investigated since it offers high lipid yield; however, this method needs high power consumption and suffers from the destruction of the lipid structure by the ultrasounds process [8]. Few researchers used the ball milling, where cell wall is ruptured by a rotating cylinder without any required preparation. Compared with other methods, the ball milling offers the advantages of shorter extraction time and minimal damage to lipids; however, this method only extracts less than 40% of lipids from microbial cells [5, 9]. Microwave-assisted lipid release is performed by heat waves, which lyses cells similar to the ultrasound-assisted method. This method offers high lipid yield (80%), but suffers from low efficiency, volatile or non-polar solvents, and damage of neutral lipids by microwave radiation [3, 10]. The disruption of the cell walls by enzymes is an effective method, which is limited to the selection and combination of enzymes [11]. For example, the identified enzyme MAN5C can specifically degrade the cell walls of Rhodosporidium with β-1,3-glucomannanase activity [11]. In addition, enzymatic pretreatment takes more than 20 h, which is not suitable for the analysis of lipids [11, 12]. Therefore, compared with AHE, ultrasound-assisted, ball milling–assisted, microwave-assisted, and enzyme-assisted methods offer several advantages; however, every method still has its limitations [13]. Therefore, new methods need be developed to efficiently extract the whole lipid content from oleaginous yeasts. These methods need to have a simple procedure and short extraction time and cause minimal damage of lipids to enable process tracking and lipid analysis.
Automated pressurized liquid extraction (APLE) uses elevated temperatures and pressures to achieve extractions within a short time without the requirement for severe pretreatment [14]. Currently, APLE is applied to environmental biotic samples and for the analysis of contaminants with environmental affability, since it features a simple procedure and short extraction time [15, 16]. APLE is used for the extraction of environmental biotic samples where it offers more than 90% recovery, and can extract pesticides and lipophilic marine toxins from the sediment [17, 18]. Moreover, dibenzo-p-dioxins/dibenzofurans and organophosphate esters can be obtained from soil [19, 20]. Furthermore, more than 95% of dibenzo-p-dioxins/dibenzofurans and organochlorine were extracted from fish via APLE [21,22,23]. Therefore, APLE can effectively extract various compounds from different samples via a simple process and short time; however, few reports describe the extraction of bioactive compounds from cells [24,25,26].
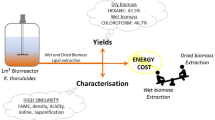
This study is the first to use APLE for the lipid extraction process from oleaginous yeast. Figure 1 summarizes the extraction process of the APLE method and compares its features to the AHE method. The two typical oleaginous yeasts R. toruloides and C. curvatus were selected for this test. By optimizing temperature, extraction time, and dispersion, the highest lipid extraction yield of more than 95% was achieved with a simple procedure and short extraction time. This extraction method its convenient for the tracking of the lipid production process and for lipids analysis.
Materials and Methods
Strain, Media, and Chemicals
The yeast C. curvatus ATCC 20509 was obtained from the American Type Culture Collection and R. toruloides CGMCC 2.1389 was purchased from the China General Microbiological Culture Collection Center, and both were maintained at 4 °C on YEPD agar slant (containing 20 g/L glucose, 10 g/L yeast extract, and 10 g/L peptone, agar 10 g/L, pH 6.0) and subcultured every month; further, the strains were grown in YEPD liquid medium (containing 20 g/L glucose, 10 g/L yeast extract, and 10 g/L peptone, pH 6.0) at 30 °C and 200 rpm.
The medium for R. toruloides batch culture contained 60 g/L glucose, 15.7 g/L yeast extract, and 15.7 g/L peptone. The batch culture medium for C. curvatus was obtained via enzymatic hydrolysis of corn stover. In brief, corn stover was mixed with 10% (w/w) sodium carbonate solution (100 mg sodium carbonate/1 g corn stover), which reacted at 131 °C for 3 h. After 3 h, the pH was adjusted to 4.8 and 5 mg/g cellulase (purchased from Novozymes, China), 2.5 mg/g xylanase (purchased from ncmchem, China) [27], and 50 μg/mL AMP were added to prevent bacterial contamination. The sample was placed in a reactor at 50 °C and 200 rpm for 72 h to obtain hydrolysates for the culture medium [4, 28].
