Abstract
The special features of cold-adapted lipolytic biocatalysts have made their use possible in several industrial applications. In fact, cold-active enzymes are known to be able to catalyze reactions at low temperatures, avoiding side reactions taking place at higher temperatures and preserving the integrity of products. A lipolytic gene was isolated from the Arctic marine bacterium Rhodococcus sp. AW25M09 and expressed in Escherichia coli as inclusion bodies. The recombinant enzyme (hereafter called RhLip) showed interesting cold-active esterase activity. The refolded purified enzyme displayed optimal activity at 30 °C and was cold-active with retention of 50 % activity at 10 °C. It is worth noting that the optimal pH was 11, and the low relative activity below pH 10 revealed that RhLip was an alkaliphilic esterase. The enzyme was active toward short-chain p-nitrophenyl esters (C2–C6), displaying optimal activity with the butyrate (C4) ester. In addition, the enzyme revealed a good organic solvent and salt tolerance. These features make this an interesting enzyme for exploitation in some industrial applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esterases (EC 3.1.1.1) are hydrolytic enzymes that catalyze the hydrolysis of esters into alcohol and acid. They generally differ from lipases (EC 3.1.1.3) regarding the substrate specificity. By now, it is well recognized that esterases catalyze the hydrolysis and the synthesis of short-chain esters (less than 10 carbon atoms), while lipases act on substrates with long chains of carbon atoms (more than 10) [1]. These enzymes belong to the α/β-hydrolase superfamily and possess a highly conserved catalytic triad formed by Ser, His, and Asp [2]. Lipolytic enzymes are employed in a wide range of industrial applications including the food industry and detergent production as well as biocatalysts for chemical synthesis [3]. In particular, cold-active lipolytic enzymes are extremely appealing for industrial uses. It is generally accepted that cold-active biocatalysts have a more flexible structure [4] compared to mesophilic and thermophilic counterparts, and this high flexibility enables increased complementarity between active site and substrates, resulting in high specific activity at low temperatures [5]. Thus, the use of cold-adapted biocatalysts can minimize undesirable side reactions taking place at higher temperatures and allow reactions involving heat-sensitive substrates [6]. These properties are important for exploitation in the food industry, where the preservation of the nutritional value and flavor of the food is fundamental. Furthermore, thanks to their relatively low thermostability, these enzymes can often easily be inactivated.
Therefore, bioprospecting, looking for cold-active lypolitic enzymes from Arctic regions, has become an active and expanding discipline. In fact, organisms living in an extremely cold habitat have adopted several strategies to survive and thrive in these challenging environments, and these include the expression of enzymes able to efficiently catalyze reactions at temperatures close to 0 °C [7].
In this paper, we present the purification and characterization of a cold-active esterase from the marine psychrophilic actinobacterium Rhodococcus sp. AW25M09. This bacterium was isolated from the intestines/stomach of an Atlantic hagfish (Myxine glutinosa) captured on the cold seafloor during sampling performed in Hadsel Fjord, North Norway. Its genome was recently published [8]. The lip3 gene was selected for its unique amino acid sequence and its homology with other lipases/esterases. The gene was amplified by PCR then cloned and recombinantly expressed in Escherichia coli, and the protein, aggregated as inclusion bodies, was refolded and extensively characterized.
Materials and Methods
Isolation of a Lipolytic Gene
The genome of the cold-adapted Rhodococcus sp. AW25M09 has been sequenced and has been deposited at DDBJ/EMBL/GenBank under accession number CAPS00000000. The genome was analyzed using the Artemis [9] with the aim to identify new genes encoding for esterases and lipases to be expressed in E. coli. Sequence analysis revealed an open reading frame (ORF) of 1,056 bp, the lipR lipolytic gene that encodes for a protein of 352 amino acids.
lipR Gene Cloning
Rhodococcus sp. AW25M09 was grown in Marine 2216 broth (Difco, Sparks, USA) at 20 °C, and the genomic DNA was purified with Sigma’s GenElute Bacterial Genomic Kit according to the manufacturer’s instruction and was used as template for the lipR gene amplification by PCR.
Two primers containing NdeI and NotI restriction sites were designed: 5′-AATACATATGTACCGCAGCAACGACTCCAACG-3′ and 5′-AATAGCGGCCGCGCAGTTGGACGGTGCAGGCACT-3′.
