Abstract
Cellulolytic bacteria from cattle rumen with ability to hydrolyze cellulose rich biomass were explored. The study selected Paenibacillus polymyxa ND24 from 847 isolates as the most potent strain, which can efficiently produce cellulase by utilizing sugarcane bagasse, rice straw, corn starch, CMC, and avicel as a sole carbon source. On annotation of P. polymyxa ND24 genome, 116 members of glycoside hydrolase (GH) family from CAZy clusters were identified and the presence of 10 potential cellulases was validated using protein folding information. Cellulase production was further demonstrated at lab-scale 5-L bioreactor exhibiting maximum endoglucanase activity up to 0.72 U/mL when cultivated in the medium containing bagasse (2% w/v) after 72 h. The bagasse hydrolysate so produced was further utilized for efficient biogas production. The presence of diverse hydrolytic enzymes and formidable cellulase activity supports the use of P. polymyxa ND24 for cost-effective bioprocessing of cellulosic biomass.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exploitation of agricultural residues is an important factor for the progress of bio-based economy. In this context, big efforts are engaged to improve the use of agricultural wastes as widely available renewable sources [1]. However, hydrolysis of cellulosic component remains a major challenge for rapid and economic utilization of biomass. Cellulases, the key players of enzymatic hydrolysis comprising endo-1, 4-β-glucanase (EC 3.2.1.4), exo-1, 4-β-glucanase or cellobiohydrolase (EC 3.2.1.91), and β-glucosidase (EC 3.2.1.21), act in synergy to depolymerize cellulose into soluble fermentable sugars for further conversion into value-added bio-products [2]. The need for economical production of bio-product using cellulose-rich waste has led to the exploration of more efficient cellulase-producing microorganisms. During the past decade, breakdown of complex cellulosic polymer structures was restricted to a limited number of microorganisms, chiefly fungi [3, 4]. However, research is now been shifting towards bacterial cellulase systems as they have unique cellulolytic mechanisms along with high enzyme specificity in their arsenal. Cellulose-degrading bacteria have multi-enzyme system working in synergy to provide increased function and efficiency. These multi-enzyme complexes provide higher rate of enzymatic hydrolysis, increased product recovery as they are adaptable, and prone to facilitative genetic manipulation [5, 6]. Bacteria responsible for degradation of cellulosic polysaccharides have been isolated, characterized, and identified from various sources [7,8,9]. Ruminant animals host a diverse range of microbial flora from strictly anaerobic to some facultative anaerobic microorganisms responsible for efficient digestion of complex biomass rich in cellulose and hemicellulose in their rumen. Few rumen bacteria demonstrating cellulose biomass degrading capacity have already been discovered [10, 11]. However, the potential of rumen bacteria to efficiently degrade lignocellulosic substrates is largely overlooked.
In this work, production of extracellular cellulase by Paenibacillus polymyxa ND24 was evaluated using sugarcane bagasse (SB) as sole carbon source. Cellulase production was demonstrated in lab-scale reactor and SB hydrolysate so generated was further employed for biogas production. Also, the draft genome sequence of the P. polymyxa ND24 was analyzed for the presence of diverse and significant cellulolytic, hemicellulolytic, and amylolytic enzymes. In silico, molecular characterization of annotated cellulases was performed with their three-dimensional protein structure prediction to classify their diversity within the strain. These results will contribute in promoting the exploitation of agricultural wastes in the perspective of growing demand for biofuels and other valuable bio-products.
Materials and Methods
Sample Collection
Rumen fluid samples were collected from healthy adult cow and buffalo as described by Khampa et al. [12] using a flexible stomach tube coupled with suction pump at Swargiya Moropant Pingle Gourakshan Kendra, Nagpur, India, under sterile conditions. Samples were immediately brought to the laboratory and maintained at 4 °C for further study.
Enrichment and Isolation of Diverse Bacterial Population From Rumen Habitat
For enrichment, 5 mL rumen sample of cow and buffalo was inoculated in a flask containing 50 mL Berg minimal salt (BMS) media [13] amended with 1% (w/v) carboxymethyl cellulose (CMC) or SB as a sole carbon source and incubated at 120 rpm, 37 °C for 3 days. Afterwards, 1 mL of culture from each of these flasks was transferred to 50 mL of fresh BMS with 1% (w/v) CMC or sugarcane bagasse and 5 mL from leftover volume was used for screening of bacterial isolates. The process of sub-culture and isolation was continued for 4 weeks. Bacteria were isolated from both fresh and enriched rumen sample. For isolation, 100 μL of 10−3–10−6 times diluted sample in phosphate-buffer saline was spread on 24 different media plate including 1% CMC amended BMS agar plate and media for isolating facultative anaerobes. The media plates were incubated at 37 °C for 48 h. Based on colony characteristics such as shape, margin elevation, and color, 847 morphologically distinct colonies were obtained. To differentiate microbial populations at genetic level, isolates were subjected for RAPD profiling [14] with the primer 60S (5′-CAGCAGCAGCAG-3′) as described by Saxton et al. [15]. Bionumerics v7.1 was used to identify distinct bacterial isolates based on UPGMA method. Distinct isolates were further screened for enzyme activity through zymography.
