Abstract
Actinobacteria is a prolific producer of complex natural products; we isolated a potential marine Streptomyces sp. PM49 strain from Bay of Bengal coastal area of India. The strain PM49 exhibited highly efficient antibacterial properties on multidrug-resistant pathogens with a zone of inhibition of 14–17 mm. SSF was adopted for the production of the secondary metabolites from PM49 with ISP2; utilizing agricultural wastes for compound extraction was also attempted. Bioactive fraction of Rf value 0.69 resolved using chloroform and ethyl acetate (1:1, v/v) was obtained and subjected to further analysis. Based on UV, IR, ESI-MS, and 1H and 13C NMR spectral analysis, it was revealed that the compound is closely similar to cyslabdan with a molecular mass of 467.66 corresponding to the molecular formula C25H41NO5S. ESBL and MBL production was screened in the hospital test isolates of Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae, and Staphylococcus aureus. PCR amplification in the phenotypically positive strains was positive for bla IMP, bla SHV, bla CTX-M, and mec genes. The β-lactamase enzyme from tested strains had cephalosporinase activity with a 31-kDa protein and isolated compound from the strain possessing β-lactamase inhibitory potential. MIC of the active fraction was 16–32 μg/ml on ATCC strains; the ceftazidime and meropenem sensitive and resistant test strains showed MIC of 64–256 μg/ml. The Streptomyces sp. PM49 aerial mycelium was rectiflexibile; the 16S rRNA showed 99 % identity with Streptomyces rochei and submitted at Genbank with accession no JX904061.1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infectious diseases are the second major cause of death worldwide, and the propensity to tolerate drying and resist multiple classes of antibiotics is the key factor in enabling pathogenic organisms to survive and spread in the nosocomial environment. The treatment of infections with Pseudomonas aeruginosa and Acinetobacter baumannii is extremely difficult because of the widespread resistance to the major groups of antibiotics with multiple mechanisms of resistance and which are associated with serious infections [1]. Klebsiella pneumoniae is an important human pathogen associated with many nosocomial outbreaks, and the level of extended spectrum beta-lactamase (ESBL)-positive Escherichia coli started to become increasingly harder to treat. Occurrence of CTX-M, SHV, and the TEM genes among the extended spectrum β-lactamase-producing isolates of E. coli and K. pneumoniae are reported in India [2]. The production of β-lactamases is the most prevalent mechanism of bacterial resistance to β-lactam antibiotics amino acid substitutions in the β-lactamases rendering resistance to penicillins and cephalosporins [3]. Gram-negative bacteria are inherently more resistant to antimicrobials than Gram-positive organisms, and this has been ascribed to the combined exclusion of antimicrobial compounds by double membrane barrier and transmembrance efflux [4].
There is an urgent need for a sustainable supply of new, potential, and safer antibacterial drugs having no cross-resistance to those currently used as antibiotics; unfortunately since the 1970s, only one new class of antibiotics has been introduced. Natural products have led to the discovery of new compounds and drug leads especially from unexplored marine resources [5]. Marine actinobacteria have yielded numerous novel secondary metabolites, and discovering new Streptomyces spp. are proving to be a valuable source of new bioactive metabolites. They have been reported to produce beta-lactam, carbapenems, and many other antibiotics and still remain as a promising resource of new antibacterial compound [6]. Streptomyces is known for the production of many β-lactam antibiotics; they have produced novel inhibitor molecules as cyslabdan, a potentiator of carbapenem antibiotic [7]. In this article, we describe the isolation and identification of a potential marine Streptomyces sp. (PM49) and seek to determine the biological activities and elucidate the structure of the antibacterial compound.
Materials and Methods
Isolation and Screening of a Potential Marine Actinobacteria
Marine sediment samples were collected from Bay of Bengal, Parangipettai coastal area (Lat.11 29′ N; Long. 79 47′ E) of India. After pretreatment, 100 μl of aliquot from 10−3, 10−4, and 10−5 dilution of the sample was spread-plated on starch casein agar supplemented with nalidixic acid 20 μg/ml and cycloheximide 100 μg/ml to inhibit bacterial and fungal contaminants. Colonies with actinobacterial morphology were purified with yeast extract malt extract agar (ISP-2) and stored in 20 % glycerol stock at −20 °C. Preliminary screening for antimicrobial strain was done with cross-streak method against E. coli ATCC 25922, P. aeruginosa ATCC 27853, B. subtilis NCIM 2063, and Staphylococcus aureus ATCC 33591 and was also confirmed using the control strains by agar plug method. MDR strains of E. coli (2531), K. pneumoniae (551), P. aeruginosa (2030), A. baumannii (3473), and S. aureus (15140) with the specified identification numbers isolated from hospital were also tested for antagonistic activity [8]. Inhibition of bacterial growth was observed after 24 h of incubation at 37 °C and was further tested for the confirmation of antibacterial activity using disc and well diffusion methods with the crude extract of the strain PM49 grown on ISP-2 media [9].
