Abstract
Exogenous application of synthetic and natural elicitors of plant defence has been shown to result in mass production of secondary metabolites with nutraceuticals properties in cultured cells. In particular, salicylic acid (SA) treatment has been reported to induce the production of phenylpropanoids, including cinnamic acid derivatives bound to quinic acid (chlorogenic acids). Centella asiatica is an important medicinal plant with several therapeutic properties owing to its wide spectrum of secondary metabolites. We investigated the effect of SA on C. asiatica cells by monitoring perturbation of chlorogenic acids in particular. Different concentrations of SA were used to treat C. asiatica cells, and extracts from both treated and untreated cells were analysed using an optimised UHPLC-QTOF-MS/MS method. Semi-targeted multivariate data analyses with the aid of principal component analysis (PCA) and orthogonal projection to latent structures-discriminant analysis (OPLS-DA) revealed a concentration-dependent metabolic response. Surprisingly, a range of chlorogenic acid derivatives were found to be downregulated as a consequence of SA treatment. Moreover, irbic acid (3,5-O-dicaffeoyl-4-O-malonilquinic acid) was found to be a dominant CGA in C. asiatica cells, although the SA treatment also had a negative effect on its concentration. Overall SA treatment was found to be an ineffective elicitor of CGA production in cultured C. asiatica cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The biotechnological manipulation of cultured cells with plant signalling molecules appears to be the most technically feasible means to enhance secondary metabolite production [1]. The use of cell cultures as bio-reactors for production of these natural products can be easily manipulated and also offers a cost-effective and environmentally friendly alterative to using full-grown plants [2]. Plants have developed a sophisticated mechanism to defend themselves against biotic and abiotic stressors. As part of the early mechanism of defence, phytohormones such as jasmonic acid (JA) and salicylic acid (SA) and their methyl ester derivatives, methyl jasmonate (MeJA) and methyl salicylate (MeSA), are produced and released [3, 4]. These phytohormones elicit the biosynthesis of plant defensive proteins and secondary metabolites [5, 6].
Exogenous application of such phytohormones has been shown to induce metabolic changes in plants and can provide insights into the biosynthesis of secondary metabolites [7]. For instance, exogenous application of SA has been used to trigger the production of bio-active compounds in plants [7, 8]. SA has been reported to activate the phenylpropanoid pathway by enhancing the induction of phenylalanine ammonia-lyase (PAL), a key enzyme for the production of cinnamic acid, a metabolic precursor in several phenylpropanoid metabolic pathways [9].
Centella asiatica is a perennial herb classified under the Apiaceae family found in the tropical and sub-tropical areas [3]. This medicinally relevant herb has received much attention from the bio-pharmaceutical industry due to its bio-active metabolites [10–12], including the pentacyclic triterpenoids asiatic acid, madecassic acid, asiaticoside and madecassoside [13–15]. Its pharmacological relevance is largely attributed to these centelloids [3, 4, 10]. However, other terpenoid and phenolic metabolites with different bio-activities are also found in addition to the centelloids [11, 16–19]. Among the therapeutic properties reported from studies performed on animal models, cultured cells and in vitro analyses, are anti-ageing, anti-cellulite, wound healing, anti-inflammatory, anti-bacterial, antioxidant activity and recently, neuroprotection [20, 21]. In Indian Ayurvedic medical texts, C. asiatica was described as a cure for dementia, and it is still used for memory improvement in modern day age [22, 23]. The antioxidant activity of C. asiatica ethanolic extracts seems to be responsible for the positive effect on neurodegenerative diseases [20], whereas water extracts of C. asiatica on β-amyloid-associated behavioural abnormalities [21]. As such, these results show the potential of the extracts in the treatment of Alzheimer’s disease [22, 23]. Thus, there has been interest in determining the bio-active compounds in C. asiatica responsible for these interesting therapeutic properties. Recent reports have identified chlorogenic acids (CGAs) from C. asiatica as beneficial in treatment/prevention of Alzheimer’s disease [22, 23], and the effect of CGAs on neurodegenerative diseases including Alzheimer’s, Parkinsonism, Huntington chorea and multiple sclerosis was reviewed [24].
CGAs are important constituents of plant secondary metabolites and possess numerous bio-medical and pharmacological properties [25, 26]. CGAs are a family of esters consisting of (−)-quinic acid (QA) esterified to different cinnamic acid (CA) derivatives [27, 28]. The predominant naturally occurring CGAs result from the acylation of QA with either p-coumaric acid, caffeic acid or ferulic acid to form either p-coumaroylquinic acid (pCoQA), caffeoylquinic acid (CQA) or feruloylquinic acid (FQA), respectively. The number and identity of the constituent cinnamic acids defines a subgroup, and there are usually several isomers within each subgroup. Many plants produce chlorogenic acids in which esterification occurs at positions 3, 4 and 5 of the quinic acid moiety [25, 29].
Seemingly, C. asiatica possess a distinct CGA profile, some of which are characterised by unusual chemical modification. In particular, irbic acid is a CGA derivative with a malonyl group attached to its quinic acid moiety [16] that has mostly (if not only) been reported in C. asiatica species [16, 22, 30]. In vitro, irbic acid has been reported to inhibit collagenase activity [30, 31] and exhibit radical scavenging activity [30], thus showing a potential of using C. asiatica as a natural source for anti-ageing and antioxidant products, respectively.