Plate Zymography for Selection of Potent Cellulolytic Bacterial Population
For zymography, 5 μL of overnight grown isolates was spot cultured on CMC-amended BMS agar plates. After 48 h of incubation, plates were stained with 10 mL of 1% (w/v) solution of Congo red for 15 min and afterward decolorized with 1 M NaCl [16] to visualize the halo of cellulolytic activity around each colony. The cellulose hydrolyzing ability of the bacterial isolates was semi-quantitatively estimated by calculating enzyme activity index (EAI) − ratio of hydrolysis zone diameter to the colony diameter as mentioned by Dar et al. [17].
Molecular Identification of Potent Cellulolytic Strain Using 16S rDNA Sequencing
Molecular identification of isolate exhibiting high cellulolytic efficiency (EAI > 3) was achieved by amplifying 16S rDNA sequence using bacteria universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′). Fifty microliters of PCR reaction mix contained 20 ng template DNA, 5 μL 10× PCR buffer, 2.5 mmol of each dNTPs, 1 μmol of each primer, and 2.5 U of Taq polymerase, all provided by TaKaRa Bio Inc., Japan. The thermal profile for PCR was set as specified by Dafale et al. [18]. PCR products were checked for size and purity and sequenced using Sanger di-deoxy method. The sequences were analyzed using Chromas lite software. The similarity search for the sequence was carried out using the BLAST program of the National Center of Biotechnology Information.
Quantification of Endoglucanase, Exoglucanase, and β-glucosidase Activities
The potential of bacterial isolate to exhibit cellulolytic efficiency was evaluated by estimating endoglucanase, exoglucanase, and β-glucosidase activities. Supernatant obtained after centrifuging the cultures at 8000 rpm for 10 min at 4 °C was used as crude enzyme for various enzyme assays. Endoglucanase (CMCase) and exoglucanase (avicelase) assay were performed as described by Nitisinprasert et al. [19]. The reaction mixture comprising 500 μL of respective substrate (1.0% soluble CMC or avicel in 1.0 M phosphate buffer pH 7.0) and 500 μL of the enzyme was incubated at 37 °C for 30 min (for CMCase) and 60 min (for avicelase). Reaction was terminated by adding 1 mL 3,5-dinitrosalicylic acid (DNSA) reagent and incubated in boiling water bath for 10 min and cooled. Reducing sugars generated during the course of reaction was estimated spectrophotometrically at 540 nm with glucose as standard [20]. β-Glucosidase assay was done using para-nitrophenyl-β-d-glucopyranoside (pNPG) as a substrate. Reaction mixture containing 500 μL pNPG (5 mM p-nitrophenyl β-glucopyranoside in 50 mM sodium phosphate buffer pH 7) and 500 μL of the enzyme was incubated for 10 min at 37 °C, and the reaction was stopped by adding 2 mL of 1% (w/v) solution of sodium carbonate. The amount of p-nitrophenol released during the reaction was estimated by measuring the absorbance at 400 nm [21].
One unit of enzyme activity corresponds to the amount of enzyme that will release 1 μmol glucose or p-nitrophenol from the respective substrates per minute under the described conditions.
Effect of Different Carbon Sources on Cellulase Production
Based on high endoglucanase production in BMS broth amended with CMC as sole carbon source, P. polymyxa ND24 was selected for further study out of 45 isolates exhibiting EAI > 3. Endoglucanase production by P. polymyxa ND24 was studied on various carbon sources including CMC, corn powder (CP), avicel, rice straw (RS), and SB. For this, 0.1 OD cells/mL of P. polymyxa ND24 was inoculated into BMS media containing one of the carbon sources and incubated at 37 °C for 48 h. Endoglucanase activity was measured after 48 h.
P. polymyxa ND24 Genome Sequencing, Assembly, and Annotation
Fast DNA Soil Kit (MP Biomedicals, CA, USA) was used for DNA isolation from P. polymyxa ND24 following manufacturer’s protocols. The genomic DNA was quantified using NanoDrop-8000. One microliter of each sample was loaded in NanoDrop for concentration and A260/280 determination. The genome of P. polymyxa ND24 capable of hydrolyzing cellulose was sequenced using Illumina MiSeq. The 5.5-Mb high-quality data was assembled into 75 contigs using Velvet on optimized k-mer. Genome annotation was done using Rapid Annotations using Subsystems Technology (RAST) v4.0 server [22]. The genome was deposited at NCBI under the accession number LZCE00000000. Graphical comparison of P. polymyxa ND24 with P. polymyxa ATCC 842 and P. polymyxa SC2 at genomic scale was done by CGview server. Analysis of sequenced genome was done using dbCAN carbohydrate-active enzymes (CAZy) annotation algorithm from dbCAN pipelines (http://csbl.bmb.uga.edu/dbCAN/index.php) [23] against the Carbohydrate-Active Enzymes database (http://www.cazy.org/) [24] for identification of genes responsible in lignocellulose degradation. CDSs annotated by dbCAN as glycoside hydrolase (GH) were functionally classified by BLASTP algorithm using the public database KEGG and NR. Positional locations of the endoglucanase, exoglucanase, and β-glucosidase were mapped in the genome by using NCBI server.