Fermentation and Extraction of Metabolites
Solid-state fermentation was adopted for the production of metabolites, and the crude extract was prepared by streaking on the yeast extract malt extract agar (ISP2 medium) plates and incubating at 28 °C for 10 days. After scraping the mycelial growth, the metabolite was extracted using ethyl acetate and quantified. The antimicrobial activity was determined using 50-μl discs of PM49 crude compound (1 % DMSO) in the concentration of 100 μg/ml, which was prepared and allowed to dry. Efficiency was tested against S. aureus ATCC 33591, B. subtilis NCIM 2063, and E. coli ATCC 25922 by disc diffusion method [9]. For large-scale production of crude extract from PM49, agricultural wastes of soybean, bran of wheat and rice, and tapioca were dried, and 10 g of different substrates were added with 100 ml of sea water and boiled [10]. The filtrate was extracted, and the volume was doubled with distilled water. Other variable factors such as 1.5, 2, 2.5, and 3 % of NaCl, 100, 75, and 50 % of sea water, organic nitrogen sources as yeast extract, malt extract, beef extract, and peptone were optimized at pH 7 by adopting the classical one-factor-at-a-time method. Inoculum of the strain was prepared using ISP-2 broth for 48 h of incubation at 28 °C in a rotary shaker, then streaked onto plates with different substrates, and incubated for 10 days at 28 °C.
Purification of Streptomyces sp. PM49 Compound
The crude extract from the strain PM49 was subjected to thin layer chromatography using silica gel (60 F, 254 nm, Merck). After optimizing the organic solvent system in order of their polarity with n-hexane, n-butanol, diethyl ether, chloroform, ethyl acetate, methanol, and water in different proportions, the crude extract was successfully fractionated. TLC sheets were air-dried, observed in UV light, and developed in iodine chamber; Rf value of the separated fractions were calculated and recorded [11]. Bioautography for antimicrobial activity was determined by inverting the chromatogram on top of culture nutrient agar plate seeded with test strains. E. coli ATCC 25922, S. aureus ATCC 33591, and the mentioned MDR strains were checked and observed for zone of inhibition. To further confirm the inhibitory activity, semisolid MHA with 12 % agar inoculated with the test organisms was poured onto TLC plates. After incubation for 24 h at 37 °C, bacterial growth inhibition was estimated; sensitivity of the test was enhanced with the addition of 0.1 % triphenyl tetrazolium chloride (TTC). The crude extract was further purified by column chromatography and preparative TLC using silica gel (60 F, 254 nm, Merck); the compound was checked for its antibacterial potential on the abovementioned strains. It was also synergistically tested along with 10 μg of meropenem and 30 μg of ceftazidime discs.
Characterization by Spectroscopic Measurements
The active fraction purified by chromatography was subjected to all spectral analyses. UV spectra were obtained with a CARY 2E UV-VIS-NIR spectrophotometer at room temperature with resolution between 200 and 500 nm range [12]. The IR spectrum was recorded with a PERKIN ELMER Spectrum FT-IR Spectrometer (KBr plate, ν max in cm−1), and the spectrum was recorded in the range 4000–400 cm−1. The 1H- and 13C-NMR spectrum were measured with a Bruker AVANCE III 500 MHz (AV 500) spectrometer operating at 500 MHz using CDCl3 and TMS as an internal reference at 500 MHz for 1H and 125 MHz for 13C; the chemical shifts were expressed in parts per million (ppm) with reference to (TMS) [13]. The HRESI mass spectra were obtained on a Finnigan MAT 8230 MS Mass spectrometer; the identification of the compound was based on the peak area, molecular weight, and molecular formula.
Characterization of the Test Pathogens
The phenotypically positive meropenem- and ceftazidime-resistant E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii isolates screened from endotracheal secretions, cerebrospinal fluid, blood, catheter, peritoneal fluid, pus, and surgical swabs were characterized. The test strains of 0.5McFarland standards were inoculated on MHA plate; disc approximation method was adopted for identifying of extended-spectrum beta-lactamase (ESBL). For metallo-β-lactamase (MBL) production, a 5-mm Whatman filter paper disc with 0.5 M ETDA of 5 μl (930 μg per disk) and a 10-mg meropenem disc constituted with 10 μl of 50 mM zinc sulphate was dispensed at 15 mm and tested using combined disc test (CDT) and double-disk synergy test (DDST) [2]. Template DNA was extracted from the selected multidrug-resistant E. coli (2531), K. pneumoniae (551), P. aeruginosa (2030), A. baumannii (3473), and S. aureus (15140) and few other isolates by rapid alkali lysis method using primers specific for common ESBL and MBL genes bla SHV, bla IMP, bla OXA-58, bla CTX-M, and mec genes [14–16]. PCR conditions were 3 min at 93 °C, 40 cycles of 1 min at 93 °C, 1 min at 55 °C, 1 min at 72 °C, and finally 7 min at 72 °C. Amplification was carried out in a thermocycler (Eppendorf), and the amplicon was separated in 1% agarose gel, visualized with ethidium bromide (0.5 μg /ml), and compared with molecular marker.