Analytical grade methanol and chloroform from Lichrosolv (Darmstadt, German) and Duksan (Ansan, South Korea) were used for lipid extraction. All other reagents and chemicals were of analytical grade and were purchased locally.
Fed-Batch Cultivation of R. toruloides and C. curvatus
Fed-batch cultivation of R. toruloides was performed in a 15-L bioreactor (Shanghai Bailun Bio-Technology Co. Ltd, Shanghai) according to previously described conditions [4]. Briefly, a 7-L batch culture was initiated with 10 vol.% inoculum culture at 30 °C with an internal tank pressure of 0.05 MPa and 0.678 vvm via ventilation. The pH was auto controlled at 5.6 by flow plus 4 mol/L NaOH and 4 mol/L HCl. When the glucose concentration in the bioreactor decreased below 10 g/L, 500 mL sterilized glucose solution (1000 g/L) was added. After four cycles of substrate feeding, the final cell mass and lipid content were 62.5 g/L and 50.0%, respectively.
Fed-batch cultivation of C. curvatus was also performed in a 15-L bioreactor. In brief, a 9-L batch culture was initiated with 10 vol.% inoculum culture at 30 °C. The pH was auto controlled at 5.6 by flow plus 4 mol/L NaOH and 4 mol/L HCl, with 0.05 MPa internal tank pressure and 0.678 vvm via ventilation. After 58 h of incubation, a final cell density and lipid content of 14.0 g/L and 51.4%, respectively, were obtained.
For all samples of cell mass, cells from yeast culture were washed twice with deionized water and were then lyophilized.
Lipid Extraction with the Acid-Heating Extraction Method
AHE was used to estimate the lipid extraction yield of total lipids [4]. Briefly, dried yeast cells were digested at 78 °C for 1 h with 4 M HCl and were extracted twice with chloroform/methanol (1:1, vol/vol). The extracts were washed with 0.1% NaCl, dried over anhydrous Na2SO4, evaporated in vacuo, and the residue was dried at 105 °C for 24 h to obtain the total lipids.
where LE is the lipid yield; WAHE, lipids obtained from acid-heating extraction method (AHE); and WAPLE, lipids obtained from automated pressurized liquid extraction method (APLE).
Lipid Extraction with the Automated Pressurized Liquid Extraction Method
The APLE system, as shown in Fig. 1 (APLE-3500), provided by Titan Instruments Co., Ltd, Beijing, China, was used for the lipid extraction. The main lipid extraction steps of the APLE process are shown in Fig. 2. Samples were mixed with diatomite and were loaded in a stainless steel chamber (11 mL) that was placed in the APLE system. This system could maintain various extraction parameters (pressure, temperature, volume of extraction solvent, and extraction time). The adopted pressure (10 MPa) maintained the solvents as liquids at temperatures above their boiling points and forward improving the pressure exerted little impact on lipid recovery (data not shown). The boiling point of solvents at the ratio of chloroform:methanol = 1:1 (v/v) was 138 °C under at 10 MPa pressure, which was calculated via the Peng Robinson thermodynamics equation [29]. Ethyl acetate, n-hexane, and petroleum ether were used as solvents; however, these had low extraction efficiency. Methanol and chloroform, at a set ratio, were pumped through a stainless steel frit at the outlet and a filter membrane (LM3500/3000) and diffused in the loaded sample. Two extraction cycles with this chloroform/methanol mixture (1/1, v/v) were used, which provides a modified AHE method, which was adapted for yeast lipid extraction. For each experiment, 0.3–0.4 g of lyophilized cell mass was subjected to APLE with up to 2 g of diatomite (dried at 500 °C), which was used as a dispersant in the extraction cells. The extracts were washed with 0.1% NaCl, dried over anhydrous Na2SO4, evaporated in vacuo, and the residue was dried at 105 °C for 24 h to provide the total lipids.