Protein Structure Prediction of Annotated Cellulases (Endoglucanase, Exoglucanase, and β-glucosidase)
The 3D structures of annotated cellulases were predicted by Swiss-Model workplace (http://swissmodel.expasy.org/) via selection of the most identical template as described by Pal et al. [25] and visualized as ribbon diagrams using UCSF chimera viewer 1.11 (http://www.rbvi.ucsf.edu/chimera) [26]. Structural comparison with respect to residue conservation and presence of potential active sites in the selected predicted protein structures were also performed using UCSF Chimera. Protein sequence alignment of respective enzymes was accomplished using MUSCLE alignment tool. This alignment file was then linked with UCSF Chimera for rendering protein structure where each residue was colored as per their conservational status [27]. Endoglucanase CelX (accession no. AAP04424) from Pseudoalteromonas sp. DY3 [28], for endoglucanase and cellobiohydrolase Cel6B (accession no. B2AC20) from Thermobifida fusca [29]; ExgS (accession no. AAC38571) from Clostridium cellulovorans [30], for exoglucanase; and β-glucosidase BglB (accession no. AAA25311) from P. polymyxa 209P [31], for β-glucosidase, were taken as reference enzyme for structure and active site comparison.
Cellulase Production in 5-L Bioreactor Using P. polymyxa ND24
Various concentrations 10, 20, 30, and 40 g/L of SB were examined for enzyme production owing to efficiency of P. polymyxa ND24 to utilize SB more efficiently over other carbon source. Subsequently, optimal concentration of SB was used as sole carbon source for cellulase production at lab-scale reactor. The experiment was carried out in the batch reactor of volume 5.0 L having working volume capacity of 4.0 L. The reactor was filled with 3.6 L of BMS medium supplemented with 20 g/L SB and was inoculated with 400 mL P. polymyxa ND24 biomass grown for 48 h. The reactor was maintained at an ambient temperature of 37 °C ± 0.2, pH 7.0, and 120 rpm throughout the experiment (96 h). Samples were withdrawn after interval of every 12 h, and endoglucanase, exoglucanase, and β-glucosidase activities and soluble chemical oxygen demand (sCOD) of the crude supernatant were determined.
Biomethane Production Using Hydrolyzed Substrates
For biochemical methane potential (BMP) assay, SB hydrolysates containing deconstructed SB were harvested after 48 h of incubation. Equal volume of hydrolysate was mixed with anaerobic sludge (20 mL) and made up to 100 mL using autoclaved distilled water. The mixture was then transferred to 125-mL serum bottle subsequently sealed with an aluminum crimp to maintain anaerobic conditions. BMP assay was carried out for 4 days, and biogas volume and composition were monitored at regular interval of 24 h. Frictionless glass syringe was used for measuring biogas volume and gas chromatography (Agilent 7890A) for determining its composition. Gas chromatograph was operated using thermal conductivity detector and packed column (Restek; 6 ft. × 2 mm ID) with hydrogen as carrier gas. The conditions used for analysis of biogas composition were as described by Gulhane et al [32].
Results and Discussion
Exploration of Cattle Rumen Samples to Select the Cellulolytic Potent Strain
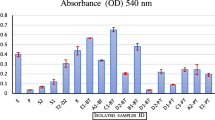
Fresh rumen sample and enriched rumen sample of both cow and buffalo were used for isolation of rumen bacteria. Initially, 847 isolates (409 from cow rumen sample and 438 from buffalo rumen sample) were obtained from 23 different media plates based on colony morphology. RAPD profiling of these isolates and band pattern analysis using bionumeric software revealed 239 and 234 distinct isolates from cow and buffalo, respectively. Of these, only 53.7 % (129 isolates) from cow and 62.5% (146 isolates) from buffalo showed cellulolytic efficiency on plate zymography. Thus, from both the ruminant organisms, a total of 275 cellulolytic isolates were obtained. The enzyme activity index ranged between 1.2 and 5.8 among all the cellulolytic isolates of which 14% show high activity (EAI ≥ 3), 30.86% medium activity (EAI 2–3), and 16.27% show low activity (EAI 1–2). Result reflects the ability of multiple bacterial isolates from the rumen of cow and buffalo to produce cellulase with different enzymatic efficiency. Members exhibiting EAI ≥ 3 were subsequently tested for cellulase production in broth medium, which resulted in selection of strain CS1 exhibiting enzyme activity 0.237 U/mL, for further study. Based on BLAST search using 16S rDNA gene sequence retrieved in this study, the strain CS1 displays highest identity (99%) with P. polymyxa SC2. Given that bacteria with 97% similarity in their 16S rDNA gene sequences can be considered as the members of the same species, the strain was designated as P. polymyxa ND24. The 16S rDNA partial sequence of strain P. polymyxa ND24 was deposited in GenBank and assigned accession number KX404920.1.