SDS PAGE Analysis of β-Lactamase Enzyme
The MDR non-lactose fermenters P. aeruginosa (2030) and A. baumannii (3473) cultures were grown from a single colony in LB broth, and the expression of β-lactamase enzymes was analyzed with the induction of ceftazidime (20 μg and 40 μg/ml). The flasks were incubated at 37 °C until the cultures reached OD600–0.7–0.8); the cells were disrupted using an ultrasonic disintegrator at 4 °C until complete visible cell lysate was obtained. Cell debris was removed by centrifugation (14,000 rpm, 20 min at 4 °C), and the enzyme was purified by ammonium sulphate precipitation up to 80 % saturation at pH 8 and left undisturbed overnight at 4 °C. The precipitated protein was collected by centrifugation at 14,000 rpm, 30 min at 4 °C, and re-dissolved in 30 ml of ice-cold 30 mM Tris-HCl, pH 7.6 [17]. The supernatant was loaded into a 20 × 200-mm Q-Sepharose column, and the bound proteins were eluted with 0–400 mM NaCl in 30 mM Tris-HCl, pH 7.6, at a flow rate of 2 ml/min. The fractions were separated by 12 % denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis with TGS electrophoresed for 2 h, stained with Coomassie brilliant blue, and destained.
Mechanism of Activity by Streptomyces sp. PM49 Active Fraction
The mode of action of Streptomyces sp. PM49 active fraction was tested for efflux and β-lactamase inhibition using the purified β-lactamase enzymes. The bioactive fractions of 40, 80, and 160 μg/ml were tested by agar dilution method in the presence of 1,3,5-triphenyltetrazolium chloride/reserpine [18]. Decrease in MIC was observed on each of the strains of E. coli (2531), K. pneumoniae (551), P. aeruginosa (2030), A. baumannii (3473) and S. aureus (15140); S. aureus ATCC 25923 (G+) and P. aeruginosa ATCC 27853 (G−) were used as control. Beta-lactamase inactivation using 50 μl nitrocefin and ceftazidime (30 μg + 160 μl active fraction) was observed for the color change from yellow to red [19]. β-Lactamase potentiating activity was also performed with the bioactive fraction of Streptomyces sp. PM49.
Determination of the MIC and Time Kill Assay
The above test strains were tested for MIC with the active fraction of stock 6.5 mg/ml by agar dilution method using 0.5 McFarland standards [20]. The MIC of ceftazidime and meropenem was used as positive control, and each experiment was duplicated. Negative control tubes were devoid of antibiotic and compound; ethylacetate and DMSO (used to dissolve compound) were tested to ensure that solvents had no influence on MIC. The concentration was ranging from 0.5 to 256 μg/ml; standard ATCC cultures were used for quality control to find the lowest concentration that is able to inhibit any visible growth [12]. The rate of kill assay with the active fraction on the pathogenic strains inoculum were prepared following the described guidelines of the EUCAST with bioactive fraction of doubling dilutions of concentration from MIC 0.5 to 64 μg/ml [21]. The resultant cell suspension was diluted 1:100 with fresh sterile broth and used to inoculate 5-ml volume of nutrient broth incorporated with the metabolite at multiples of the MIC to a final cell density of 5 × 105 cfu/ml. The flasks were then incubated with shaking at 37 °C on an orbital shaker at 120 rpm. About 100 μl of samples was withdrawn at 6- and 12-h intervals, diluted appropriately, plated on MHA plates, and then incubated at 37 °C for 24 h [22]. The numbers of surviving cells were enumerated inclusive of control (minus extract) nutrient broth inoculated with test organism.
Taxonomy and Phylogenetic Analysis
The selected strain Streptomyces sp. PM49 was morphologically characterized and phylogenetically determined. The mycelium structure and arrangement of spores were examined under bright field and scanning electron microscope (JEOL model 6390). Various physiological characteristics and biochemical tests were performed for the isolate. Chemical and sugar analysis of the strain was executed using TLC. Genomic DNA was extracted from Streptomyces sp. PM49, and sequencing was performed using consensus 16S rRNA primers (ABI 3500 XL Genetic Analyzer with Big Dye Terminator version 3.1″ Cycle sequencing kit). The amplified product after purification was examined by agarose gel electrophoresis. Multiple sequence alignment was performed to identify closely related homologs with the help of BLASTN search tool available at NCBI (http://www.nih.nov.ncbi). Sequences were aligned using the MUSCLE software (multiple sequence comparison by log-expectation), and phylogenetic tree was constructed using Mega 5.0 [23].