PCR was performed by using Mastercycler personal (Eppendorf, New York, USA). The reaction conditions used were as follows: 1 cycle (94 °C for 3 min), 30 cycles (94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min), and a final cycle of 72 °C for 7 min. The amplified PCR product of 1,056 bp was cloned into pET-22b expression vector previously digested by NdeI and NotI restriction enzymes (New England BioLabs, Ipswich, MA, USA) including an in-frame C-terminal fusion purification 6×His-Tag. E. coli DH5-α competent cells were first transformed through the ligation reaction, and the construct was verified by bidirectional DNA sequencing. The isolated plasmid was then used to transform E. coli strain BL21(DE3) competent cells.
RhLip Recombinant Production in E. coli Cells
E. coli BL21(DE3) carrying pET-22b-lipR vector was grown in a shake flask containing Luria Bertani broth (LB) medium supplemented with 100 μg/mL ampicillin at 37 °C for 16 h. Growing culture was diluted to a cell density of about 0.05 OD600 in a 1-L shake flask containing 200 mL of LB medium supplemented with 100 μg/mL ampicillin. RhLip induction was performed when the culture density reached 0.5–0.6 at OD600 by the addition of filter-sterilized isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. Culture was carried out at constant agitation (220 rpm) at 20 °C for 16 h post-induction. Cells were then harvested by centrifugation at 6,000 rpm for 20 min at 4 °C, divided into 0.5-g aliquots, and frozen at −20 °C.
RhLip Purification and Refolding from Inclusion Bodies
The bacterial pellet (0.5 g) was frozen and thawed twice, and resuspended in 4 mL of Tris-EDTA (TE) buffer (20 mM Tris-HCl pH 8.0, 5 mM EDTA pH 8.0), and 0.6 mg of lysozyme and 0.75 g sucrose were added to the suspension; the suspension was incubated at 37 °C. After 30 min, 4 mL ice-cold TE buffer was added to the suspension and the suspension was incubated for 30 min at 37 °C; then, cells were subjected to sonication. Sonicated cells were centrifuged at 6,700 rcf for 20 min at 4 °C; then, the pellet was washed with 4 mL TE buffer and centrifuged and washed with 2 mL of 20 mM Tris-HCl pH 8.0. The extract was finally centrifuged for 10 min at 4 °C at 10,000 rcf. The pellet was resuspended in 10 mL of 6 M urea, 20 mM Tris-HCl pH 8.0, 15 mM β-mercaptoethanol, and 5 mM EDTA pH 8.0 at 4 °C with gentle shaking for 2 h. The insoluble material was removed by centrifugation at 15,000 rcf for 30 min at 4 °C. Renaturation of the supernatant containing the RhLip was achieved by a 30-fold dilution of the denaturant in 20 mM Tris-HCl pH 8.0, 500 mM arginine, 0.6 mM GSH, and 12 mM GSSG, and the solution was concentrated to about 10 mL using an Amicon ultrafiltration cell (Millipore, Billerica, USA) equipped with a 10-kDa membrane and abundantly dialyzed against 20 mM Tris-HCl pH 8.0. The protein was finally aliquoted and stored at −20 °C in the presence of 20 % glycerol.
Electrophoretic Analysis
Electrophoretic runs were performed with a Mini-Protean II cell (Bio-Rad, Hercules, CA) unit at room temperature. Twelve percent SDS-PAGE was made as described by Laemmli [10]. Marker XL-OPTI Protein 2.8 (ABM, Richmond, BC, Canada) was used as molecular weight standard.
RhLip Determination of pH and Temperature Optima
The esterase activity was monitored at 348 nm (the pH-independent isosbestic point of p-nitrophenol and p-nitrophenoxide ion) with pNP-pentanoate (100 μM) as substrate. The kinetic measurements were performed at 25 °C, and the buffers used were 0.1 M Na-phosphate over the pH range of 7.0–7.5, 0.1 M Tris-HCl over the pH range of 7.5–9.5, and 0.1 M CAPS over the pH range of 9.5–12.0. The assays were carried out in duplicate or triplicate, and the results were the means of two or three independent experiments. Due to the high self-degradation rate of the pNP-esters at high pH values, all further characterizations were performed at pH 10.0. The dependence of activity on temperature was studied over the range of 10–60 °C, with pNP-pentanoate (100 μM) as substrate, in a reaction mixture 0.1 M CAPS pH 10.0, containing 3 % acetonitrile (standard conditions).