In our recent study, we found that tobacco cells treated with SA produced certain CGA molecules in higher concentration [32]. In this communication, exogenous SA was used as a chemical elicitor to trigger CGAs production in C. asiatica cells. A semi-targeted metabolomics approach was used to monitor the levels and responses of CGA molecules due to SA treatment.
Materials and Methods
Cell Culture and Elicitation
Cell suspensions of C. asiatica were established and maintained as previously described [12, 17]. Murashige and Skoog (MS) medium containing 1 μM 2,4-dichlorophenoxyacetic acid and 1 μM benzyl aminopurine, 1 g/L casein hydrolysate and 30 g/L sucrose (pH 5.8) was used to grow C. asiatica cell cultures in Erlenmeyer flasks of approximately 100 mL. After 3 days of subculture, cells from three flasks were combined in a 1000-mL flask and gently shaken to obtain a homogenous culture. The culture was divided into 20-mL aliquots into sterile 50-mL Falcon tubes. The required amount of the sodium salicylate (100 mM, pH 5.8) (Merck, Johannesburg, South Africa) stock solution used as inducing agent was added to reach final concentrations of 0, 100, 200 and 300 μM, respectively (each condition in triplicate). The tubes were then capped and placed horizontally on an orbital shaker at 130 rpm with a light/dark cycle of 12 h/12 h, and low light intensity of 30 μmol/m2/s in a plant tissue culture room at 23 °C for 24 h [33]. The experimental design consisted of three independent biological replicates.
Metabolite Extraction and Concentration
After the incubation time, the cell suspensions were collected by filtration using a mild vacuum filtration system (Millipore, Billerica, MA, USA) and washed with 50 mL with cold, sterile, MS salts medium to wash the cells free of any original culture medium. Two grammes of the air dried cells were weighed and re-suspended in 100 % methanol at a ratio of 1 g:10 mL in a Falcon tube. A probe sonicator (Bandelin Sonopuls, Germany) set at 55 % power for 15 s with 4 cycles was used to sonicate the cells. The homogenates were centrifuged in a benchtop centrifuge at 5100×g for 15 min at 25 °C. The supernatants were transferred to a clean 50-mL round bottom flask, and the methanol evaporated to approximately 1 mL using a rotary evaporator. The 1-mL crude extracts were transferred to Eppendorf tubes and dried to completeness overnight at 55 °C in a dry bath. The amount of dried extract and the yield per 2 g of dried cells were determined. The dried residues were reconstituted with 400 μL 50 % aqueous methanol (Romil, Cambridge, UK) for all the samples and placed in pre-labelled UHPLC glass vials fitted with unslitted caps (Separations, Johannesburg, South Africa).
UHPLC-MS Analyses
LC-MS analyses were performed on a Waters Acquity UHPLC system coupled in tandem to a Waters photodiode array (PDA) detector and a SYNAPT G1 HDMS QTOF mass spectrometer (Waters Corporation, Milford, MA, USA). Chromatographic separation of the extracts was achieved on a Waters Acquity UHPLC column (CSH C18 150 × 2.1 mm, 1.7 μm) thermostatted at 60 °C with gradient elution. A binary solvent was utilised, consisting of water (eluent A) and acetonitrile, both with 0.1 % formic acid (Romil, Cambridge, UK) (eluent B). The flow rate and injection volume was set at 0.4 mL/min and 5 μL, respectively. The initial conditions were kept constant for 0.1 min at 95 % A before gradient elution was introduced to change chromatographic conditions to 10 % A over 0.1–16 min and held for 1 min. The analytical column was restored to the initial conditions at 18 min for 2 min, resulting in a run time of 20 min. Chromatographic elution was monitored with a PDA detector operating from 200 to 500 nm (1.2-nm resolution) with a sampling rate of 20 points/s. Post-PDA detection, the SYNAPT G1 HDMS (Waters Corporation, Manchester, UK) was used in V-optics and operated in positive and negative electrospray ionisation (ESI) modes to detect the compounds of interest. The MS settings were as follows: capillary voltage of 2.5 kV, sample cone voltage of 60 V, extraction cone voltage of 5 V, collision energy of 3 eV, detector voltage of 1660 V, source temperature of 120 °C, m/z range of 100–1100, scan time of 0.2 s and interscan time of 0.02 s, in centroid data mode. High purity nitrogen was used as desolvation gas at 450 °C and cone gas at 50 L/h. To achieve mass accuracy, leucine enkephalin (556.2771 Da, ESI+, 554.2615 Da, ESI−) was used as calibrant at a flow rate of 0.1 mL/min and a mass window of 0.5 Da. For analyses, the samples were randomised, and each sample was analysed in triplicate.