Main steps of the lipid extraction via the APLE process. Firstly, oleaginous yeasts were fully mixed with diatomite in a mortar. Secondly, the sample was transferred into a stainless steel chamber through a funnel. Thirdly, the chamber was placed in the APLE system, where solvent and collection bottle were prepared in advance. Fourthly, the extraction parameters (temperature, time, and pressure) were set and the extraction was performed. Finally, the fatty acid and neutral compositions of the lipids in the collection bottle were analyzed for lipids using GC and HPTLC, respectively
Analysis Methods
The lipid products were transmethylated following previously described methods [28]. Briefly, 70 mg of lipids was treated with 0.5 mL of 5% KOH solution in methanol at 65 °C for 50 min, followed by addition of 0.2 mL BF3 diethyletherate and 0.5 mL methanol. The mixture was refluxed for 10 min, cooled, and extracted with n-hexane. The organic layer was washed twice with distilled water, and was used for the analysis of the fatty acid composition. Finally, the compositional profiling of fatty acids was measured by a 7890F gas chromatograph (GC; Techcomp Scientific Instrument Co. Ltd., Shanghai, China), equipped with a cross-linked capillary FFAP column (30 m × 0.25 mm × 0.25 mm) and a flame ionization detector. The flow rates for N2, H2, and air were 720 mL/min, 30 mL/min, and 100 mL/min, respectively. The temperatures of injection port, oven, and detector were set to 250 °C, 190 °C, and 280 °C, respectively. The injection volume was 0.5 μL. Fatty acids were identified by comparing with the retention time of standards and quantified by the respective peak areas.
To assess the different neutral lipid concentrations with both methods, different lipid samples were analyzed by high-performance thin-layer chromatography (HPTLC; Darmstadt, Germany) on 60F254 silica gel plates (10 cm × 20 cm; Darmstadt, Germany). The analytic procedure was adapted from a published procedure with minor modifications [30]. Total lipids were separated on a silica gel plates using petroleum ether/diethyl ether/acetic acid (80:40:1, v/v/v) as solvent system. For the visualization of separated lipids, the developed silica gel plates were air-dried, uniformly sprayed with 8% (w/v) H3PO4 containing 10% (w/v) copper(II) sulfate pentahydrate, and charred at 160 °C for 10 min. The lipids were quantified using densitometry and image analysis (HPTLC Densitometer, DESAGA, German), scaled to a dilution series of the corresponding lipid standard. All values show the averages of two independent experiments.
Results
Effects of Temperature on Lipid Extraction Yield
In this test, 0.4 g of dry yeast cells and 2.5 g of diatomite were thoroughly mixed and added to a sample chamber with a volume of 11 mL. The sample cell was heated for 5 min to the selected temperature. After that, a mixture of methanol and chloroform, at a blending ratio of 1:1 (v/v), was added to the sample cells and lipid extraction was started and held for 15 min at 10 MPa. Finally, the sample was washed by the chloroform/methanol mixture described above, and the amount of the mixture was 80% volume of the extraction chamber. The above steps were performed twice to complete the whole process of lipid extraction.
As shown in Fig. 3a, the lipid extraction yield was only 58.0% at a temperature of 50 °C and reached 89.0% after increasing the temperature to 105 °C. However, when the temperature was increased to 130 °C, lipid extraction yield is only 57.3%. A similar trend is shown in Fig. 3b for C. curvatus, where 65.1% of the lipid extraction yield was obtained at 60 °C and 89.5% could be reached when the temperature was increased to 105 °C. Higher temperatures are accompanied by an increased capacity of solvent solubility, which indicated that analytes could transfer quicker from intracellular matrix, leading to the extraction of the majority of analytes. Within a specific range, the higher temperature lowered the viscosity of the solvent and increased the movement of molecules. But the temperature cannot increase without limitation, because according to under the pressure of 10 MPa and the chloroform:methanol ratio of 1:1 (v/v), the boiling point of solvents was 138 °C, according to the Peng Robinson thermodynamics equation [29]. This resulted in easier yeast cell wall destruction and thus enhanced the lipid extraction. From both R. toruloides and C. curvatus, the highest lipid extraction yields were achieved at a temperature of 105 °C; therefore, the following experiments were carried out at 105 °C.
