Abstract
Exo-xylanases are a class of glycosyl hydrolases and play an important role in hydrolysis of xylan to xylose. They belong to glycosyl hydrolase (GH) family 8 with a characteristic (α/α)6 barrel fold in their molecular structures. These enzymes are generally produced by bacteria. Exceptionally, the endo-xylanases from Trichoderma reesei Rut C-30 and a few bacterial strains also show considerable exo-xylanase activities. Exo-xylanases are active on natural xylan substances, hydrolyzing long-chain xylo-oligomers from the reducing end to produce short-chain xylo-oligomers and xylose. Exo-xylanases usually show multiple enzyme functions such as β-xylosidase, exo-glucanase, β-glucosidase, and arabinofuranosidase activities, which are helpful for more efficient hydrolysis of xylan. The combined use of exo- and endo-xylanases can increase the xylose yield compared to using either of them alone. Screening new exo-xylanase-producing microbes, mining the enzyme coding sequences, genetically engineering the enzymes, and producing them in a large scale are recommended for their commercial applications in lignocellulose-based biorefinery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant cell wall is composed of three types of polymers including cellulose, hemicellulose, and lignin. Cellulose and hemicellulose are the major carbohydrate polymers produced during photosynthesis. Cellulose is a homo-polymeric polysaccharide composed of β-D-glucopyransoyl residues interlinked by β-1,4-glyosidic bonds. Cellulose predominantly exists in crystalline form being recalcitrant to enzyme or chemical attack. Hemicellulose is an amorphous hetero-polymeric polysaccharide composed of pentose (C5) and hexose (C6) sugars. Lignin is an aromatic polymer connected with hemicellulose that forms a matrix around the cellulose microfibrils [1–4].
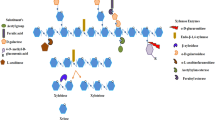
In herbaceous and hardwood biomass, D-xylose is the sugar component of hemicellulose existing as a branched xylan backbone which is substituted with L-arabinose, D-mannoase, D-galactose, and glucouronic acid through glysosidic bonds and with acetic acid and ferulic acid by ester bonds [5, 6]. While in soft or coniferous woods, mannose is the major sugar component of hemicellulose [7] (Fig. 1). The heterogeneous nature of hemicellulose makes it inaccessible for a single enzyme for complete depolymerization. During depolymerization of hemicellulose, the polymer structure changes significantly revealing new hidden bonds to recruit new enzymes for deeper depolymerization. The complete depolymerization of hemicellulose requires synergistic action of both glycosyl hydrolases and carbohydrate esterases [8, 9], including endo-xylanase (endo-1,4-β-xylanase, E.C.3.2.1.8), β-xylosidase (xylan-1,4-β-xylosidase, E.C.3.2.1.37), α-glucuronidase (α-glucosiduronase, E.C.3.2.1.139), α-L-arabinofuranosidase (E.C.3.2.1.55), arabinase (endo α-L-arabinase, E.C.3.2.1.99), acetyl xylan esterase (E.C.3.2.1.72), and feruloyl xylan esterase (E.C.3.2.1.73) [9, 10]. Among them, endo-xylanases and β-xylosidases (collectively xylanases) have been extensively investigated possibly due to xylan being the major pentose sugar component of hemicellulose [10]. However, exo-xylanases (exo-1, 4-β-xylanase, E.C.3.2.1.37), another xylan-depolymerizing enzyme, have not yet received much attention. The exo-xylanase activity was first reported in 1989 for the xylanases from the fungus Chaetomium thermophile var. coprophiles. This fungus secreted xylanases I and II. Xylanase I generated xylobiose and xylotriose whereas xylanase II released mainly xylobiose from the larch wood xylan [11]. In 2004, a paper on purification and characterization of Aeromonas caviae ME1 xylanase V was published, reporting that xylobiose was exclusively produced from oat spelt and birch wood xylans by this enzyme in the exo-xylanase mechanism[12].
Structure of xylan with different side chain substitutions. Different glyosidic linkages are also shown [6]
Exo-xylanases show a significant variation in their mode of action on xylan when compared to endo-xylanases and β-xylosidases. Endo-xylanases randomly cleave the xylan backbone from the inside (endo-fashion) yielding long-chain xylo-oligomers, on which β-xylosidases act releasing xylose monomers. Exo-xylanases act on the xylan backbone from the reducing end (exo-fashion) producing short-chain oligomers [12] or hydrolyzing short-chain xylo-oligomers with apolymerization degree of ≥2–3 to release xylose. Additionally, these two enzymes can be further distinguished from each other by nuclear magnetic resonance spectroscopic analysis for configurations of the xylose residues released during hydrolysis. Surprisingly, these enzymes are inert on pure polymeric xylan and inactive on xylobiose [13, 14]. To the best of our knowledge, this is the first review paper on exo-xylanases elucidating their different properties and synergistic functions with endo-xylanases and β-xylosidases in hydrolysis of xylan.