Multivariate Data Analysis
The raw UPLC-MS data were extracted and analysed using MassLynx XS software (Waters Corporation, Manchester, UK). Software parameters were set to analyse the 2–14-min Rt range of the chromatogram, mass range 100–900 Da, mass tolerance 0.01 Da, mass window 0.05 Da and a Rt window of 0.20 min. The resulting data matrices were exported to SIMCA 13.0.2 software (Umetrics, Umea, Sweden) for principal component analyses (PCA) and orthogonal projection to latent structures-discriminant analysis (OPLS-DA) as well as variable importance in projection (VIP) analyses as previously described [12, 17]. All models were Pareto-scaled. The OPLS-DA models were computed to complement the PCA models, and had reliable CV-ANOVA p values of < 0.01.
Metabolite Annotation
The selected mass ions from the OPLS-DA-derived S- and VIP plots were annotated and putatively identified by generating molecular formulae (MF) which were further used to search for molecule identities in databases such as the Dictionary of Natural Products (DNP) (www.dnp.chemnetbase.com) and ChemSpider (www.chemspider.com). For CGA identifications, tandem MS/MS analysis, based on the in-source collision-induced dissociation (ISCID) approach [34], was used to generate characteristic CGA fragments. Different regional isomers of CGAs were identified by generating diagnostic ions such as those of the quinic acid moiety at m/z 191 in Q1 [quinic acid-H]− and m/z 173 in Q2 [quinic acid-H2O]− at a higher cone voltage of 60 V and collision energy of 30 eV (for CQA as an example). In addition, 3,5-dicaffeoyl quinic acid (PhytoLab, Vestenberggreuth, Germany) was obtained as authentic standard.
Results and Discussion
Chromatographic and Mass Spectrometric Analyses
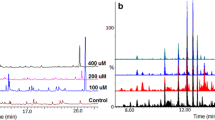
Here, a semi-targeted metabolomic approach based on UHPLC-MS was used to determine the qualitative and quantitative effects of SA treatment on C. asiatica cells. MS data acquisition was carried out on both ESI negative and positive ionisation modes. However, previous metabolite fingerprinting studies on chlorogenic acid derivatives have mostly been carried out in ESI negative mode [25, 29, 35–37]. As the focus of this paper is to report the effect of SA on chlorogenic acid profiles in C. asiatica cells, further data analyses were carried out only on ESI negative ionisation mode data. Chromatograms generated from ESI negative ionisation of the methanolic samples extracted from the different SA treatments are shown in Fig. 1. Differences in peak intensities and presence/absence of peaks in crude extracts are not always obvious and require multivariate data analysis approaches to scrutinise.
UHPLC- QTOF-MS analyses of methanolic extracts from SA-treated C. asiatica cells incubated for 24 h. ESI negative mode base peak intensity (BPI) chromatograms. The chromatograms show differences in some peak intensities whereas there is presence/absence of other peaks across different concentrations of SA treatment. The intensity of ion peak of m/z 137 at Rt 5.72 min increases from 100-μM to 300-μM SA treatment (a to c) whilst the peak is not present on the non-treated samples (d). This ion peak corresponds to the inducer, salicylic acid (C7H6O3, Mr 138). Contrary, the peak at m/z 529 at Rt 7.4 min decreases with increasing SA treatment and was characterised as described below
Multivariate Data Analysis
Multivariate data analyses (MVDA) were carried out to reveal the underlying differences of the metabolite profiles obtained from C. asiatica cells treated with different SA concentrations. As such, principal component analysis (PCA) (Fig. 2) and orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score plot (Fig. 3, Figure S1) revealed clustering of data points corresponding to the different experimental conditions. The results thus show that SA treatment induced concentration-dependent differential metabolic responses in C. asiatica cells.
Principal component analyses (PCA) generated from the ESI negative mode data of the SA-treated C. asiatica cells incubated for 24 h. a Scores plot showing variation between and within clusters corresponding to control, 100-, 200- and 300-μM SA-treated cells. The score plot, computed from PC1 and PC2, explains 54.8 % of the total variation and shows similar clustering between and within each treated sample group. b Loadings plot with each variable (m/z and Rt) representing each ion. The ions representing the statistically significant biomarkers were annotated as 1, 2, 3 and 4 as assigned in Table 1, along with their fragments as 1f, 2f, 3f and 4f, respectively
Multivariate data analysis of the ESI negative data, showing the orthogonal projection to latent structure-discriminant analysis (OPLS-DA) generated from SA-treated C. asiatica cells incubated for 24 h. a Score plot showing clear separation between control (non-treated) and 300-μM SA-treated groups. b S-plot (explaining 60,2 % of total variation) showing the selection of significant biomarkers associated with the SA-treated C. asiatica cells. The mass ions in the upper right quadrant of the S-plot are positively correlated to the SA treatment whilst the ions in the lower quadrant are negatively related to the treatment. The ions representing the statistically significant biomarkers were annotated as 1, 2, 3 and 4 as assigned in Table 1, along with their fragments as 1f, 2f, 3f and 4f, respectively. c Representative variable influence on projection (VIP) plot showing variables that are most important for both predictive and orthogonal components
The use of OPLS-DA (Fig. 3, S1) further allowed the extraction of potential biomarkers responsible for the significant separation between the non-treated and 300-μM SA-treated samples [39, 40]. The mass ions in the upper right quadrant of the S-plot (Fig. 3b) are positively correlated to the SA treatment (such as m/z 299 and 599) while the ions in the lower quadrant are negatively related to the treatment (such as m/z 367, 601, 515 and 529). To confirm the significance of these selected ions/variables as potential biomarkers, a variable importance in projection (VIP) plot (Fig. 3c) was generated where p ≥ 1.0 (y-axis) was set as the minimum value for significance [41–43]. Interestingly, upon the evaluation of the OPLS-DA loading S-plot generated from the ESI negative ionisation mode, the significant biomarkers correlating to the non-treated samples appeared to be mostly CGA-related derivatives. Marker ions positively correlated to the SA treatment included metabolites of SA (SA-glycoside, SA-acylated hexose isomers and N-salicyloyl aspartic acid) and markers that could not be identified (data not shown).