Results and Discussions
Actinobacteria are known to produce clavams (clavulanic acid), carbapenems (thienamycin), nocardicins, and monobactams. Streptomyces clavuligerus is reported for ceplalosporin β lactam antibiotics and its inhibitors [24]. The marine isolates were designated as PM1–PM100, and colonies with actinobacterial features and distinct morphology were selected. The strains were analyzed on the basis of spore mass color, aerial and substrate mycelium formation, production of diffusible pigments, and good antagonistic activity. Based on this, a potential actinobacterial strain PM49 was selected and was characterized by morphological and biochemical methods as it exhibited broadly clear antimicrobial activity. Valan et al. [25] has reported that marine samples from Bay of Bengal coastal area are rich source for novel marine Streptomyces sp. [25]. We evaluated the Streptomyces sp. PM49 compound activity against both Gram-positive and Gram-negative bacteria. Zone of inhibition with diameter 13–15 mm was obtained for S. aureus (15140) pathogenic strain and 15–17 mm for S. aureus ATCC 33591 and B. subtilis NCIM 2063 control strains. About ~21-mm inhibition zones were obtained on cephalosporin- and carbapenem-sensitive strains of E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii. The specificity for the compound was recapitulated by agar plug method; the lactose-fermenting E. coli and K. pneumoniae exhibited 16–17 mm, and non-lactose fermenters P. aeruginosa and A. baumannii showed 14–17-mm zones of inhibition. Both disc and well diffusion methods of the crude extract demonstrated inhibitory zone on E. coli ATCC 25922 and P. aeruginosa ATCC 27853 of 15 and 17 mm, respectively. Marine actinobacteria with antimicrobial potential on pathogenic strains with inhibitory zone of >13 ± 3 mm as in this study have been reported [26].
Variability in secondary metabolite production was observed with different temperatures, pH levels, and NaCl concentration but with pH 7, and 2 % of NaCl growth was enhanced and the production of secondary metabolite was high in ISP2 at 28 °C. In the preliminary studies using one-factor-at-a-time approach, the important four independent variables, temperature, pH, sea water, and NaCl, along with carbon source and nitrogen sources were found to influence the production of the compound as described [27]. Solid-state fermentation for the above set parameters gave a good yield of compound with soybean meal (6.5 μg/plate), followed by wheat bran and rice bran (6 μg/plate) growth that was slightly delayed with tapioca plates. Optimum temperature of 28 °C at pH 7 with 50 % seawater and 2 % NaCl has been observed to yield high compound by solid-state fermentation [28]. Agricultural waste extracts being prepared with marine water had been demonstrated to increase the production of antimicrobial components [29]. There are reports that antibiotics isolated from Streptomyces were found active only in agar medium and produced extracellularly. The combinatorial variables of 2%NaCl, 50 % marine water, 10 % of agricultural substrates of rice bran, wheat bran, and soybean along with 0.5 % of yeast extract produced a bioactive crude extract relatively similar to the extract from ISP2-defined media, but with chromatography there were a few impurities observed. Effect of culture conditions on the growth and antimicrobial activity of Streptomyces sp. PM49 shows that soy bean and wheat bran can be utilized for bulk compound production as a cost-effective resource. The ethyl acetate extract obtained also exhibited good antibacterial inhibition against S. aureus ATCC 33591 and E. coli ATCC 25922, but more work is necessary for standardization.
We observed increased production of compound in solid medium and extraction using ethyl acetate at pH 7.0 [30]. The crude compound obtained by solid-state fermentation using ISP2 was best resolved using chloroform and ethylacetate (1:1, v/v), and the spot separated into five bands of Rf values 0.41, 0.53, 0.69, 0.72, 0.87, and 0.92. The compound was freely soluble in chloroform, ethyl acetate, acetonitrile, DMSO ethyl alcohol, methanol, and 10 % isopropyl alcohol but insoluble in petroleum ether and n-hexane; similar results have been recorded. The active spots on exposure to naphthoresorcin– sulphuric acid developed brown color, and one clear zone of Rf value 0.69 having antagonistic activity was observed for the test isolates. Bioautography revealed that the third spot exhibited good antibacterial activity on spraying with 2 % w/v aqueous solution of 2,3,5-triphenyl tetrazolium chloride on both the ATCC and pathogenic strains. The crude extract with antimicrobial activity having Rf values of 0.53 to 0.69 and bioautography revealing the antagonistic activity have been reported [31]. The zone of inhibition on multidrug-resistant strains non-susceptible to fourth-generation cephalosporins showed increase in activity of 13 to 15 mm with the bioactive fraction. The Streptomyces sp. PM49 compound alone exhibited significant inhibitory activity of 3–5 mm zone size more than antibiotic disc on the test strains, but along with ceftazidime 30/10 μg of meropenem disc at 25 μl/disc concentration, enhanced zone of inhibition of 5–7 mm was obtained (Table 1).