Structure of Exo-xylanases
Exo-xylanases on average have a molecular mass of ~40 kDa with a simple architecture containing a catalytic domain (CD) that performs enzyme functions. The CD of this class of enzymes is a glycosyl hydrolase (GH) that brings hydrolysis of glyosidic bonds present in the xylo-oligomers [15]. The structure of exo-xylanases is explained by taking the crystal structure of Bacillus halodurans C-125 exo-oligoxylanase as an example, which is, to the best of our knowledge, the only entry for the exo-xylanase crystal structure (www. rcsb.org) according to the protein database. The coding sequence of this protein is 1,167-bp-long encoding a peptide chain with a molecular mass of 45 kDa after complete post-translational modifications (PDB ID: 1WU4). According to CAZY database (www.cazy.org), this enzyme belongs to GH family 8 and clan GH-M. Depending on the position of catalytic base residues, the GH family 8 is subdivided into GH-8a, GH-8b, and GH-8c. This enzyme belongs to subfamily GH-8a [14]. It shows the GH-8 family characteristic (α/α)6 barrel fold with a proton donor (Glu70) and a catalytic base (Asp263) residue at the N-terminal α4 and α8 helices, respectively. The structure of this enzyme clearly explains its specificity toward short-chain xylo-oligomers with a reducing end [16].
The crystal structure of this enzyme indicates the presence of a disordered (α/α)6 barrel fold structure (Fig. 2). This enzyme has four free Cys residues, a characteristic feature of intracellular enzymes. Its substrate-binding cleft (subsite +2) is short and blocked by a barrier at the top side. Binding of the substrates to the enzyme induces a slight moment in the regions of Thr62-Asn64 and Gly355-Arg357 (subsite +1). The catalytic base Asp263 holds the water molecule by hydrogen bonds with Tyr198. This water molecule acts as a nucleophile molecule activated by the catalytic base residue Asp263. The structural basis for the specificity of the enzyme for the reducing end is explained by His319 which forms a specific hydrogen bond with the hydroxyl group of a reducing end xylose at subsite +1 contributing to the discrimination of anomers at the reducing end. Leu318 blocks subsite +2 with its long side chain together with His319. Pro320 presents specificity for this enzyme allowing the protein to bend at right angle at this position. This bent loop structure seems to be intrinsically more stable as there is no difference between liganded and unliganded protein structures [16].
Ribbon diagram of reducing end xylose-releasing exo-oligoxylanase from Bacillus halodurans C-125 (PDB ID: 1WU4). Protein chains are colored from the N-terminal to the C-terminal using a rainbow (spectral) color gradient. The enzyme shows GH family 8 characteristic (α/α)6 barrel fold [14] (Color figure online)
Functionally, exo- and endo-xylanases perform the same enzyme function that is the hydrolysis of glyosidic bonds. Structurally, the active site of endo-xylanases has a β-α loop architecture forming a large tunnel-like structure for the entry of long-chain xylan polymers. The two subsite at the reducing end are relatively open, and after releasing the cleaved short-chain oligosaccharide, the xylan chain slides for the next round of processing position [10]. In contrast, the exo-oligoxylanase subsite +2 is blocked by a curve in the loop before α10 at Ser317-Pro320 inhibiting the entry of long-chain xylan polymer (Fig. 3) converting this enzyme into an exo-xylanase [16].
Molecular surface of Bacillus halodurans C-125 reducing end xylose-releasing exo-oligoxylanase showing the substrate binding cleft. Positive and negative potentials are shown in blue and red colors. Top side of the substrate binding cleft is closed preventing the entry of long-chain xylo-oligosaccharides [15] (Color figure online)
Screening Exo-xylanase-Producing Microbes
Hemicellulases are produced by a wide spectrum of microbes in nature. The reported exo-xylanases were mainly from bacteria. Native exo-xylanase-producing bacteria were isolated from the rumen of cattle, gut of silk worm, soil sample near the plants, hot water springs, and soil in the Antarctic station. Entries of the nucleotide sequences for the newly discovered exo-xylanases are very limited. Screening novel exo-xylanase-producing microbes should be done under top priority as their number is very low when compared to the number of other xylanase producers. Colony screening of exo-xylanase producers is done either using Congo Red assay or incorporating Remazol Brilliant Blue xylan (RBB-xylan) onto agar plates. Exo-xylanase-expressing microbes that have been isolated from different natural sources and their entries in NCBI and Uniprot database are listed in Table 1 [7, 17, 18–21].