Relative Concentration of Identified Biomarkers
Due to the unavailability of most authentic plant standards, integrated peak area was used to generate the relative quantification estimation for each of the represented ions as shown in Fig. 4. The ion with m/z 515 appears to be the most abundant whilst the ion at m/z 367 has the lowest peak intensity. With an increase of SA treatment of the C. asiatica cells, each identified biomarker was found to decrease in relative concentration. This affirmed the finding from the OPLS-DA S-loadings plot, where all the selected significant biomarkers appeared as downregulated. There is a distinct pattern of compound distribution across SA treatment, with all the compounds being more pronounced in the control (non-treated) group.
Relative peak intensity of the identified biomarkers in SA-treated C. asiatica cells. Shown are biomarker 1 (m/z 367) and biomarker 2 (m/z 515) (a) and biomarker 3 (m/z 601) and biomarker 4 (m/z 529) (b). These biomarkers were annotated as trans-5-feruloylquinic acid, 3,5 di-caffeoylquinic acid, irbic acid (3,5-O-dicaffeoyl-4-O-malonilquinic acid) and 3-caffeoyl, 5-feruloylquinic acid, respectively. Integrated peak area was used to generate the relative quantification estimation for each of the represented ions, and standard deviation error bars were calculated based on sample population
A possible explanation for these observations is that SA-linked responses might be diverting hydroxycinnamates (HCAs) towards other phenylpropanoid molecules such as monolignols and benzoic acids, thus depleting the pool of activated HCAs available for esterification to QA [32]. In comparison to our previous results with methyl jasmonate as elicitor [12, 13, 17], the present results point to the fact that the phenylpropanoid pathway appears not be a dominant pathway as in the case of the terpenoid pathway. Furthermore, C. asiatica cells are known to accumulate considerable amounts of CGAs, especially irbic acid [31], and it is possible that the treated cells have reached a metabolic ‘saturation’ point, beyond which further stimulation of the phenylpropanoid pathway may be limited.
Characterisation of Identified Biomarkers
The manner of validating the annotated/putatively identified significant metabolites was carried out as in our previous work [34]. Therefore, the biomarkers were putatively identified through metabolite fingerprinting where the fragmentation patterns were compared. As such, single mass extracted ion chromatograms (XIC) (Figure S2A–D) were generated from the UHPLC-QTOF-MS ISCID data and used to monitor compounds harbouring common cinnamic acid moieties in their core structure. As the identification and annotation of structurally similar isomers of cinnamic acid derivatives have proven to be a challenging task [37], an IUPAC numbering system was used in order to avoid ambiguity. In addition to the chromatographic separation and the mass spectrometric behaviour as reported before [36], all possible factors such as positional and geometrical isomerism were considered (Table 1).
Trans-5-feruloylquinic Acid
Biomarker 1 (Figure S2A), that eluted at 7.56 min, was annotated as trans-5-feruloylquinic acid of Mr 368 (C17H20O9) with a precursor ion at m/z of 367.0994 [M-H]−. This molecule produced peaks at m/z 191 [quinic acid-H] and another ion at m/z 173 [quinic acid–H2O-H]− [44]. This compound has been previously reported in C. asiatica [45]. However, this molecule was found to be of very low peak intensity relative to the other identified molecules. This is in agreement with a previous communication that found it to be a low concentration molecule [44].
3,5 Di-caffeoylquinic Acid
Biomarker 2 (Figure S2B), that eluted at 8.99 min, was annotated as 3,5 di-caffeoylquinic acid of Mr 516 (C25H23O12). This molecule produced a precursor ion at m/z 515.1201 [M-H]− and a base peak at m/z 353 [M-H-162]−, corresponding to the loss of a caffeoyl moiety, and has product ions at m/z 191 [M-H-162-162]− corresponding to the loss of the second caffeoyl moiety, and at m/z 179 [caffeic acid-H]− as well as m/z 135 [caffeic acid-CO2]−. With caffeoylquinic acids, the presence of a prominent ion peak at m/z 191 is characteristic of a 5-CQA whereas an ion at m/z 179 at 1:2 ratio relative to m/z 191 is indicative of a 3-CQA [29, 36, 46, 47]. The 1,5 di-CQA and 3,5 di-CQA isomers are distinguished from each other by the presence of an ion at m/z 335 which is detectable only in the former CQA [29, 36, 46, 47]. Comparison of the fragmentation pattern observed for this molecule with an authentic standard further identified it as 3,5 di-CQA. Interestingly, this molecule was reported as the active constituent of C. asiatica extracts to possess in vitro and ex vivo inhibitory effects on thrombosis [48].