Effect of Different Carbon Sources on Endoglucanase Production
Cellulases are inducible enzymes and their production enhances on the presence of cellulose rich substrate; therefore, cellulase production medium usually consists cellulose-rich substrates as a carbon source [33]. Present study examines the effect various carbon source on cellulase production by potent strain under similar conditions, inoculum concentration, 0.1 OD cells/ mL; temperature, 37 °C; rotation speed, 120 rpm; and incubation time, 48 h. The result indicates potential of selected bacteria to utilize diverse carbon sources—CMC, RS, CP, avicel, and SB at different degrees. The maximal level of endoglucanase, 0.32 U/mL, was achieved when sugarcane bagasse was employed as sole carbon source (Fig. 1). SB is highly abundant cellulose-rich biomass in nature and economical raw material source for cellulolytic enzyme production [34]. Ability to hydrolyze diverse biomass by P. polymyxa ND24 led to the genomic analysis for better understanding of cellulolytic machinery.
Annotation of CAZy Clusters in P. polymyxa ND24
The genome of P. polymyxa ND24 comprises of 5.3078-Mb data assembled into 75 contigs that are annotated by NCBI PGAAP into 4602 protein coding sequences (CDS) (detailed results in supplementary file, Table S1). dbCAN carbohydrate-active enzymes (CAZy) annotation algorithm identified 328 genes belonging to CAZy family, comprising of 116 GHs, 43 carbohydrate esterases (CEs), 9 polysaccharide lyases (PLs), 65 glycosyl transferases (GTs), and 5 proteins with auxiliary activities (AAs). Cazymes often display associated modular structure which help in adhesion to the carbohydrates, additionally covering 55 carbohydrate binding modules (CBMs) and 35 surface layer homology domain (SLH) (Table S2). Circular representation of genome by CGView server (Fig. 2) shows nearly 90% regions of the genome being similar to P. polymyxa SC2 and P. polymyxa ATCC 842. The figure also reveals that there are large regions which are unique to only P. polymyxa ND24 and not found in both P. polymyxa SC2 and P. polymyxa ATCC 842. CDS comparison of P. polymyxa ND24 with other two P. polymyxa species shows similar trends as obtained by CG View result. From 115 GHs, 10 cellulase genes were found including four endoglucanases (GH5) gene, two exoglucanases (GH6, GH48), and four β-glucosidase genes (GH1, GH3). Carbohydrate-binding module was found to be associated with three of the cellulases including one endoglucanase and two exoglucanases. Additionally, 21 GHs responsible for hydrolysis of hemicelluloses, one 1,4-β-xylanase, three β-xylosidase, one α-glucuronidase, three α-N-arabinofuranosidase, two α-arabinosidase, five α-galactosidase, one β-mannanase, one β-mannosidase, two 1,3-β-glucanase, and one xyloglucanase, were also annotated. Moreover, 11 esterase genes, including two acetyl esterase, two acyl-CoA thioesterase, two carboxylesterase, and five esterases, responsible for complex carbohydrate hydrolysis have also been annotated. GHs responsible for hydrolysing starch, five α-amylase and two α-glucosidase, have also been found in P. polymyxa ND24 genome (Table 1). The presence of wide ranges of GH in the genome depicts the great potential of P. polymyxa for hydrolysis of various types of cellulosic and hemicellulosic polysaccharides.
Molecular Characterization of Different Cellulase Annotated in P. polymyxa ND24
Endoglucanase
Four endoglucanase protein product WP_064795906.1, WP_064796645.1, WP_063210412.1, and WP_063211222.1 were annotated at locus positions A9P44_RS05395, A9P44_RS11815, A9P44_RS15720, and A9P44_RS14040, respectively, based on NCBI annotation. Predicted proteins were assessed for existence and conservation of potential catalytic amino acid residue by comparing with already reported CelX endoglucanase. The arrangement of key residues in the active site was found to be in a similar orientation in all the endoglucanases from P. polymyxa ND24 with respect to amino acids Arg57, His100, His194, Glu135, and Glu222 in CelX endoglucanase. Further, the catalytic domain of all the endoglucanase folded as a (β/α)8 barrel with two glutamate residues, on strands IV and VII, that acts as catalytic acid/base and nucleophile, respectively (Fig. 3a). Representation of residue conservation in structure also reveals that in spite of having very low amino acid residue conservation, the overall structure of the enzyme remains the same.