Enzyme Activity Assay
Exo-xylanases are produced in relatively smaller amounts when compared to other class of xylanases. Their substrate specificity is not yet very clear. Activity assays are conducted using either natural or synthetic substrates.
In the category of natural substrates, birch wood xylan, beech wood xylan, oat spelt xylan, larch wood xylan, Palmaria palmata xylan, hardwood xylan with reduced content of 4-O-methyl-D-glucuronic acid, 4-O-methylglucuronoxylan from birch wood, glucuronoxylan, rhodymenan, wheat arabinoxylan, and Xylan isolated from the empty fruit bunch of palm oil trees were used as natural substrates to detect the exo-xylanase activity. The amounts of reducing sugars released were analyzed by DNS method or high-performance liquid chromatography (HPLC). To detect the natural specificity of newly discovered enzymes for different xylo-oligomers, xylo-oligomers of different lengths (2–6) were used as substrates. The cleavage products from the oligomers after enzymatic hydrolysis were analyzed by high-performance liquid chromatography (HPLC) or thin-layer chromatography (TLC) [14, 19, 21].
The synthetic substrates for exo-xylanases are classified into chromogenic, fluorogenic, radioactive, and xylo-oligosaccharide derivatives. Cleavages of 4-nitrophenyl-β-D-xyloside and 4-nitrophenyl-β-D-xylobioside release 4-nitrophenol which is detected at 410 nm. Remazol Brilliant Blue R-D-xylan (RBB-xylan) on cleavage releases brilliant blue which is monitored at 595 nm. Cleavages of 4-methylumbelliferyl-β-D-xyloside and 4-methylumbelliferyl-β-D-xylobioside release 4-methylumbelliferone which is detected at 354 nm. Radiolabelled xylo-oligomers (Xyl-4Xyl-4Xyl-Me; Xyl-Xyl-Xyl-de) were used for 1H-NMR study of stereo-chemical course of hydrolysis of glycosidic linkages, and xylo-oligosaccharide derivatives (modified and non-linear) were used as substrates (Glu-Xyl2, Xyl2-Glu, Glu-Xyl-Glu, Xyl-Glu2, Glu2-Xyl, Xyl-Glu-Xyl, Hex-A3Xyl3, MeGlc-A3Xyl3, Ara2Xyl3, Xyl-4Xyl-Me, Xyl-4Xyl-4Xyl-Me, MeGlcA3Xyl3, MeGlcA3Xyl4, Ara2Xyl4, and Ara2Xyl3) with their cleavage products being detected by TLC [13, 26].
Exo-xylanases from Bacteria
Prevotella ruminicola B14 holds an exo-xylanase (XynB) of 319-amino-acid-long peptide coded by a 960-bp-long nucleotide. After complete post-translational modifications, the peptide chain moulds into a functional protein with a molecular mass of 36.40 kDa. This protein lacks native N-terminal signal peptide so it is expected to be a cytoplasmic protein. The enzyme activity decreased exponentially at 50 °C in 0.05 M Na phosphate buffer–2 mM at pH 6.5. The half-life of the enzyme activity toward 4-nitrophenyl β-D-xylopyranoside and 4-nitrophenyl α-L-arabinopyranoside at 50 °C was 4 min. It is O2 sensitive and shows a full activity only in anaerobic conditions. XynB is active against birch wood xylan releasing xylobiose and xylose but completely inactive against oat spelt xylan. It can release xylose progressively from the non-reducing ends of longer xylo-oligosaccharides and xylobiose. Aldotetraouronic acid and 4-O-methyl glucuronic acid substitutions of xylan inhibit the enzyme activity. This was the first report on the putative bacterial exo-xylanase with its complete nucleotide sequence [17].
A new exo-xylanase, xylanase IV, was discovered from A. caviae ME-1 in the culture supernatant. The purified protein has a molecular mass of 41 kDa. Enzymatic hydrolysis of 1 % oat spelt xylan at 40 °C and pH 6.8 yielded xylotetraose as the only short-chain xylo-oligosaccharide. This enzyme is active on oat spelt xylan (100 %) and larch wood xylan (69 %) but inactive on 4-nitrophenyl-β-D-xyloside and cellulose substrates, indicating that it is a true exo-xylanase. Congo Red staining of the enzyme zymogram shows a clear hollow zone around the protein, indicating that the enzyme hydrolyzed the xylan backbone. This was the first report on the bacterial exo-xylanase producing predominantly xylotetraose [18].