Irbic Acid (3,5-O-dicaffeoyl-4-O-malonilquinic Acid)
Biomarker 3 (Figure S2C), that eluted at 9.75 min, was annotated as irbic acid (3,5-O-dicaffeoyl-4-O-malonilquinic acid) of Mr 602 (C28H26O15). This molecule produced a precursor ion at m/z 601.1159 [M-H]−. The identity of this molecule was originally proposed by Antognoni (2011) [31]. The fragments observed in the spectra of this molecule at m/z 515, 395, 353, 233 have been reported to be characteristic for 3,5-O-dicaffeoyl-4-O-malonilquinic acid [16]. As mentioned, irbic acid has been only identified in C. asiatica species, and our results also suggest that it is present in relatively large amounts in cell cultures of this plant.
3-Caffeoyl, 5-feruloylquinic Acid
Biomarker 4 (Figure S2D), that eluted at 10.46 min, was annotated as 3-caffeoyl, 5-feruloylquinic acid (C26H26O12, Mr 530) [47, 49]. This molecule produced a precursor ion at m/z 529.1414 [M-H]−, a base peak at m/z 353 [M-ferulic acid-H2O]− representing the loss of ferulic acid (194 Da) and another product ion at m/z 367 [M-caffeic acid-H2O]− representing the loss of a caffeic acid (180 Da). Thus, the ions at m/z 353 and 367 represent the caffeoylquinic acid (Mr 354) and feruloylquinic acid (Mr 368) moieties, respectively whereas the product ions at m/z 179, 191 and 193 are characteristic for [caffeoyl-H]−, [quinic acid-H]− and [ferulic acid-H]−, respectively [29, 36, 46, 47, 50, 51]. The assignment as 3-caffeoyl, 5-feruloylquinic acid was based on the presence of the two peaks at m/z 179 and 191 at an approximate ratio of 1:2, indicative of 3-CQA [36, 47, 50], while the presence of the fragment peak at m/z 191 and a minor m/z at 193 (Figure S2) could then be indicative of a 5-feruloyl moiety. According to the best of our knowledge, this is the first report on the presence of this molecule in C. asiatica.
Conclusion
The treatment of C. asiatica cell suspensions with SA was found to have a negative effect on the biosynthesis of chlorogenic acid derivatives. The CGA derivatives that were found to be downregulated as a consequence of the treatment were 5-feruloylquinic acid, 3,5 di-caffeoylquinic acid, irbic acid (3,5-O-dicaffeoyl-4-O-malonilquinic acid) and 3-caffeoyl, 5-feruloylquinic acid. CGA molecules from C. asiatica have been previously reported to possess various pharmacological properties, including therapeutic effects against Alzheimer’s disease and other neurodegenerative diseases. The upregulation of these CGA derivatives in cultured cells of this plant would therefore contribute to their application in this bio-medical context. However, an attempt to increase the biosynthesis of these molecules by SA treatment proved to be ineffective and counter-productive. Irbic acid, a molecule confined to Centella species, was also reported in this study whilst 3,5-feruloyl-caffeoylquinic acid was reported for the first time in this plant. Interestingly, no glycosylated chlorogenic acid derivatives were found to be affected by the SA treatment. Further studies could involve the treatment of C. asiatica cells with different biotic or abiotic elicitor in an attempt to increase the upregulation of CGA derivatives.
References
Verpoorte, R., Contin, A., & Memelink, J. (2002). Biotechnology for the production of plant secondary metabolites. Phytochemistry Reviews, 1, 13–25. doi:10.1023/A:1015871916833.
Kolewe, M. E., Gaurav, V., & Roberts, S. C. (2008). Pharmaceutically active natural product synthesis and supply via plant cell culture technology. Molecular Pharmaceutics, 5, 243–256. doi:10.1021/mp7001494.
Gallego, A., Ramirez-Estrada, K., Vidal-Limon, H. R., Hidalgo, D., Lalaleo, L., Khan Kayani, W., Cusido, R. M., & Palazon, J. (2014). Biotechnological production of centellosides in cell cultures of Centella asiatica (L) Urban. Engineering in Life Sciences, 14, 633–642. doi:10.1002/elsc.201300164.
Gupta, A., Verma, S., Kushwaha, P., Srivastava, S., & Aks, R. (2014). Quantitative estimation of asiatic acid, asiaticoside & madecassoside in two accessions of Centella asiatica (L) Urban for morpho-chemotypic variation. Indian Journal of Pharmaceutical Education and Research, 48, 75–79. doi:10.5530/ijper.48.3.9.
Mangas, S., Bonfill, M., Osuna, L., Moyano, E., Tortoriello, J., Cusido, R. M., Teresa Piñol, M., & Palazón, J. (2006). The effect of methyl jasmonate on triterpene and sterol metabolisms of Centella asiatica, Ruscus aculeatus and Galphimia glauca cultured plants. Phytochemistry, 67, 2041–2049. doi:10.1016/j.phytochem.2006.06.025.