Predicted three-dimensional structure of (a) Endoglucanase: key residues in the active site (corresponding to Arg57, His100, His194, Glu135, and Glu222 in CelX from Pseudoalteromonas sp. DY3) are highlighted with green. (b) Exoglucanase: three aspartate residues responsible for the catalytic activity in WP_064795544.1 (GH6) and Glu88 (proton donor) and Asp267 (nucleophile) located in the catalytic domain of WP_064797024.1 (GH48) are highlighted with green. (c) β-glucosidase: catalytic nucleophile/base and catalytic proton donor glutamate residue are highlighted with green in P. polymyxa ND24 visualized and labeled through UCSF chimera viewer 1.11. Highly conserved regions are shown in red whereas less-conserved region in blue
Exoglucanase
Two exoglucanase protein products WP_064795544.1 (GH6) and WP_064797024.1 (GH48) were annotated in draft genome of P. polymyxa ND24 located at A9P44_RS02675 and A9P44_RS14680, respectively. Structure prediction of the two exoglucanases WP_064795544.1 and WP_064797024.1 with exoglucanase Cel6B and ExgS, respectively, shows a similar structure around the catalytic site in spite of differences in their protein size. The arrangement of catalytic residues in the active site was found to be in a similar orientation in both the exoglucanases (Fig. 3b). The three aspartate residues which were predicted to be accountable for the catalytic activity in Cel6B were found to be at the similar position in Exg1 exoglucanase. Similarly, Glu88 (proton donor) and Asp267 (nucleophile) located in catalytic domain of Exg 2 were also found at the similar location in ExgS.
β-Glucosidase
From the 4 β-glucosidase protein product, two β-glucosidase proteins WP_064797241.1 and WP_064797468.1 that were present at locus positions A9P44_RS16545 and A9P44_RS18720, respectively, belong to GH1 family. In spite of variation in residue length, structure of β-glucosidase WP_064797241.1 and WP_064797468.1 showed similar catalytic domain folded as a (β/α)8 barrel structure except for some distortion in WP_064797241.1. Arrangement of catalytic nucleophile/base and catalytic proton donor glutamate residue in the active site was found to be in a similar orientation in both the β-glucosidase as compared with β-glucosidase Bgl B (Fig. 3c). Remaining two β-glucosidase protein WP_064795501.1 and WP_064795646.1 annotated at locus position A9P44_RS02365 and A9P44_RS03290 belongs to GH3 family. Structural comparison of all the cellulases suggests a very high similarity in terms of active/catalytic sites which are present in all the respective types of cellulases at specific position in spite of variations in their residues composition.
Cellulase Production in 5-L Bioreactor Using P. polymyxa ND24
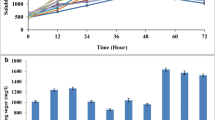
Different concentrations of SB were also examined for the optimum concentration of carbon source. The result indicates maximum cellulase production—0.373 U/mL at 2% w/v SB concentration (Fig. 4a). Therefore, easily available, inexpensive sugarcane bagasse is utilized as suitable carbon source for achieving high cellulase activity at lab-scale reactor. Annotation result for the presence of complete cellulolytic arsenal comprising endoglucanase exoglucanase and β-glucosidase in P. polymyxa ND24 genome was further demonstrated in lab-scale reactor using SB. Endoglucanase activity increased logarithmically during initial 72 h with maximal activity of 0.72 U/mL and reached steady state thereafter. Whereas exoglucanase and β-glucosidase activity increased up till 96 h with maximum of 0.37- and 0.72-U/mL enzyme activity, respectively. This result fits with the cascade of enzyme action where endoglucanase is responsible for cleavage of the long cellulose chains followed by exoglucanase and β-glucosidase that act on the smaller fragments. P. polymyxa ND24 shows higher enzyme production when compared with other cellulolytic bacteria (Table 2). Treatment of SB using P. polymyxa ND24 caused significant rise of sCOD in the hydrolysate from 593 to 1544 mg/L after 48 h with no significant change in sCOD of control variant. The observed rise in sCOD concentration reflects hydrolysis of insoluble organic components of SB into soluble fermentable sugars. After 48 h, subsequent decrease in sCOD of hydrolysate was observed (Fig. 4b).
(a) Endoglucanase production using different concentration of sugarcane bagasse by P. polymyxa ND24 after 48 h of incubation at 37 °C and pH 7. (b) Change in endoglucanase, exoglucanase, and β-glucosidase activity (inset: change in soluble COD) during hydrolysis of sugarcane bagasse with time in 5-L reactor using sugarcane bagasse (2%) as sole carbon source by Paenibacillus polymyxa ND24 at an ambient temperature of 37 °C ± 0.2, pH 7.0, and 120 rpm
Effect of Hydrolyzed Substrate on Escalation of Biogas Production
High sCOD concentration does not necessarily signify efficient generation of biomethane, as complex plant biomass is not easily used by methanogenic archaea. Therefore, it is essential to determine the effect of isolates on SB hydrolyzate which has direct impact on biogas production. Thus, serum bottle experiments were carried out in anaerobic condition for 96 h. The SB hydrolyzate derived after 48 h was sole source of organic compounds to carryout anaerobic digestion. The daily biogas production for 96 h is shown in Fig. 5. The biogas production was enhanced using previously hydrolyzed SB as substrate for anaerobic digestion in comparison to controls. Maximum biogas production was detected within 48 h and reached 86 mL and decreased further with cumulative biogas volume of 145 mL in 96 h. During the time course, methane content increased from 44 to 52% resulting in corresponding methane yield of 75 mL, whereas no significant biogas production (34 mL) was observed using untreated SB as a control. Results indicate 4.2-fold elevation in biogas production with increased methanization efficiency of SB hydrolysate derived using P. polymyxa ND24.