Effects of Different Extraction Times on Lipid Yield
In the theory, increasing the static extraction time at elevated temperatures can allow these compounds to diffuse into the extraction solvent [14, 31, 32]. Therefore, long static time would cause more lipids transferred to the extraction solvent.
As shown in Fig. 4a, increasing the lipid static extraction time of R. toruloides improved the lipid extraction yield within a certain range. At a lipid extraction time of 10 min, the lipid extraction yield reached 82.6%, and at a time of up to 40 min, the lipid extraction yield reached 95.2%. Longer static time may also increase risk of unexpected reactions in R. toruloides. In theory, the static extraction time increase can allow more lipids to diffuse into solvent during certain time. Actually, lipid yield was 94.4% at a static extraction time of 20 min, while lipid yield was 95.2% at that time of 40 min. Thus, the lipid yield was almost unchanged during static extraction time from 20 to 40 min. Similar phenomena were found for C. curvatus, and Fig. 4 b illustrates that the lipid extraction yield was only 42.4% within 5 min of static extraction time, while it increased to 89.5% with a static extraction time of 15 min. However, further increasing the static extraction time to 20 min caused a decrease of the lipid extraction yield to 83.8%. The more static time meant an increased risk of unexpected reactions at such a high temperature, especially in such complex samples as yeast cells. Therefore, increasing the static lipid extraction time within a certain range could lead to full contact of the solvent with the yeast cells toward improving the lipid extraction yield.
Effects of Dispersion on Lipid Extraction Yield
Diatomite prevented the yeast cells from adhering to each other when under pressure. Diatomite can absorb the water released from yeast cells. Moreover, diatomite enhanced the dispersion of yeast cells in the sample chamber. Diatomite addition improved the dispersion of yeast cells, which is beneficial to increase the contacting area between cells and the extraction solvent, which facilitates the lipid extraction of yeast cells.
As shown in Fig. 5a, for R. toruloides, the lipid yield reached 90.0% at a 5 (g/g) ratio of diatomite to yeast. If this ratio was increased to 8.3 (g/g), the lipid extraction yield reached up to 94.5%. For R. toruloides, ratio of diatomite to yeast had little impact on lipid yield. As shown in Fig. 5b, for C. curvatus, the lipid yield reached 82.0% at a 5 (g/g) ratio of diatomite to yeast, while the complete lipids were extracted at 8.3 (g/g) ratio. These results illustrated that a higher ratio of diatomite to yeast resulted in higher dispersion and lipid extraction yields. A higher diatomite ratio led to a more even dispersion, which promoted lipid recovery, consistent with the theory [14].
Comparison of the Lipids Extracted via APLE and AHE
The lipids from both APLE and AHE methods all consisted of free fatty acid (FFA), monoacylglycerol (MAG), diacylglycerol (DAG), and triglyceride (TAG), the contents of which were analyzed by HPTLC. GC was used to analyze the composition of fatty acids.
As shown in Table 1, the main lipid component was TAG, which exceeded 90% in R. toruloides and C. curvatus, showing a similar trend than previously reported [11]. The lipid yield reached 95.6% and 98.5% in R. toruloides and C. curvatus, respectively, which was consistent with the above data. FFA, MAG, DAG, and TAG are similar between the APLE and AHE in the R. toruloides and C. curvatus. This indicated that the profile of neutral lipids was same between the APLE and the AHE methods.
As shown in Fig. 6, the fatty acids obtained by both strains were mainly composed of myristic acid (c14:0), palmitic acid (c16:0), palmitoleic acid (c16:1), stearic acid (c18:0), oleic acid (C18:1), and linoleic acid (C18:2), which was consistent with previous literature [28, 33]. By analyzing the lipids extracted from R. toruloides by the APLE method, the fatty acid composition was found to be similar with that extracted by the AHE method. The same phenomena were found for C. curvatus. Therefore, these results demonstrated that the APLE method exerted little effects on the fatty acid composition obtained with the AHE method.