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., & Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nature Chemical Biology, 5, 308–316. doi:10.1038/nchembio.164.
Hayat, Q., Hayat, S., Irfan, M., & Ahmad, A. (2010). Effect of exogenous salicylic acid under changing environment: a review. Environmental and Experimental Botany, 68, 14–25. doi:10.1016/j.envexpbot.2009.08.005.
Ali, M. B., Hahn, E. J., & Paek, K. Y. (2007). Methyl jasmonate and salicylic acid induced oxidative stress and accumulation of phenolics in Panax ginseng bioreactor root suspension cultures. Molecules, 12, 607–621. doi:10.3390/12030607.
Rodas-Junco, B. A., Cab-Guillén, Y., Muñoz-Sánchez, J. A., Vázquez-Flota, F., Monforte-González, M., & Hernández-Sotomayor, S. M. T. (2013). Salicylic acid induces vanillin synthesis through the phospholipid signaling pathway in Capsicum chinense cell cultures. Plant Signaling & Behavior, 8, e26752–1–8. doi:10.4161/psb.26752.
James, J. T., & Dubery, I. A. (2009). Pentacyclic triterpenoids from the medicinal herb, Centella asiatica (L.) Urban. Molecules, 14, 3922–3941. doi:10.3390/molecules14103922.
Oyedeji, O. A., & Afolayan, A. J. (2005). Chemical composition and antibacterial activity of the essential oil of Centella asiatica growing in South Africa. Pharmaceutical Biology, 43, 249–252. doi:10.1080/13880200590928843.
James, J. T., Meyer, R., & Dubery, I. A. (2008). The occurrence of triterpenoid saponins in callus, cell suspensions and leaves of two Southern African phenotypes of Centella asiatica. Plant Cell, Tissue and Organ Culture, 94, 91–99.
James, J. T., Tugizimana, F., Steenkamp, P. A., & Dubery, I. A. (2013). Metabolomic analysis of methyl jasmonate-induced triterpenoid production in the medicinal herb Centella asiatica (L.) Urban. Molecules, 18, 4267–4281. doi:10.3390/molecules18044267.
Kim, O. T., Um, Y., Jin, M. L., Kim, Y. C., Bang, K. H., Hyun, D. Y., Lee, H. S., & Lee, Y. (2013). Analysis of expressed sequence tags from Centella asiatica (L.) Urban hairy roots elicited by methyl jasmonate to discover genes related to cytochrome P450s and glucosyltransferases. Plant Biotechnology Reports, 8, 211–220. doi:10.1007/s11816-013-0311-2.
Raoseta, O., Rodier-goud, M., Rivallan, R., Lussert, A., Danthu, P., Lamotte, F. D., Ramavovololona, P., Noyer, J.-L., & Baurens, F. (2013). Insight into the biology, genetics and evolution of the Centella asiatica polyploid complex in Madagascar. Industrial Crops and Products, 47, 118–125. doi:10.1016/j.indcrop.2013.02.022.
Maulidiani, Abas, F., Khatib, A., Shaari, K., & Lajis, N. H. (2014). Chemical characterization and antioxidant activity of three medicinal Apiaceae species. Industrial Crops and Products, 55, 238–247. doi:10.1016/j.indcrop.2014.02.013.
Tugizimana, F., Ncube, E. N., Steenkamp, P. A., & Dubery, I. A. (2015). Metabolomics-derived insights into the manipulation of terpenoid synthesis in Centella asiatica cells by methyl jasmonate. Plant Biotechnology Reports, 9, 125–136. doi:10.1007/s11816-015-0350-y.
Govarthanan, M., Rajinikanth, R., Kamala-Kannan, S., & Selvankumar, T. (2015). A comparative study on bioactive constituents between wild and in vitro propagated Centella asiatica. Journal of Genetic Engineering and Biotechnology, 13, 25–29. doi:10.1016/j.jgeb.2014.12.003.
Bylka, W., Znajdek-Awiżeń, P., Studzińska-Sroka, E., Dańczak-Pazdrowska, A., & Brzezińska, M. (2014). Centella asiatica in dermatology: an overview. Phytotherapy Research, 28, 7–1124. doi:10.1002/ptr.5110.
Omar, N. S., Akmal, Z., Zakaria, C., & Mian, T. S. (2011). Centella asiatica modulates neuron cell survival by altering caspase-9 pathway. Journal of Medicinal Plants Research, 5, 2201–2209.
Soumyanath, A., Zhong, Y. P., Henson, E., Wadsworth, T., Bishop, J., Gold, B. G., & Quinn, J. F. (2012). Centella asiatica extract improves behavioral deficits in a mouse model of Alzheimer’s disease: investigation of a possible mechanism of action. International Journal of Alzheimers Disease, 2012, 381974. doi:10.1155/2012/381974.
Gray, N. E., Morré, J., Kelley, J., Maier, C. S., Stevens, J. F., Joseph, F., & Soumyanath, A. (2014). Caffeoylquinic acids in Centella asiatica protect against β-amyloid toxicity. Journal of Alzheimers Disease, 40, 359–373. doi:10.3233/JAD-131913.