The second exo-xylanase, xylanase V, was isolated from the culture supernatant of A. caviae ME-1. The purified protein has a molecular mass of 46 kDa and PI of 5.4. The enzyme showed its maximal activity at pH 6.8 and 30–37 °C. It acted on birch wood xylan with a Km and Vmax of 2 mg ml−1 and 182 μmol min−1 mg−1 of protein, respectively. This enzyme produces exclusively xylobiose from both oat spelt and birch wood xylan. The smallest substrate that this enzyme can cleave is xylotriose, indicating that it cleaves xylo-oligosaccharides by the exo-mechanism [7].
The third exo-xylanase, xylanase X, from A. caviae ME-1 was heterologously expressed in Escherichia coli P2 392. The enzyme coding sequence is 1,002 bp long encoding a 333-amino-acid-long peptide. After complete post-translational modifications, the peptide chain moulds into a functional protein with a molecular mass of 38.61 kDa. Xylanase X hydrolyzed birch wood xylan (0.5 %) at 40 °C producing predominantly xylobiose and tetraose. Details of this enzyme hydrolyzing xylan chain either from its reducing or non-reducing end are not yet available. This enzyme did not show any transglycosylation activity. Homology modeling of its peptide sequence indicated that this enzyme belongs to GH family 10 with a characteristic (α/β)8 Tim barrel fold and a long loop between seventh α-helix and eighth β-sheet, which is thought to play a pivotal role in hydrolyzing xylo-oligomers into xylobiose and tetraose [7]. We found that along with its exo-xylanase activity, this enzyme also showed considerable β-xylosidase and exo-glucanase activities on 4-nitrophenyl β-D-xylopyranoside and 4-nitrophenyl β-D-cellobioside, respectively (unpublished data).
Paenibacillus sp. BP-23 produced cell-associated family 10 xylanase. Heterologous expression of the xylanase in Bacillus subitilis in the secretory mode ended up in the cell-associated protein. Exo-xylanase showed high specific activity toward birch wood, beech wood, and oat spelt xylans, wheat arabinoxylan, rye arabioxylan, and methylglucuronoxylan. It hydrolyzes xylan in the exo-fashion. Along with the exo-xylanase activity, this enzyme also showed β-xylosidase activity on 4-nitrophenyl β-D-xylopyranoside and 2-nitrophenyl β-D-xylopyranoside; β-glucosidase activity on 4-nitrophenyl β-D-glucopyranoside; exo-glucanase activity on 4-nitrophenyl β-D-cellobioside, 4-nitrophenyl β-D-cellotrioside, 4-nitrophenyl β-D-cellotetraoside, and 4-nitrophenyl β-D-cellopentaoside; and arabinofuranosidase activity on 4-nitrophenyl α-L-arabinopyranoside and 4-nitrophenyl α-L-arabinofuranoside [11].
A putative exo-xylanase gene was cloned from Geobacillus thermoleovorans IT-08 and heterologously expressed in E. coli DH5α cells, which is a 1,815-bp nucleotide encoding a 604-amino-acid-long peptide chain. After complete post-translational modifications, the peptide forms a functional protein with a molecular mass of 70 kDa. On the genomic DNA, this exo-xylanase gene is present first followed by β-xylosidase and α-arabinofuranosidase genes [20].
Alkalophilic bacterium B. halodurans C-125 was discovered to express an exo-xylanase that hydrolyzes xylo-oligosaacharides exclusively from the reducing end. A 1,167-bp-long enzyme coding sequence was cloned into pET28b and expressed in E. coli BL21 (GOLD) (DE3) cells. The His-tagged protein has a molecular mass of 45 kDa, belonging to GH family 8. The purified enzyme was inactive on cellulose substrates, birch wood xylan, 4-nitrophenyl-β-D-xyloside, and 4-nitrophenyl-β-D-xylobioside. The enzyme showed its maximal activity at 50 °C and pH 6.2–7.3. It hydrolyzed xylopentaose liberating xylose and xylotetraose initially, and xylose and xylobiose finally. There were no oligosaccharides larger than the initial substrates in the enzymatic reaction system, indicating the lack of a trans-glycosylation activity. As this enzyme exclusively hydrolyzes xylo-oligosaccharides from its reducing end liberating xylose, this exo-xylanase was renamed as exo-oligoxylanase [13].