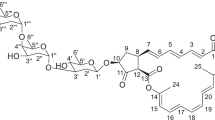
The compound was orange colored amorphous powder, and the spectroscopic characteristics of the antibacterial compound using ultraviolet and IR absorption spectrum of the active fraction were recorded (Fig. 1). The spectral data revealed the presence of two maximum absorption peaks in UV at 205 (0.927) and 257 (0.596); infra-red absorption spectrum was represented by 15 peaks. IR absorption bands indicated the presence of hydroxyl and other functional groups as tabulated; the proton NMR spectrum also showed one proton singlet peak attributed to the hydroxyl group (Table 2). The molecular formula was calculated as C25H41NO5S from its elemental and mass spectral analyses according to m/z (pos.ions) [M-H + 2Na]+ = 467.66 (Fig. 2). ESI-MS spectral measurement taken together with 1H and 13C NMR spectrum showed signals for all the carbons of the molecule. The structure was elucidated, and this sulphur-containing compound was found closely being similar to cyslabdan based on all the tabulated physico-chemical properties and literature survey (Table 2). By complete analysis of spectroscopic data and comparison with known antibiotics, the metabolite had β-lactam-potentiating activity of carbapenem drug similar to cyslabdan [7]. The compound from our study had both synergistic and β-lactamase enzyme inhibitory capability on both Gram-positive and Gram-negative MDR strains. It was isolated from a marine Streptomyces sp. PM49 unlike Streptomyces sp. K04-0144 that was a potentiator of imipenem against Gram-positive MRSA only [7].
The clinical isolates screened were commonly found to be multidrug resistant and exhibited resistance to eight to 11 antibiotics; E. coli was predominant with 71.28 % followed by K. pneumoniae 34.5 %, P. aeruginosa 34.1 %, and A. baumannii 4.8 %. ESBL production was screened in 30/96 (31.2 %) of E. coli, 29/53 (35.8 %) of K. pneumoniae, 19/87 (21.8 %) of P. aeruginosa, and 5/24 (20.8 %) of A. baumannii. In case of CDT and DDST, with ceftazidime and meropenem discs, an increase in the inhibitory zone of <5 mm was observed. About 40.4 % of P. aeruginosa, 12 % A. baumannii, 0.16 % of K. pneumoniae, and 0.06 % of E. coli were phenotypically MBL positive. The production of inducible β-lactamases and the rapid resistance acquisition of these mutant genes are responsible for the development of strong hydrolysis of beta-lactam antibiotics [3]. The frequency of MDR strains prevailed high in the non-lactose fermenters P. aeruginosa and A. baumannii. MBL production was detected more in phenotypic tests, but genotypic analysis using PCR was not proportional to CDT and DDST. On PCR amplification, two P. aeruginosa strains expressed a bla IMP gene of 188 bp and one strain bla SHV; in A. baumannii, three strains had bla OXA-58-like gene of 599 bp. There was amplification of bla CTX-M 550 bp in K. pneumoniae and mec 540 bp in S. aureus (Fig. 3). The non-lactose fermenter P. aeruginosa carrying bla IMP and bla VIM has been previously demonstrated in India [32]. The CTX-M type of β-lactamases has become a more important type of enzyme which constitutes a distinct lineage of the molecular class A β-lactamases [2]. These types of β-lactamases, which is a rapidly growing group, are the major ESBLs reported in K. pneumoniae strains across many countries. The selected P. aeruginosa (2030) and A. baumannii (3473) strains showed resistance to a series of betalactams and carbapenems antibiotics. β-Lactamase enzyme was obtained with an effective yield of 20.1 mg/l medium; the enzyme was active and stable at pH 8 for 48 h. This extracellular enzyme had cephalosporinase activity and got induced with ceftazidime at concentrations of 20 and 40 μg/ml at the same level, increasing the concentration twofolds more than MIC increased the activity. The enzyme was efficiently induced in the presence of ceftazidime than cefepime and was inhibited in the presence of clavulunic acid, confirming the β-lactamases potential; a single band of 31-kDa purified protein was observed on SDS-PAGE (Fig. 3).
PCR amplification for drug-resistant genes. On PCR amplification, two P. aeruginosa strains expressed bla IMP gene of 188 bp and one strain bla SHV. Lanes 1, 2 and 3 bla OXA-58-like gene of 599 bp, lane 4 100-bp ladder in A. baumannii. Lane 1 100-bp ladder, lanes 2, 3, 4 and 5 bla CTX-M 550 bp in K. pneumoniae, lane1 100-bp ladder, lanes 2 and 3, mec 540 bp in S. aureus. SDS-PAGE analysis of β-lactamase enzyme, lane 1 crude enzyme extract, lane 2 and 3 precipitated protein, lane 4 molecular mass marker (kDa)
There was no reduction in MIC of the compound observed in the presence of 1,3,5-triphenyltetrazolium chloride TTC/reserpine and bioactive fraction of 40, 80, and 160 μg/ml concentrations. The active fraction was not effective with the tested S. aureus ATCC 25923 and P. aeruginosa ATCC 27853 strains and MDR E. coli (2531), K. pneumoniae (551), P. aeruginosa (2030), A. baumannii (3473), and S. aureus (15140). This indicated that the bioactive fraction did not harbor an efflux system mode of inhibitory mechanism. We observed the absence of efflux system mode of inhibition by our bioactive compound; the lack of actinobacteria as an efflux inhibitor is cited in literature. There are many natural phyto-chemical compounds known for efflux inhibitory mechanism [33]. The β-lactamase inactivation using nitrocefin suggests the possibility for the β-lactamase inhibition; this chromogenic substance has been used for studying the specific interaction between β-lactams and penicillin-binding protein from methicillin-resistant S. aureus [34]. Active fraction on testing for β-lactamase inactivation in the presence of the β-lactam antibiotic ceftazidime and the chromogen nitrocefin was showing no color change to red and remained yellow similar to the control tubes devoid of β-lactamase where negative tube minus compound changed from yellow to red in color. If β-lactamase is not inhibited, it changes in color from yellow to red due to the interaction of the enzyme with nitrocefin as already demonstrated with S. aureus [35]. This suggests the possibility for the presence of β-lactamase inhibition and potentiating activity but lack of efflux inhibitory mechanism by the bioactive fraction.