Discussion
After adjusting dispersion, temperature, and time, more than 95% of lipids could be directly extracted from the oleaginous yeasts R. toruloides and C. curvatus. Table 2 shows a comparison of the latest development during ultrasound-assisted, microwave treatment–assisted, ball milling–assisted, and enzyme-assisted lipid extraction methods, in addition to the two conventional methods, Soxhlet extraction and AHE. The lipid yield of the APLE method was prominently higher than yields achieved via microwave-assisted, ball milling–assisted, and enzyme-assisted extraction methods, and reached a level equal to both Soxhlet extraction and AHE. Table 2 shows that the total extraction times of the APLE method for R. toruloides and C. curvatus were 0.67 h and 0.5 h, respectively, which were both clearly shorter than the extraction times of other extraction methods (except for the ball milling–assisted extraction method). The timeliness of lipid analysis is key for microbial lipid production, since only timely data analysis enables proper control of the fermentation process [4]. For example, the AHE method requires 3 h to complete an extraction; however, this long time renders the analysis of data obtained at this time for the fermentation process meaningless. Therefore, the sample analysis time during the process must be controlled to remain within 1 h, so that the resulting data can guide the fermentation process control. The APLE method decreased 80% of extraction time, which is significant for the microbial lipid production process.
The APLE method is similar to Soxhlet extraction, making it simpler than other extraction methods. When high-power input methods are used, such as ultrasound-assisted extraction and microwave-assisted extraction, a number of neutral lipids may decompose into low–weight molecular lipids due to harsh pretreatment. As shown in Table 1, the similar neutral lipids were extracted via APLE within short time. Therefore, APLE is well suited for the rapid analysis of neutral lipids due to its quick and simple procedure.
Table 3 summarizes the features of the APLE method in comparison with other extraction methods. The APLE method efficiently extracts lipids from oleaginous yeasts and offers high lipid yield, shorter extraction time, simple procedure, and minimal damage of lipids. This is a lipid extraction method with good potential.
Conclusions
Without any pretreatment (either chemical or physical), the APLE method can directly extract more than 95.2% of lipids from R. toruloides and 100% of lipids from C. curvatus. The fatty acid composition of the lipids obtained from the APLE method was consistent with that obtained with the AHE method. Under optimal conditions, HPTLC analysis showed that samples from the APLE method had similar amounts of FFA, MAG, DAG, and TAG, which was coincident with ball milling–assisted and enzyme-assisted methods. Hence, the APLE method is a promising technology for the effective lipid extraction of oleaginous yeasts, offering short extraction time and minimal damage to lipids. APLE enables fast and accurate evaluation of the lipid production process and lipid analysis.
References
Spagnuolo, M., Yaguchi, A., & Blenner, M. (2019). Oleaginous yeast for biofuel and oleochemical production. Current Opinion in Biotechnology, 57, 73–81.
Sanchez i, N., Violeta, B., Brenna, A., Kruger, J. S., Singer, C. A., Ramirez, K. J., Reed, M. L., Cleveland, N. S., Singer, E. R., Yi, X., Yeap, R. Y., Linger, J. G., & Beckham, G. T. (2018). Integrated diesel production from lignocellulosic sugars via oleaginous yeast. Green Chemistry, 20, 4349–4365.
Dong, T., Knoshaug, E. P., Pienkos, P. T., & Laurens, L. M. L. (2016). Lipid recovery from wet oleaginous microbial biomass for biofuel production: a critical review. Applied Energy, 177, 879–895.
Li, Y., Zhao, Z., & Bai, F. (2007). High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme and Microbial Technology, 41(3), 312–317.
Menegazzo, M. L., & Fonseca, G. G. (2019). Biomass recovery and lipid extraction processes for microalgae biofuels production: a review. Renewable and Sustainable Energy Reviews, 107, 87–107.
Sati, H., Mitra, M., Mishra, S., & Baredar, P. (2019). Microalgal lipid extraction strategies for biodiesel production: a review. Algal Research, 38, 101413.