A psychrophilic exo-xylanase-producing bacterium Pseudoalteromonas haloplanktis was discovered from an Antarctic soil sample. The enzyme belongs to GH family 8. A 1,278-bp-long nucleotide coding sequence encodes a 426-amino-acid-long peptide chain. After complete post-translational modifications, the peptide moulds into a functional protein with a molecular mass of 45.98 kDa. The native enzyme has a 21-amino-acid-long signal peptide making it secreted into the external medium. The enzyme shows its maximal activity at 25 °C and pH 6.5. It hydrolyzes long-chain xylo-oligosaccharides to xylobiose and tetraose. The enzyme is inactive over synthetic substrates (4-nitrophenyl, 4-methylumbelliferyl), cellulose, and aryl-β-glycoside xylo-oligomers of Xyl1, Xyl 2, and Xyl 3. The psychrophilic xylanase attacks the β-1,4 bond that is preceded by a β-1,3 bond at the non-reducing end but cleaves only the β-1,4 linkage into two xylosyl residues from the reducing end of a β-1,3 bond. It catalyzes the hydrolysis from the β-1,3 linkage via a double-displacement mechanism retaining its anomeric configuration. As this bacterium produces only a single xylanase with a catalyticability of generating xylotriose from xylohexaose, it is presumed that this enzyme has a catalytic site in the centre of its molecule with six substrate binding subsites accommodating the long-chain xylo-oligosaccharides [21].
The exo-oligoxylanase from B. halodurans C-125 and P. haloplanktis exo-xylanase both are classified under GH family 8. In spite of belonging to the same family, these two enzymes show differences in their enzyme activity. The reducing-end xylose-releasing exo-oligoxylanase (Rex) can hydrolyze the xylo-oligomers with a polymerization degree of ≥3 producing xylose. Whereas the psychrophilic exo-xylanase exclusively acts on the β-1,4 bond preceded by a β-1,3 bond at the non-reducing end but can cleave only the β-1,4 linkage into xylo-oligodisacchrides from the reducing end of a β-1,3 bond. The active site of Rex is short and closed on one side making it inaccessible for the entry of long-chain xylo-oligomers (Fig. 3). On contrary, the active site of the psychrophilic exo-xylanase is in the centre of the enzyme molecule with two subsites being open and accessible for the long chain xylo-oligosaccharides producing xylotriose and tetraose. However, more crystallization studies need to be done to better understand the action modes of these exo-xylanases [13, 21].
Hemicelluloses from diverse plant sources have different chemical compositions, which are thought to affect the structures of hemicelluloses. This might be the reason why exo-xylanases show different affinities toward different natural xylan substrates. The evolution of multiple enzyme activities of exo-xylanases is accompanied with the co-presence of cellulose and hemicellulose in plant cell walls. The heterogeneous feature of hemicelluloses further induces the evolution of multiple enzyme activities. For example, A. caviae expressed multiple exo-xylanases with both cellulase and hemicellulase activities, which would help destroy the cellulose component leading to more efficient hydrolysis of xylan from various sources [19, 21].
Action Mechanisms of Exo-xylanases in Bacteria
Bacteria produce exo-xylanases for more efficient hydrolysis of xylan. The Xyn-X from A. caviae ME-1 does not have a native N-terminal signal peptide. It is a cytoplasmic protein both in the native strain and in E. coli when heterologously expressed in the normal or osmostatic conditions. In Gram-negative bacteria such as E. coli, the cell envelope contains the outer and inner membranes and periplasmic space between them. When cells undergo an osmotic shock, the enzymes are transported from the cytoplasm to the periplasmic space. Immunoelectron microscopy examination of osmotically down-shocked cells (both native strain and recombinant E. coli) showed that the protein was accumulated in the inner side of the membrane. Mechano-sensitive channels (MscL) are located in the inner side of the inner membrane. Through these channels, Xyn-X was transported into the periplasmic space. This was further confirmed by the protein transport inhibition studies in E. coli using gadolinium chloride, an inhibitor to MscL. As A. caviae is also a Gram-negative bacterium, the possibility for the existence of similar mechano-sensitive channels is high leading to the transport of proteins from cytoplasm to periplasm [27].
Aeromonas spp. are generally found in the gut of silkworms, fresh water, and sewage water. The osmolality of the hydrosphere is much less than that of the gut and excreta. When bacteria shuttle between these two extreme environments, they are subjected to osmotic stress. Under these conditions, Xyn-X is transported to periplasm. Outside the cells, long-chain oligosaccharides are depolymerized by endo-xylanases into short-chain oligomers which are then absorbed onto the cells and hydrolyzed to xylo-oligodisaccharides and tetrasacchrides by the exo-xylanase present in the periplasmic space. These disaccharides and tetrasacchrides are further hydrolyzed into xylose by the cytoplasmic β-xylosidase. Similarly, P. ruminicola B14 is a ruminant bacterium found in the cattle rumen. The exo-xylanase from this bacterium lacks the native N-terminal signal peptide, so the protein exists within the cytoplasm of the cells. But, it is not yet elucidated whether the protein remains in the cytoplasm or moves to periplasm upon an osmotic shock to exhibit its enzyme function. The endo-glucanase from this bacterium is cell-associated, so it is assumed that similar to the endo-glucanase, the exo-xylanase should be transported to the periplasmic space [17].