Conclusion
The cattle rumen harbors diverse range of cellulolytic bacteria which can hydrolyze the cellulosic biomass. The potent isolate, P. polymyxa ND24, studied in the present work efficiently produced extracellular cellulase with ability towards the hydrolysis of agricultural wastes which was further used for acceleration of biogas production. Genome annotation of P. polymyxa ND24 reveals the presence of various carbohydrates utilizing enzymes whose synergistic action is responsible for hydrolysis of cellulose, hemicelluloses, and starch. We believe that our findings of a novel isolate with 116 enzymes of GH family proposes further studies for its effective utilization at pilot scale for valorization of cellulosic plant biomass. The 5-L bioreactor study of P. polymyxa ND24 along with structural characterization of different types of cellulase shows the presence of an efficient cellulase system in P. polymyxa ND24, which could effectively utilize the SB. Finally, the results obtained also provided information to approach upcoming studies on enzyme production and cellulose hydrolysis by using proteomic tools, in order to reveal insight on the development of functional biocatalysts from P. polymyxa ND24.
References
FitzPatrick, M., Champagne, P., Cunningham, M. F., & Whitney, R. A. (2010). A biorefinery processing perspective: treatment of lignocellulosic materials for the production of value-added products. Bioresource Technology, 101, 8915–8922.
Lynd, L. R., Weimer, P. J., Van Zyl, W. H., & Pretorius, I. S. (2002). Microbial cellulose utilization: fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, 66, 506–577.
Jain, K. K., Kumar, S., Deswal, D., & Kuhad, R. C. (2017). Improved Production of Thermostable Cellulase from Thermoascus aurantiacus RCKK by Fermentation Bioprocessing and its application in the hydrolysis of office waste paper, algal pulp, and biologically treated wheat straw. Applied Biochemistry and Biotechnology, 181, 784–800.
Srivastava, N., Srivastava, M., Manikanta, A., Singh, P., Ramteke, P. W., Mishra, P. K., & Malhotra, B. D. (2017). Production and optimization of physicochemical parameters of cellulase using untreated orange waste by newly isolated Emericella variecolor NS3. Applied Biochemistry and Biotechnology, 183, 601–612.
Pandey, S., Singh, S., Yadav, A. N., Nain, L., & Saxena, A. K. (2013). Phylogenetic diversity and characterization of novel and efficient cellulase producing bacterial isolates from various extreme environments. Bioscience, Biotechnology, and Biochemistry, 77, 1474–1480.
Woo, H. L., Hazen, T. C., Simmons, B. A., & DeAngelis, K. M. (2014). Enzyme activities of aerobic lignocellulolytic bacteria isolated from wet tropical forest soils. Systematic and Applied Microbiology, 37, 60–67.
Ma, J., Zhang, K., Liao, H., Hector, S. B., Shi, X., Li, J., Liu, B., Xu, T., Tong, C., Liu, X., & Zhu, Y. (2016). Genomic and secretomic insight into lignocellulolytic system of an endophytic bacterium Pantoea ananatis Sd-1. Biotechnology for Biofuels, 9(1), 25.
Irfan, M., Tayyab, A., Hasan, F., Khan, S., Badshah, M., & Shah, A. A. (2017). Production and characterization of organic solvent-tolerant cellulase from Bacillus amyloliquefaciens AK9 isolated from hot spring. Applied Biochemistry and Biotechnology, 182, 1390–1402.
Wibberg, D., Al-Dilaimi, A., Busche, T., Wedderhoff, I., Schrempf, H., Kalinowski, J., & de Orué Lucana, D. O. (2016). Complete genome sequence of Streptomyces reticuli, an efficient degrader of crystalline cellulose. Journal of Biotechnology, 222, 13–14.
Dassa, B., Borovok, I., Ruimy-Israeli, V., Lamed, R., Flint, H. J., Duncan, S. H., Henrissat, B., Coutinho, P., Morrison, M., Mosoni, P., & Yeoman, C. J. (2014). Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One, 9(7), e99221.
Suen, G., Weimer, P. J., Stevenson, D. M., Aylward, F. O., Boyum, J., Deneke, J., Drinkwater, C., Ivanova, N. N., Mikhailova, N., Chertkov, O., & Goodwin, L. A. (2011). The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One, 6(4), e18814.