Ashokkumar, V., Chen, W.-H., Ngamcharussrivichai, C., Agila, E., & Ani, F. N. (2019). Potential of sustainable bioenergy production from Synechocystis sp. cultivated in wastewater at large scale – a low cost biorefinery approach. Energy Conversion and Management, 186, 188–199.
Ido, A. L., de Luna, M. D. G., Capareda, S. C., Maglinao, A. L., & Nam, H. (2018). Application of central composite design in the optimization of lipid yield from Scenedesmus obliquus microalgae by ultrasound-assisted solvent extraction. Energy, 157, 949–956.
Prabakaran, P., & Ravindran, A. D. (2011). A comparative study on effective cell disruption methods for lipid extraction from microalgae. Letters in Applied Microbiology, 53(2), 150–154.
Balasubramanian, S., Allen, J. D., Kanitkar, A., & Boldor, D. (2011). Oil extraction from Scenedesmus obliquus using a continuous microwave system - design, optimization, and quality characterization. Bioresource Technology, 102(3), 3396–3403.
Jin, G., Yang, F., Hu, C., Shen, H., & Zhao, Z. K. (2012). Enzyme-assisted extraction of lipids directly from the culture of the oleaginous yeast Rhodosporidium toruloides. Bioresource Technology, 111, 378–382.
Masri, M. A., Garbe, D., Mehlmer, N., & Brück, T. B. (2019). A sustainable, high-performance process for the economic production of waste-free microbial oils that can replace plant-based equivalents. Energy & Environmental Science, 12(9), 2717–2732.
Deshmukh, S., Kumar, R., & Bala, K. (2019). Microalgae biodiesel: a review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Processing Technology, 191, 232–247.
Richter, B. E., Jones, B. A., Ezzell, J. L., Porter, N. L., Avdalovic, N., & Pohl, C. (1996). Accelerated solvent extraction: a technique for sample preparation. Analytical Chemistry, 68(6), 1033–1039.
Ali, M., & Watson, I. A. (2015). Microwave treatment of wet algal paste for enhanced solvent extraction of lipids for biodiesel production. Renewable Energy, 76, 470–477.
Ligor, M., Ratiu, I.-A., Kielbasa, A., Al-Suod, H., & Buszewski, B. (2018). Extraction approaches used for the determination of biologically active compounds (cyclitols, polyphenols and saponins) isolated from plant material. Electrophoresis, 39(15), 1860–1874.
Rodrigues, E. T., Pardal, M. A., Salgueiro-Gonzalez, N., Muniategui-Lorenzo, S., & Alpendurada, M. F. (2016). A single-step pesticide extraction and clean-up multi-residue analytical method by selective pressurized liquid extraction followed by on-line solid phase extraction and ultra-high-performance liquid chromatography-tandem mass spectrometry for complex matrices. Journal of Chromatography A, 1452, 10–17.
Wang, Y., Chen, J., Li, Z., Wang, S., Shi, Q., Cao, W., Zheng, X., Sun, C., Wang, X., & Zheng, L. (2015). Determination of typical lipophilic marine toxins in marine sediments from three coastal bays of China using liquid chromatography-tandem mass spectrometry after accelerated solvent extraction. Marine Pollution Bulletin, 101(2), 954–960.
Klees, M., Bogatzki, C., & Hiester, E. (2016). Selective pressurized liquid extraction for the analysis of polychlorinated biphenyls, polychlorinated dibenzo-p-dioxins and dibenzofurans in soil. Journal of Chromatography A, 1468, 10–16.
Luo, Q., Wang, S., Sun, L.-n., & Wang, H. (2018). Simultaneous accelerated solvent extraction and purification for the determination of 13 organophosphate esters in soils by gas chromatography-tandem mass spectrometry. Environmental Science and Pollution Research, 25(20), 19546–19554.
Augusti, D. V., Magalhaes, E. J., Nunes, C. M., dos Santos, E. V., Dardot Prates, R. G., & Pissinatti, R. (2014). Method validation and occurrence of dioxins and furans (PCDD/Fs) in fish from Brazil. Analytical Methods, 6(6), 1963–1969.