Dhanasekaran, M., Holcomb, L., Hitt, A., Tharakhan, B., Porter, J., Young, K., & Manyam, B. (2009). Centella asiatica extract selectively decreases amyloid beta levels in hippocampus of Alzheimer’s disease animal model. Phytotherapy Research, 23, 14–19. doi:10.1002/ptr.
Santos, R. M. M., Hunter, T., Wright, N., & Andrade, L. D. R. (2013). Caffeine and chlorogenic acids in coffee and effects on selective neurodegenerative diseases. Journal of Pharmaceutical and Scientific Innovation, 2, 9–17.
Clifford, M. N. (2000). Review chlorogenic acids and other cinnamates—nature, occurrence, dietary burden, absorption and metabolism. Journal of the Science of Food and Agriculture, 80, 1033–1043. doi:10.1002/(SICI)1097-0010(20000515)80:7<1033::AID-JSFA595>3.0.CO;2-T.
Marques, V., & Farah, A. (2009). Chlorogenic acids and related compounds in medicinal plants and infusions. Food Chemistry, 113, 1370–1376. doi:10.1016/j.foodchem.2008.08.086.
Koshiro, Y., Jackson, M. C., Katahira, R., Wang, M. L., Nagai, C., & Ashihara, H. (2007). Biosynthesis of chlorogenic acids in growing and ripening fruits of Coffea arabica and Coffea canephora plants. Zeitschrift für Naturforschung—Section C, 62, 731–742.
Plazas, M., Andújar, I., Vilanova, S., Hurtado, M., Gramazio, P., Herraiz, F. J., & Prohens, J. (2013). Breeding for chlorogenic acid content in eggplant: interest and prospects. Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 41, 26–35.
Alakolanga, A. G. A. W., Siriwardene, A. M. D., Savitri Kumar, N., Jayasinghe, L., Jaiswal, R., & Kuhnert, N. (2014). LC-MSn identification and characterization of the phenolic compounds from the fruits of Flacourtia indica (Burm. F.) Merr. and Flacourtia inermis Roxb. Food Research International, 62, 388–396. doi:10.1016/j.foodres.2014.03.036.
Long, H. S., Stander, M. A., & Van Wyk, B. E. (2012). Notes on the occurrence and significance of triterpenoids (asiaticoside and related compounds) and caffeoylquinic acids in Centella species. South African Journal of Botany, 82, 53–59. doi:10.1016/j.sajb.2012.07.017.
Antognoni, F., Perellino, N. C., Crippa, S., Dal Toso, R., Danieli, B., Minghetti, A., Poli, F., & Pressi, G. (2011). Irbic acid, a dicaffeoylquinic acid derivative from Centella asiatica cell cultures. Fitoterapia, 82, 950–954. doi:10.1016/j.fitote.2011.05.008.
Mhlongo, M. I., Piater, L. A., Steenkamp, P. A., Madala, N. E., & Dubery, I. A. (2014). Priming agents of plant defence stimulate the accumulation of mono- and di-acylated quinic acids in cultured tobacco cells. Physiological and Molecular Plant Pathology, 88, 61–66. doi:10.1016/j.pmpp.2014.09.002.
Sanabria, N. M., & Dubery, I. A. (2006). Differential display profiling of the Nicotiana response to LPS reveals elements of plant basal resistance. Biochemical and Biophysical Research Communications, 344, 1001–1007. doi:10.1016/j.bbrc.2006.03.216.
Ncube, E. N., Mhlongo, M. I., Piater, L. A., Steenkamp, P. A., Dubery, I. A., & Madala, N. E. (2014). Analyses of chlorogenic acids and related cinnamic acid derivatives from Nicotiana tabacum tissues with the aid of UPLC-QTOF-MS/MS based on the in-source collision-induced dissociation method. Chemistry Central Journal, 8, 1–10. doi:10.1186/s13065-014-0066-z.
Clifford, M. N., Knight, S., & Kuhnert, N. (2005). Discriminating between the six isomers of dicaffeoylquinic acid by LC-MSn. Journal of Agricultural and Food Chemistry, 53, 3821–3832. doi:10.1021/jf050046h.
Clifford, M. N., Kirkpatrick, J., Kuhnert, N., Roozendaal, H., & Salgado, P. R. (2008). LC-MSn analysis of the cis isomers of chlorogenic acids. Food Chemistry, 106, 379–385. doi:10.1016/j.foodchem.2007.05.081.
Jaiswal, R., Müller, H., Müller, A., Karar, M. G. E., & Kuhnert, N. (2014). Identification and characterization of chlorogenic acids, chlorogenic acid glycosides and flavonoids from Lonicera henryi L. (Caprifoliaceae) leaves by LC–MSn. Phytochemistry, 108, 252–263. doi:10.1016/j.phytochem.2014.08.023.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG), Metabolomics Standards Initiative (MSI). Metabolomics, 3, 211–221. doi:10.1007/s11306-007-0082-2.
Trygg, J., Holmes, E., & Lundstedt, T. (2007). Chemometrics in metabonomics. Journal of Proteome Research, 6, 469–479. doi:10.1021/pr060594q.