The MIC of the active fraction was 16–32 μg/ml on ATCC strains, and the test pathogens inclusive of ceftazidime and meropenem sensitive and resistant strains showed MIC at 64—256 μg/ml. Lowest MIC was obtained with A. baumannii (3473) at 64 μg/ml with strongest inhibitory activity and no hetero-resistant subcolonies. Time kill studies revealed that the Streptomyces sp. PM49 active fraction had bacteriostatic effects on all the test strains. In A. baumannii, reduction on cell counts was observed within 6 h of exposure, but in 12 h the test strains increased in growth beyond the initial cell density. But in P. aeruginosa and S. aureus, the increase was slightly delayed (8 h) in all concentrations with the exposure time; it was however weakly bactericidal at 8 × MIC of (8 μg/ml). The extract showed limited bactericidal activity against both K. pneumoniae and E. coli at MIC levels of 32 μg/ml after 6 h of exposure but showed bacteriostatic effects at the same MIC levels after 12 h of exposure. Low-level resistance to carbapenems MIC has been reported; the MIC of marine Actinobacteria novel compound from PM49 was effective, but the concentration required was high on comparison with meropenem [36]. The isolated compound was efficient in counteracting the drug-resistant pathogens by interacting with the β-lactamases, thus enhancing the probability in the usage of even older cephalosporins and also carbapenems. The number of surviving cells during time kill assay for P. aeruginosa was only obtainable at higher concentration; similar results have been evaluated in other studies [37].
The colonies of Streptomyces sp. PM49 were slow growing, pale gray, and powdery; it was acid fast negative and gram positive, micromorphology of the strain was examined by SEM and the spores were rectiflexibiles having smooth sporophores. It was characterized to have LL-DAP; marine Streptomyces producing bioactive compounds with broad spectrum of activity have been reported. The functional group of the active fraction should be a sugar molecule as it developed brown-colored spot on exposure to naphthoresorcin–sulphuric acid. The strain showed 99 % identity with 16S rRNA sequences of Streptomyces rochei; however, secondary metabolite production is strain specific [38]. Taxonomic position of the strain PM49 using in silico sequence analyzer phylogeny.fr along with comparative biological analysis data of the closely related microbial strains confirms the new isolate to be very efficient in harboring antimicrobial potential. The vegetative mycelia grew abundantly on both synthetic and complex media; the aerial mycelia grew abundantly on yeast extract–malt extract agar (ISP-2) and starch–nitrate agar medium and oatmeal agar medium (ISP-3). Total nucleotide of 1457 bp showed 99 % identity with 16S rRNA sequences of S. rochei; the new isolate Streptomyces sp. PM49 was submitted to Genbank with accession no. JX904061.1 (Fig. 4). Xinghaiamine A, a sulfoxide compound isolated from marine Streptomyces xinghaiensis in China had potential activity against P. areuginosa, A. baumannii, and S. aureus [39]. Marine sediments are resourceful for the isolation of potential novel strains that can be tampered for novel antimicrobial compounds.
Scanning electron microscopic image and phylogenetic tree of Streptomyces sp. PM49. SEM of Streptomycetes sp. PM49 showing colony morphology and chain of smooth spores ×10,000 grown on yeast extract–malt extract agar (ISP2) for 7 days at 28 °C, 1 μm. Phylogenetic relationship of the new marine strain Streptomyces sp. PM49 based on 16S rRNA analysis using Mega 5.0
Conclusion and Future Prospects
This study illustrates sulphur and sulfoxide compounds having broad spectrums of biological activities which have yet to be explored from marine sediments and demonstrate experiments to determine the mode of action of the isolated compound. It is probably to the best of our knowledge the first sulfanyl cyslabdan-like compound from a marine Streptomyces sp. PM49 having efficient β-lactamase inhibitory and potentiating activity against notorious pathogens. Further, the compound not only potentiated the activity of imipenem unlike cyslabdan; it had β-lactamase inhibitory capability and enhanced the activity of third-generation cephalosporins and other carbapenems as meropenem. It was efficient on inhibiting the β-lactamase enzymes of Gram-negative pathogens and also MRSA; the compound was able to hydrolyze ceftazidime and meropenem. Sequence comparison of specific genes for the metabolite produced, structure elucidation, and functional group analysis of the beta lactamase inhibitory compound will reveal the novelty of the strain. The Streptomyces sp. PM49 is an interesting candidate for development as a new antimicrobial against MDR bacteria for future drug formulation, but further studies are required to ascribe the exact structure and precise mechanism of action of the isolated compound.