The B. halodurans C-125 exo-oligoxylanase lacks the signal peptide so it is an intracellular protein. It is assumed that the endo-xylanases and acetyl xylan esterases are secreted outside the cells, which brings the hydrolysis of xylan to a smaller size α-arabinofuranosyl and/or α-glucuronyl side chain xylo-oligosaccharides that are then transported into the cells by unknown transporter proteins. In the cytoplasm of the cells, exo-xylanase, β-xylosidase, α-glucouronidase, and α-arabinofuranosidase are present. α-Glucouronidase and α-arabinofuranosidase cleave the side chains of the transported xylo-oligosaccharides releasing xylo-oligomers, which are attacked by exo-xylanase and β-xylosidase both from their reducing and non-reducing ends releasing xylose (Fig. 4) [13].
Proposed mechanism of xylose metabolism in Bacillus halodurans C-125. Extracellular endo-xylanase and acetyl xylan esterase hydrolyze xylan to short-chain xylo-oligomers, which are transported into cells and further hydrolyzed to monomer sugars by the intracellular enzymes [13]
Generally, endo-xylanases are secreted outside the cells. In contrast, exo-xylanases are usually cytoplasmic. It is presumed that endo-xylanases hydrolyze hemicelluloses to long-chain oligomers with different side chain substitutions. Then, these oligomers with substitutions are transported into the cells where the hemicellulases other than xylanases hydrolyze these substitutions to release xylo-oligomers without any substituions. These xylo-oligomers are hydrolyzed by exo-xylanases to give short-chain xylo-oligomers, which are the substrates for the intracellular β-xylosidase to produce xylose. This suggests that endo-xylanases and exo-xylanases act successively to bring efficient hydrolysis of xylan [13, 17, 22]. It has been shown that short-chain cellulose oligomers can induce the production of cellulases and hemicellulases in Hypocrea jecorina [23]. It is worth investigating whether short-chain xylo-oligomers such as xylobiose generated by exo-xylanases are able to induce cellulase and hemicellulase expressions similarly.
Principal Xylanases Exhibiting Exo-xylanase Activity
Principal xylanases are known to exhibit dual enzyme functions, which are considered to be evolved due to the heterogenous nature of hemicellulose. Xylanases with dual enzyme functions bring more efficient hydrolysis of xylan releasing monomer sugars for the metabolisms of host cells [24, 25].
Paenibacillus xylanilyticus KJ-03 was isolated from the soil samples obtained from the fields of Amorphophallus konjac plants. This strain produces a 345-amino-acid-long endo-1,4 β-xylanase A with a molecular mass of 40 kDa. It belongs to GH family 10. It has been reported that along with its endo-xylanase activity, this enzyme also shows significant exo-xylanase activity, which is deduced by its ability to hydrolyze 4-nitrophenyl-β-D-xylopyranoside. In the enzymatic reaction catalyzed by XynA, xylobiose was cleaved into xylose by its exo-xylanase activity. The produced xylose was re-polymerized to form long-chain oligosaccharides larger than xylobiose due to the trans-glycosylation activity of the exo-xylanase [26].
A xylanase (XYN IV) with a molecular mass of 43 kDa was isolated from the fungus Trichoderma reesei Rut C30. This enzyme was discovered based on its unique ability to hydrolyze aldotetraohexenuronic acid xylo-oligosaccharides (HexA-2Xyl-4Xyl-4Xyl, HexA3Xyl3) from their reducing ends releasing xylose. This property was not exhibited by other xylanases I–III and xylan-hydrolyzing endo-β-1,4-glucanase EG I from this fungus. This enzyme shows both exo- and endo-xylanase properties. Linear xylo-oligosaccharides are attacked by this enzyme exclusively on their first glyosidic linkage from the reducing end. The coding nucleotide sequence of this enzyme was amplified, cloned, and heterologously expressed in Pichia pastoris. The purified recombinant protein showed similar properties with the native one. The exo-xylanase property of this enzyme is thought to be helpful for liberating xylose from its reducing end of branched oligosaccharides that are resistant to enzymatic hydrolysis of endo-xylanases and β-xylosidases [28].