Wiklund, S., Johansson, E., Sjöström, L., Mellerowicz, E. J., Edlund, U., Shockcor, J. P., Gottfries, J., Moritz, T., & Trygg, J. (2008). Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical Chemistry, 80, 115–122. doi:10.1021/ac0713510.
Galindo-Prieto, B., Eriksson, L., & Trygg, J. (2014). Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). Journal of Chemometrics, 28, 623–632. doi:10.1002/cem.2627.
Madala, N. E., Steenkamp, P. A., Piater, L. A., & Dubery, I. A. (2014). Metabolomic insights into the bioconversion of isonitrosoacetophenone in Arabidopsis thaliana and its effects on defense-related pathways. Plant Physiology and Biochemistry, 84, 87–95. doi:10.1016/j.plaphy.2014.08.023.
Saccenti, E., Hoefsloot, H. C. J., Smilde, A. K., Westerhuis, J. A., & Hendriks, M. M. W. B. (2014). Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics, 10, 361–374. doi:10.1007/s11306-013-0598-6.
Zhang, J. Y., Zhang, Q., Li, N., Wang, Z. J., Lu, J. Q., & Qiao, Y. J. (2013). Diagnostic fragment-ion-based and extension strategy coupled to DFIs intensity analysis for identification of chlorogenic acids isomers in Flos Lonicerae Japonicae by HPLC-ESI-MSn. Talanta, 104, 1–9. doi:10.1016/j.talanta.2012.11.012.
Shukri, M. M., Alan, C., & Noorzuraini, R. S. (2011). Polyphenols and antioxidant activities of selected traditional vegetables. Journal of Tropical Agriculture and Food Science, 39, 69–83.
Alonso-Salces, R. M., Guillou, C., & Berrueta, L. A. (2009). Liquid chromatography coupled with ultraviolet absorbance detection, electrospray ionization, collision-induced dissociation and tandem mass spectrometry on a triple quadrupole for the on-line characterization of polyphenols and methylxanthines in green coffee beans. Rapid Communications in Mass Spectrometry, 23, 363–383. doi:10.1002/rcm.3884.
Jaiswal, R., Deshpande, S., & Kuhnert, N. (2011). Profiling the chlorogenic acids of Rudbeckia hirta, Helianthus tuberosus, Carlina acaulis and Symphyotrichum novae-angliae leaves by LC-MSn. Phytochemical Analysis, 22, 432–441. doi:10.1002/pca.1299.
Satake, T., Kamiya, K., An, Y., Oishi Nee Taka, T., & Yamamoto, J. (2007). The anti-thrombotic active constituents from Centella asiatica. Biological and Pharmaceutical Bulletin, 30, 935–40. doi:10.1248/bpb.30.935.
Plazonić, A., Bucar, F., Maleŝ, Z., Mornar, A., Nigović, B., & Kujundẑić, N. (2009). Identification and quantification of flavonoids and phenolic acids in burr parsley (Caucalis platycarpos L.), using high-performance liquid chromatography with diode array detection and electrospray ionization mass spectrometry. Molecules, 14, 2466–2490. doi:10.3390/molecules14072466.
Nandutu, A. M., Clifford, M., & Howell, N. Z. (2007). Analysis of phenolic compounds in Ugandan sweet potato varieties (NSP, SPK and TZ). African Journal of Biochemistry Research, 1, 29–36.
Jaiswal, R., Patras, M. A., Eravuchira, P. J., & Kuhnert, N. (2010). Profile and characterization of the chlorogenic acids in green Robusta coffee beans by LC-MSn: identification of seven new classes of compounds. Journal of Agricultural and Food Chemistry, 58, 8722–8737. doi:10.1021/jf1014457.
Acknowledgments
The authors would like to thank the South African National Research Foundation and the University of Johannesburg for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Figure S1
Multivariate data analysis of the ESI negative data, showing the orthogonal projection to latent structure-discriminant analysis (OPLS-DA) generated from SA-treated C. asiatica cells incubated for 24 h. Scores plot (A) and S-plot (B) comparing the non-treated cells vs. the 100-μM SA treatment and the scores plot (C) and S-plot (D) comparing the non-treated cells vs. the 200-μM SA treatment. The mass ions in the upper right quadrant of the S-plot are positively correlated to the SA treatment whilst the ions in the lower quadrant are negatively related to the treatment. (GIF 517 kb)
Figure S2
UHPLC-QTOF-MS fragmentation spectra. Showing (A) trans-5-feruloylquinic acid, (B) 3,5 di-caffeoylquinic acid, (C) irbic acid (3,5-O-dicaffeoyl-4-O-malonilquinic acid) and (D) 3-caffeoyl, 5-feruloylquinic acid. (GIF 47 kb)
Rights and permissions
About this article
Cite this article
Ncube, E.N., Steenkamp, P.A., Madala, N.E. et al. Chlorogenic Acids Biosynthesis in Centella asiatica Cells Is not Stimulated by Salicylic Acid Manipulation. Appl Biochem Biotechnol 179, 685–696 (2016). https://doi.org/10.1007/s12010-016-2024-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2024-9