Khampa, S., Wanapat, M., Wachirapakorn, C., Nontaso, N., & Wattiaux, M. (2006). Effects of urea level and sodium DL-malate in concentrate containing high cassava chip on ruminal fermentation efficiency, microbial protein synthesis in lactating dairy cows raised under tropical condition. Asian-Australasian Journal of Animal Sciences, 19, 837–844.
Pawar, K. D., Dar, M. A., Rajput, B. P., & Kulkarni, G. J. (2015). Enrichment and identification of cellulolytic bacteria from the gastrointestinal tract of giant African snail, Achatinafulica. Biotechnology and Applied Biochemistry, 175, 1971–1980.
Dafale, N. A. (2011). Exploration of genetic information from dynamic microbial populations for enhancing the efficiency of azo-dye-degrading systems. Environmental Reviews, 9, 310–332.
Saxton, M. A., Naqvi, N. S., Rahman, F., Thompson, C. P., Chambers, R. M., Kaste, J. M., & Williamson, K. E. (2016). Site-specific environmental factors control bacterial and viral diversity in storm water retention ponds. Aquatic Microbial Ecology, 77, 23–36.
Bohra, V., Dafale, N. A., & Purohit, H. J. (2018). Paenibacillus polymyxa ND25: candidate genome for lignocellulosic biomass utilization. 3 Biotech, 8(5), 248.
Dar, M. A., Pawar, K. D., Jadhav, J. P., & Pandit, R. S. (2015). Isolation of cellulolytic bacteria from the gastro-intestinal tract of Achatinafulica (Gastropoda: Pulmonata) and their evaluation for cellulose biodegradation. International Biodeterioration and Biodegradation, 98, 73–80.
Dafale, N., Agrawal, L., Kapley, A., Meshram, S., Purohit, H., & Wate, S. (2010). Selection of indicator bacteria based on screening of 16S rDNA metagenomic library from a two-stage anoxic–oxic bioreactor system degrading azo dyes. Bioresource Technology, 101, 476–484.
Nitisinprasert, S., & Temmes, A. (1991). The characteristics of a new non-spore-forming cellulolytic mesophilic anaerobe strain CM126 isolated from municipal sewage sludge. The Journal of Applied Bacteriology, 71, 154–161.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
An, C. L., Lim, W. J., Hong, S. Y., Kim, E. J., Shin, E. C., Kim, M. K., Lee, J. R., Park, S. R., Woo, J. G., Lim, Y. P., & Yun, H. D. (2004). Analysis of bgl operon structure and characterization of β-glucosidase from Pectobacterium carotovorum subsp. carotovorum LY34. Bioscience, Biotechnology, and Biochemistry, 68, 2270–2278.
Kapley, A., Tanksale, H., Sagarkar, S., Prasad, A. R., Kumar, R. A., Sharma, N., Qureshi, A., & Purohit, H. J. (2016). Antimicrobial activity of Alcaligenes sp. HPC 1271 against multidrug resistant bacteria. Functional & Integrative Genomics, 16, 57–65.
Huang, L., Zhang, H., Wu, P., Entwistle, S., Li, X., Yohe, T., Yi, H., Yang, Z., & Yin, Y. (2017). dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Research, 46(D1), D516–D521.
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., & Henrissat, B. (2014). The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research, 42, 490–495.
Pal, R. R., Khardenavis, A. A., & Purohit, H. J. (2015). Identification and monitoring of nitrification and denitrification genes in Klebsiella pneumoniae EGD-HP19-C for its ability to perform heterotrophic nitrification and aerobic denitrification. Functional & Integrative Genomics, 15, 63–76.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25, 1605–1612.
Tikariha, H., Pal, R. R., Qureshi, A., Kapley, A., & Purohit, H. J. (2016). In silico analysis for prediction of degradative capacity of Pseudomonas putida SF1. Gene, 591(2), 382–392.
Zeng, R., Xiong, P., & Wen, J. (2006). Characterization and gene cloning of a cold-active cellulase from a deep-sea psychrotrophic bacterium Pseudoalteromonas sp. DY3. Extremophiles, 10, 79–82.
Sandgren, M., Wu, M., Karkehabadi, S., Mitchinson, C., Kelemen, B. R., Larenas, E. A., Ståhlberg, J., & Hansson, H. (2013). The structure of a bacterial cellobiohydrolase: the catalytic core of the Thermobifidafusca family GH6 cellobiohydrolase Cel6B. Journal of Molecular Biology, 425, 622–635.
Tsai, L. C., Amiraslanov, I., Chen, H. R., Chen, Y. W., Lee, H. L., Liang, P. H., & Liaw, Y. C. (2015). Structures of exoglucanase from Clostridium cellulovorans: cellotetraose binding and cleavage. Acta Crystallographica Section F Structural Biology Communications, 71(10), 1264–1272.