References
Lee, K., Chong, Y., Shin, H. B., Kim, Y. A., Yong, D., & Yum, J. H. (2001). Modified Hodge and EDTA disk synergy test to screen metallo beta lactamases producing strains of Pseudomonas spp and Acinetobacter spp. Clinical Microbiology and Infection, 7, 88–91.
Maninder, K., & Aruna, A. (2013). Occurrence of CTX-M, SHV and the TEM genes among the extended spectrum â-lactamase producing isolates of Enterobacteriaceae in a tertiary care hospital of North India. Journal of Clinical Diagnostic Research, 7(4), 642–645.
Andrea, M. H., Kristine, M. H., Marion, S. H., Vernon, E. A., & Robert, A. B. (2002). Amino acid substitutions at Ambler position Gly238 in the SHV-1 β-lactamase: exploring sequence requirements for resistance to penicillins and cephalosporins. Antimicrobial Agents and Chemotherapy, 46(12), 3971–3977.
Wright, G. D. (2005). Bacterial resistance to antibiotics: enzymatic degradation and modification. Advanced Drug Delivery Review, 57, 1451–1470.
Fenical, W., & Jensen, P. R. (2006). Developing a new resource for drug discovery: marine actinomycete bacteria. Nature Chemical Biology, 2, 666–673.
Wu, S. J., Fotso, S., Li, F., Qin, S., & Laatsch, H. (2007). Amorphane sesquiterpenes from a marine Streptomyces sp. Journal of Natural Products, 70, 304–306.
Atsushi, F., Yong, P. K., Hideaki, H., Kazuro, S., Hiroshi, T., & Satoshi, O. (2008). Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp. K04-0144 II. Biological activities. Journal of Antibiotics, 61(1), 7–10.
Eccleston, G. P., Brooks, P. R., & Kurtboke, D. I. (2008). The occurrence of bioactive Micromonosporae in aquatic habitats of the Sunshine Coast in Australia. Marine Drugs, 6, 243–261.
Yilmaz, E. B., Yavuz, M., & Kizil, M. (2008). Molecular characterization of rhizosphere soil Streptomycetes isolated from indigenous Turkish plants and their antimicrobial activity. World Journal of Microbiology and Biotechnology, 24, 1461–1470.
Kelman, D., Kashman, Y., Rosenberg, E., Kushmaro, A., & Loya, Y. (2006). Antimicrobial activity of red sea corals. Marine Biology, 149, 357–363.
Usha, R., Ananthaselvi, P., Venil, C. K., & Palaniswamy, M. (2010). Antimicrobial and antiangiogenesis activity of Streptomyces parvulus KUAP106 from mangrove soil. European Journal of Biological Sciences, 2, 77–83.
Sahin, N., & Ugur, A. (2003). Investigation of the antimicrobial activity of some Streptomyces isolates. Turkish Journal of Biology, 27, 73–78.
Augustine, S., Bhavasar, S. P., & Kapadnis, B. P. (2005). A non polyene antifungal antibiotic from Streptomyces albidofalvus PU 23. Journal of Bioscience, 30, 201–211.
Lee, K., Lee, W. G., Uh, Y., Ha, G. Y., Chong, Y., et al. (2003). VIM and IMP type metallo-beta-lactamase producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emergency Infectious Diseases, 9(7), 868–871.
Zhang, Z., Li, M., Zhou, D., Ruan, F., Lu, Y., et al. (2006). Detection of extended-spectrum b-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrobial Agents Chemotherapy, 50(9), 2990–2995.
Perez-Perez, F. J., & Hanson, N. D. (2002). Detection of plasmid-mediated AmpC ß-lactamase genes in clinical isolates by using multiplex PCR. Journal of Clinical Microbiology, 40, 2153–2162.
Wang, Z., & Benkovic, S. J. (1998). Purification, characterization, and kinetic studies of a soluble Bacteroides fragilis metallo-β-lactamase that provides multiple antibiotic resistance. Journal of Biological Chemistry, 273(35), 22402–22408.
Falagas, M. E., & Bliziotis, I. A. (2007). Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? International Journal of Antimicrobial Agents, 29, 630–636.
Feng, Y. C., Siu, L. K., Chang, P. F., Min Hua, H., & Monto, H. (2001). Diversity of SHV and TEM β-lactamases in Klebsiella pneumoniae: gene evolution in Northern Taiwan and two novel β-lactamases, SHV-25 and SHV-26. Antimicrobial Agents and Chemotherapy, 45(9), 2407–2413.
Andrews, J. M. (2001). Determination of minimum inhibitory concentration. Journal of Antimicrobial and Chemothereapy, 48(Suppl. 1), 5–16.
Sibanda, T., & Okoh, A. I. (2008). In vitro antibacterial regimes of crude aqueous and acetone extracts of Garcinia kola seeds. Journal of Biological Science, 8, 149–154.