Choi, M., Lee, I.-S., & Jung, R.-H. (2016). Rapid determination of organochlorine pesticides in fish using selective pressurized liquid extraction and gas chromatography-mass spectrometry. Food Chemistry, 205, 1–8.
Zhao, X., Cui, T., Guo, R., Liu, Y., Wang, X., An, Y.-X., Qiao, X., & Zheng, B. (2019). A clean-up method for determination of multi-classes of persistent organic pollutants in sediment and biota samples with an aliquot sample. Analytica Chimica Acta, 1047, 71–80.
Cescut, J., Severac, E., Molina-Jouve, C., & Uribelarrea, J. L. (2011). Optimizing pressurized liquid extraction of microbial lipids using the response surface method. Journal of Chromatography A, 1218(3), 373–379.
He, Y., Huang, Z., Zhong, C., Guo, Z., & Chen, B. (2019). Pressurized liquid extraction with ethanol as a green and efficient technology to lipid extraction of Isochrysis biomass. Bioresource Technology, 293, 122049.
Milanesio, J., Hegel, P., Medina-Gonzalez, Y., Camy, S., & Condoret, J.-S. (2013). Extraction of lipids from Yarrowia lipolytica. Journal of Chemical Technology and Biotechnology, 88(3), 378–387.
Dai, X., Shen, H., Li, Q., Rasool, K., Wang, Q., Yu, X., Wang, L., Bao, J., Yu, D., & Zhao, Z. (2019). Microbial lipid production from corn stover by the oleaginous yeast Rhodosporidium toruloides using the PreSSLP process. Energies, 12(6), 1053–1063.
Gong, Z., Shen, H., Wang, Q., Yang, X., Xie, H., & Zhao, Z. K. (2013). Efficient conversion of biomass into lipids by using the simultaneous saccharification and enhanced lipid production process. Biotechnology for Biofuels, 6, 1–10.
Wei, Y. S., & Sadus, R. J. (2000). Equations of state for the calculation of fluid-phase equilibria. AICHE Journal, 46(1), 169–196.
Yoon, K., Han, D., Li, Y., Sommerfeld, M., & Hu, Q. (2012). Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell, 24(9), 3708–3724.
Schafer, K. (1998). Accelerated solvent extraction of lipids for determining the fatty acid composition of biological material. Analytica Chimica Acta, 358(1), 69–77.
Giergielewicz-Mozajska, H., Dabrowski, L., & Namiesnik, J. (2001). Accelerated solvent extraction (ASE) in the analysis of environmental solid samples - some aspects of theory and practice. Critical Reviews in Analytical Chemistry, 31(3), 149–165.
Yang, X., Jin, G., Gong, Z., Shen, H., Bai, F., & Zhao, Z. K. (2014). Recycling biodiesel-derived glycerol by the oleaginous yeast Rhodosporidium toruloides Y4 through the two-stage lipid production process. Biochemical Engineering Journal, 91, 86–91.
Acknowledgments
We are indebted to Prof. Zongbao K. Zhao for expert opinion and consultation of Dr. Hongwei Shen for help on experimental design, Mr. Xiao Liu for assistance on APLE, and Mr. Xiaozan Dai for providing the raw material. We would like to express thanks to Energy Biotechnology Platform of Dalian Institute of Chemical Physics, CAS.
Funding
This work was supported by the National Natural Science Foundation of China (No. 51761145014).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
1. Automated pressurized liquid extraction was used to extract lipids directly.
2. Almost 100% lipids were extracted from oleaginous yeasts within 40 min.
3. Fatty acid profiles of lipids are identical to those reported in the literature.
4. This method is valuable for microbial lipid analysis.
Rights and permissions
About this article
Cite this article
Li, Q., Kamal, R., Chu, Y. et al. Automated Pressurized Liquid Extraction of Microbial Lipids from Oleaginous Yeasts. Appl Biochem Biotechnol 192, 283–295 (2020). https://doi.org/10.1007/s12010-020-03331-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03331-9