A deep-sea thermophilic bacterium Geobacillus stearothermophilus produces β-xylosidase. The coding sequence of this β-xylosidase was cloned and expressed in E. coli. The recombinant protein has a molecular mass of 79.8 kDa with its maximal activity being shown at pH 5.5 and 70 °C. This β-xylosidase belongs to GH family 52. A single change in the amino acid at the position 509 by replacing tyrosine (aromatic and polar) with glutamic acid (acidic and negatively charged) introduced additional exo-xylanase activity for this enzyme without affecting its native β-xylosidase function. The mutant enzyme showed its maximal xylanase activity at pH 6.5 and 55 °C. A single change in the amino acid widens the substrate binding site, making it accessible for the long-chain xylo-oligomers onto its active site [28].
Application of Exo-xylanases in Lignocellulose Biorefinery
The applications of exo-xylanases were hardly addressed in literature. Recently, we overexpressed the exo-xylanase from A. caviae ME1 in E. coli and used it for hydrolysis of xylan to get xylose. We conducted the enzymatic hydrolysis of beech wood xylan with 50 mg l−1 of exo-xylanase X (A. caviae ME1) obtaining 150 g l−1 of xylose. In contrast, the endo-xylanase from P. ruminicola produced only 20 g l−1 xylose at the same concentration. The combined use of exo- and endo-xylanases (50 mg l−1 each) yielded 330 g l−1 of xylose, indicating a synergistic effect of the two xylanases favoring the efficient hydrolysis of xylan to xylose. When 100 mg l−1 of exo-xylanase or endo-xylanase was used alone, 270 and 150 g l−1 of xylose were produced, respectively (unpublished data). This indicates that exo-xylanase is more efficient than endo-xylanase in xylan hydrolysis in terms of releasing xylose. The exo-xylanase showed considerable β-xylosidase activity in addition to its exo-xylanase activity, which might be the reason why using exo-xylanase alone could give a higher xylose yield than using endo-xylanase alone. Therefore, the applications of exo-xylanases in lignocellulose-based biorefinery are worth developing.
Conclusions
The complete hydrolysis of xylan requires synergistic action of two principle xylanases, i.e., endo-xylanase and β-xylosidase. However, a class of existing but largely neglected xylan hydrolyzing exo-xylanases has not yet been well studied. Exo-xylanases, mainly found from bacteria, can bring the hydrolysis of xylan or xylo-oligomers from their reducing end, producing predominantly short-chain oligomers as substrates for other xylanases such as β-xylosidase. The detailed action mechanism of exo-xylanases is still unclear and needs further investigation.
The nucleotide entries for exo-xylanases are very limited compared to other xylanases. Identification and screening of novel exo-xylanase producers with robust properties from the diverse microbial sources [32–34] and mining their coding sequences for heterologous expression [36] are worth doing their ability of bringing more efficient hydrolysis of xylan in combination with other xylanases [36]. Rational engineering and directed evolution of the enzyme coding sequences to produce tailor-made enzymes with improved properties and large scale production of these enzymes are needed for their applications in lignocellulose-based biorefinery.
References
Pérez, J., Muñoz-Dorado, J., de la Rubia, T., & Martínez, J. (2002). International Microbiology, 5, 53–63.
Corral, O.L.,&Villaseñor-Ortega, F.(2006).Advances in Agricultural and Food Biotechnology Research, ed by G-G Ramón Gerardo and T-P Irineo. Research Signpost, Kerala, pp 305-322
Sticklen, M. B. (2008). Nature Reviews Genetics, 9, 433–443.
Kumar, R., Singh, S., & Singh, O. V. (2008). Journal of Industrial Microbiology and Biotechnology, 35, 377–391.
Bastawde, K. B. (1992). World Journal of Microbiology and Biotechnology, 8, 353–368.
de Vries, R. P., & Visser, J. (2001). Microbiology and Molecular Biology Reviews, 65, 497–522.
Shallom, D., & Shoham, Y. (2003). Current Opinion in Microbiology, 6, 219–228.
Juturu, V., & Wu, J. C. (2013). Journal of Chemical Technology and Biotechnology, 88, 353–363.
Juturu, V., & Wu, J. C. (2011). Biotechnology Advances, 30, 1219–1227.
Ganju, R. K., Vithayathil, P. J., & Murthy, S. K. (1989). Canadian Journal of Microbiology, 35, 836–842.
Kubata, B. K., Suzuki, T., Horitsu, H., Kawai, K., & Takamizawa, K. (1994). Applied and Environmental Microbiology, 60, 531–535.
Honda, Y., & Kitaoka, M. A. (2004). Journal of Biological Chemistry, 279, 55097–55103.