Sathish Kumar, S.R., & Kokati, V.B.R. (2012). In-vitro antimicrobial activity of marine actinobacteria against multidrug resistance Staphylococcus aureus. Asia Pacific Journal of Tropical Biomedicine, S1802–S1807.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739.
De Pestel, D. D., Benninger, M. S., Danziger, L., LaPlante, K. L., May, C., Luskin, A., Pichichero, M., & Hadley, J. A. (2003). Cephalosporin use in treatment of patients with penicillin allergies. Journal of American Pharmaceutical Association, 48, 530–540.
Valan Arasu, M., Duraipandiyan, V., & Ignacimuthu, S. (2013). Antibacterial and antifungal activities of polyketide metabolite from marine Streptomyces sp. AP-123 and its cytotoxic effect. Chemosphere, 90(2), 479–487.
Zhonghui, Z., Wei, Z., Yaojian, H., Zhiyuan, Y., Jun, L., Huirong, C., & Wenjin, S. (2000). Detection of antitumor and antimicrobial activities in marine organism associated actinomycetes isolated from the Taiwan Strait, China. FEMS Microbiology Letters, 188, 87–91.
Gupta, M. D., & Kulkarni, P. R. (2002). A study of antifungal antibiotic production by Streptomyces chattanoogensis MTCC 3423 using full factorial design. Letters in Applied Microbiology, 35, 22–26.
El-Naggar, M. Y., EL-Assar, S. A., & Abdul Gawad, S. M. (2009). Solid state fermentation for the production of meroparamycin by Streptomyces sp. strain MAR01. Journal of Microbiology and Biotechnology, 19(5), 468–473.
Remya, M., & Vijayakumar, R. (2008). Isolation and characterization of marine antagonistic actinomycetes from west coast of India. Journal of Medicine and Biology, 15, 13–19.
Sekiguchi, M., Shiraish, N., Kobinata, K., Kudo, T., Yamaguchi, I., Osada, H., & Isono, K. (2007). RS-22A and C: new macrolide antibiotics from Streptomyces violaceusniger, taxonomy, fermentation, isolation and biological activities. Journal of Antibiotics, 48(4), 289–292.
Joseph, G. P., Stephane, B., Jean, M. F., Pierre, N., Bathelemy, N., Anatole, A., et al. (2007). Screening of some medicinal plants from cameroon for beta-lactamase inhibitory activity. Phytotherapy Research. Phytotherapy Research, 21(3), 284–287.
Masashi, T., Masaaki, Y., Seiko, O., Yasushi, T., Yasutake, H., Yuzuru, M., Ayumi, S., Hironori, F., Yasushi, O., & Junichi, K. (2005). Brasilibactin A, a cytotoxic compound from actinomycete Nocardia brasiliensis. Journal of Natural Products, 68(3), 462–464.
Siddhartha, R. C., Raymond, E. K., David, N. B., & Wu Kuand, Y. (1996). Methicillin-resistant Staphylococcus aureus: S. Antimicrobial Agents and Chemotherapy, 40(9), 2075–2079.
Mark, S. B. (2005). Natural products to drugs: natural product derived compounds in clinical trials. Natural Products Reports, 22, 162–195.
Venkata, R., Murali, K., Murali, Y. N., & Sri Rami, R. D. (2012). Novel pyridinium compound from marine actinomycete, Amycolatopsis alba var. nov. DVR D4 showing antimicrobial and cytotoxic activities in vitro. Microbiology Research, 167(6), 346–351.
Sathish Kumar, S.R., & Kokati, V.B.R. (2012). In-vitro antimicrobial activity of marine actinobacteria against multidrug resistance Staphylococcus aureus. Asia Pacific Journal of Tropical Biomedicine, S1802-S1807.
Ekrem, K., & Mettem, Y. C. (2006). Comparison of staphylococcal beta-lactamase detection. FABAD Journal of Pharmaceutical Science, 31, 79–84.
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., Dufayard, J. F., Guindon, S., Lefort, V., Lescot, M., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research, 36, 465–469.
Julain, D., & Dorothy, D. (2010). Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews, 74(3), 417–433.
Acknowledgments
The authors are indebted to the microbiology staff members and physicians of the tertiary care hospital, Bangalore, for helping us in collecting the samples and providing ATCC strains, and they also acknowledge the technicians for their help. The authors also thank the Vice-Chancellor and Registrar of Periyar University, Salem, for their support and encouragement. This work was supported by the Indian Council of Medical Research [ICMR] New Delhi, India (ICMR letter no. 5/8/5/24/9/2011—ECD-I, dt.19.12.11).
Conflict of Interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shanthi, J., Senthil, A., Gopikrishnan, V. et al. Characterization of a Potential β-Lactamase Inhibitory Metabolite from a Marine Streptomyces sp. PM49 Active Against Multidrug-Resistant Pathogens. Appl Biochem Biotechnol 175, 3696–3708 (2015). https://doi.org/10.1007/s12010-015-1538-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1538-x