Isorna, P., Polaina, J., Latorre-García, L., Cañada, F. J., González, B., & Sanz-Aparicio, J. (2007). Crystal structures of Paenibacillus polymyxa β-glucosidase B complexes reveal the molecular basis of substrate specificity and give new insights into the catalytic machinery of family I glycosidases. Journal of Molecular Biology, 371, 1204–1218.
Gulhane, M., Khardenavis, A. A., Karia, S., Pandit, P., Kanade, G. S., Lokhande, S., Vaidya, A. N., & Purohit, H. J. (2016). Biomethanation of vegetable market waste in an anaerobic baffled reactor: Effect of effluent recirculation and carbon mass balance analysis. Bioresource Technology, 215, 100–109.
Lee, Y. J., Kim, B. K., Lee, B. H., Jo, K. I., Lee, N. K., Chung, C. H., Lee, Y. C., & Lee, J. W. (2008). Purification and characterization of cellulase produced by Bacillus amyoliquefaciens DL-3 utilizing rice hull. Bioresource Technology, 99, 378–386.
Gao, Y., Xu, J., Zhang, Y., Yu, Q., Yuan, Z., & Liu, Y. (2013). Effects of different pretreatment methods on chemical composition of sugarcane bagasse and enzymatic hydrolysis. Bioresource Technology, 144, 396–400.
Sethi, S., Datta, A., Gupta, B. L., & Gupta, S. (2013). Optimization of cellulase production from bacteria isolated from soil. ISRN Biotechnology, 2013, 985685. https://doi.org/10.5402/2013/985685.
Padilha, I. Q. M., Carvalho, L. C. T., Dias, P. V. S., Grisi, T. C. S. L., Silva, F. L., Santos, S. F. M., & Araújo, D. A. M. (2015). Production and characterization of thermophilic carboxymethyl cellulase synthesized by Bacillus sp. growing on sugarcane bagasse in submerged fermentation. Brazilian Journal of Chemical Engineering, 32, 35–42.
Waghmare, P. R., Kshirsagar, S. D., Saratale, R. G., Govindwar, S. P., & Saratale, G. D. (2014). Production and characterization of cellulolytic enzymes by isolated Klebsiella sp. PRW-1 using agricultural waste biomass. Emirates Journal of Food and Agriculture, 26, 44–59.
Goyal, V., Mittal, A., Bhuwal, A. K., Singh, G., Yadav, A., & Aggarwal, N. K. (2014). Parametric optimization of cultural conditions for carboxymethyl cellulase production using pretreated rice straw by Bacillus sp. 313SI under stationary and shaking conditions. Biotechnology Research International, 2014, 651839. https://doi.org/10.1155/2014/651839.
Dantur, K. I., Enrique, R., Welin, B., & Castagnaro, A. P. (2015). Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express, 5(1), 15.
Ladeira, S. A., Cruz, E., Delatorre, A. B., Barbosa, J. B., & Leal Martins, M. L. (2015). Cellulase production by thermophilic Bacillus sp: SMIA-2 and its detergent compatibility. Electronic Journal of Biotechnology, 18, 110–115.
Gastelum-Arellanez, A., Paredes-López, O., & Olalde-Portugal, V. (2014). Extracellular endoglucanase activity from Paenibacillus polymyxa BEb-40: production, optimization and enzymatic characterization. World Journal of Microbiology and Biotechnology, 30, 2953–2965.
Ghio, S., Insani, E. M., Piccinni, F. E., Talia, P. M., Grasso, D. H., & Campos, E. (2016). GH10 XynA is the main xylanase identified in the crude enzymatic extract of Paenibacillus sp. A59 when grown on xylan or lignocellulosic biomass. Microbiological Research, 186, 16–26.
Ekperigin, M. M. (2007). Preliminary studies of cellulase production by Acinetobacter anitratus and Branhamella sp. African Journal of Biotechnology, 6, 028–033.
Deka, D., Bhargavi, P., Sharma, A., Goyal, D., Jawed, M., & Goyal, A. (2011). Enhancement of cellulase activity from a new strain of Bacillus subtilis by medium optimization and analysis with various cellulosic substrates. Enzyme Research, 2011, 151656. https://doi.org/10.4061/2011/151656.
Acknowledgements
Miss Varsha Bohra acknowledges Department of Science and Technology of India for awarding Junior Research Fellowship (IF150088) for carrying out the work. The manuscript has been checked for plagiarism by Knowledge Resource Centre, CSIR-NEERI, Nagpur, India, and assigned KRC No. CSIR-NEERI/KRC/2017/AUG/EBGD/2.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
ESM 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Bohra, V., Tikariha, H. & Dafale, N.A. Genomically Defined Paenibacillus polymyxa ND24 for Efficient Cellulase Production Utilizing Sugarcane Bagasse as a Substrate. Appl Biochem Biotechnol 187, 266–281 (2019). https://doi.org/10.1007/s12010-018-2820-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-2820-5