Ghose, T. K., & Bisaria, V. S. (1987). Pure and Applied Chemistry, 59, 1739–1752.
Henrissat, B., & Davies, G. J. (1997). Current Opinion in Structural Biology, 7, 637–644.
Fushinobu, S., Hidaka, M., Honda, Y., Wakagi, T., Shoun, H., & Kitaoka, M. (2005). Journal of Biological Chemistry, 280, 17180–17186.
Gasparic, A., Martin, J., Daniel, A. S., & Flint, H. J. (1995). Applied and Environmental Microbiology, 61, 2958–2964.
Kubata, B. K., Takamizawa, K., Kawai, K., Suzuki, T., & Horitsu, H. (1995). Applied and Environmental Microbiology, 61, 1666–1668.
Usui, K., Ibata, K., Suzuki, T., & Kawai, K. (1999). Bioscience, Biotechnology, and Biochemistry, 63, 1346–1352.
Puspaningsih, N. N. T., Suwanto, A., Suhartono, M. T., Achmadi, S., Yogiara, S., & Kimura, T. (2008). Journal of Basic Science, 9, 177–187.
Tenkanen, M., Vršanská, M., Siika-aho, M., Wong, D. W., Puchart, V., Penttilä, M., Saloheimo, M., & Biely, P. (2013). FEBS Journal, 280, 285–301.
Collins, T., Meuwis, M.-A., Stals, I., Claeyssens, M., Feller, G., & Gerday, C. (2002). Journal of Biological Chemistry, 277, 35133–35139.
Gallardo, O., Diaz, P., & Pastor, F. I. J. (2003). Applied Microbiology and Biotechnology, 61, 226–233.
Huang, Z., Liu, X., Zhang, S., &Liu, Z. (2013). Journal of Industrial Microbiology and Biotechnology 1-10.
Kubicek, C. P., Mikus, M., Schuster, A., Schmoll, M., & Seiboth, B. (2009). Biotechnology and Biofuels, 2, 19.
Juturu, V., & Wu, J. C. (2013). World Journal of Microbiology and Biotechnology, 29, 249–255.
Han, S. J., Yoo, Y. J., & Kang, H. S. (1995). JBC, 270, 26012–26019.
Park, D. J., Lee, Y. S., Chang, J., Fang, S. J., & Choi, Y. L. (2013). Journal of Microbiology and Biotechnology, 23, 397–404.
Usui, K., Suzuki, T., Akisaka, T., & Kawai, K. (2003). Journal of Bioscience and Bioengineering, 95, 488–495.
Ko, K. C., Han, Y., Shin, B. S., Choi, J. H., & Song, J. J. (2012). Applied Biochemistry and Biotechnology, 167, 677–684.
Tarayre, C., Brognaux, A., Brasseur, C., Bauwens, J., Millet, C., Mattéotti, C., Destain, J., Vandenbol, M., Portetelle, D., De Pauw, E., Haubruge, E., Francis, F., & Thonart, P. (2013). Applied Biochemistry and Biotechnology, 171, 225–245.
Brito-Cunha, C. C., de Campos, I. T., de Faria, F. P., & Bataus, L. A. (2013). Applied Biochemistry and Biotechnology, 170, 598–608.
Li, J., Zhang, H., Wu, M., Wang, C., Dong, Y., Zhu, L., & Zhang, P. (2014). Applied Biochemistry and Biotechnology, 172, 3476–3487.
McClendon, S. D., Mao, Z., Shin, H. D., Wagschal, K., & Chen, R. R. (2012). Applied Biochemistry and Biotechnology, 167, 395–411.
Lüthi, E., Love, D. R., McAnulty, J., Wallace, C., Caughey, P. A., Saul, D., & Bergquist, P. L. (1990). Applied and Environmental Microbiology, 56, 1017–1024.
Jankowitsch, F., Schwarz, J., Rückert, C., Gust, B., Szczepanowski, R., Blom, J., Pelzer, S., Kalinowski, J., & Mack, M. (2012). Journal of Bacteriology, 194, 6818–6827.
Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M. C., Desiere, F., Bork, P., Delley, M., Pridmore, R. D., & Arigoni, F. (2002). Proceedings of the National Academy of Sciences of the United States of America, 99, 14422–14427.
Acknowledgments
This research is supported by the Science and Engineering Research Council (SERC) of the Agency for Science, Technology and Research (A*STAR) of Singapore (SERC Grant no 1124004027).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Juturu, V., Wu, J.C. Microbial Exo-xylanases: A Mini Review. Appl Biochem Biotechnol 174, 81–92 (2014). https://doi.org/10.1007/s12010-014-1042-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1042-8