RhLip Thermostability
The thermal stability of RhLip was studied over the range of 5–50 °C. Pure enzyme (0.2 mg/mL in a 0.1 M CAPS buffer pH 10.0) was incubated in tubes at different temperatures. Aliquots were withdrawn after 30, 60, 90, and 120 min and assayed at 30 °C in standard condition described above, using pNP-pentanoate as substrate.
RhLip Esterase Activity
The time course of the esterase-catalyzed hydrolysis of pNP-esters was followed by monitoring of p-nitrophenoxide production at 405 nm, in 1-cm path-length cells with a Cary 100 spectrophotometer (Varian, Mulgrave, Australia). Initial rates were calculated by linear least-squares analysis of time courses comprising less than 10 % of the substrate turnover. Assays were performed at 30 °C in a mixture of 0.1 M CAPS pH 10, 3 % acetonitrile, containing pNP-esters (100 μM). Stock solutions of pNP-butanoate (C4), pNP-pentanoate (C5), and pNP-hexanoate (C6) (Sigma-Aldrich, MO, USA) were prepared by dissolving substrates in pure acetonitrile. Assays were performed in duplicate, and the results were the mean of two independent experiments. One unit of enzymatic activity was defined as the amount of the protein releasing 1 μM of p-nitrophenoxide/min from pNP-esters. The absorption coefficient used for p-nitrophenoxide was 19,000 at 30 °C and pH 10.
Kinetic Measurements and Analysis
Initial velocities versus substrate concentration data were fitted to the Michaelis-Menten equation using the software GraphPad Prism version 5.00 (GraphPad software, La Jolla, USA). The concentration of pNP-butanoate and pNP-pentanoate ranged from 0.05 to 2 mM, while pNP-hexanoate were varied from 0.1 to 1 mM. Assays were done in duplicate and the results were the mean of two independent experiments. The kinetic experiments were performed using acetonitrile as solvent.
Effect on Enzymatic Activity by Organic Solvents, Detergents, Metals, and NaCl
Enzyme activity was evaluated in the standard assay (0.1 M CAPS pH 10, 3 % acetonitrile, 30 °C) using pNP-pentanoate as substrate. The activity was measured using an increasing concentration of the solvents such as acetonitrile, dimethyl sulfoxide (DMSO), diethyl ether, and dimethyl formamide (DMFA) from 0 to 20 % (v/v) in the assay mixture. Results were reported as relative activity with respect to the value measured without solvents.
The effect of detergents on enzymatic activity of RhLip was evaluated by incubating 0.1 mg/mL of pure protein in the presence of 5 mM Triton X-100, Tween-20, and Tween-80 at 5 °C for 1 h. The residual enzymatic activity was measured in the standard condition as described above. Preferences for metal cations were analyzed by adding them separately to 0.1 mg/mL of pure protein at a final concentration of 5 mM and equilibrate at 5 °C for 1 h. The residual enzymatic activity was then measured in the standard condition as described above. The effect of NaCl on enzymatic activity was evaluated by increasing the salt concentration in a range of 0–1 M at 30 °C in standard assay conditions.
Modeling of RhLip
The three-dimensional model of RhLip was performed by a comparative modeling strategy using the structure of Candida antarctica lipase A as template (CAL A, PDB code 3GUU). The sequence alignment was calculated by the CLUSTALW program [11]. The MODELLER 9v11 program [12] was used to build 100 full atom models of RhLip structure setting 4.0 Å as root mean square deviation (RMSD) among initial models and by full model optimization. Structure validation was carried out using the pictorial database PDBsum [13]. The structure of the generated model was uploaded to the PDBsum server, and structural analyses, including PROCHECK plots [14], were generated. Moreover, the Z score of the RhLip model and C. antarctica lipase A (CAL A) structure was calculated by the web server WhatIf [15]. The Z score expresses how well the backbone conformations of all residues correspond to the known allowed areas in the Ramachandran plot. Furthermore, the solvent-accessible surface areas (SASAs) of RhLip model and CAL A structure were calculated using the POPS algorithm [16].
Molecular Dynamics
Molecular dynamics (MD) simulations were performed with GROMACS software package (v4.5.5) [17]. The model was inserted in a dodecahedron box filled with SPC216 water molecules using GROMOS43a1 all-atom force field. Simulations were carried out at different pH values. Imposing different protonation states according to the number of titratable groups reproduced neutral and basic pH conditions. The simulations were carried out by adding 26 sodium ions to have a value of zero for the net electrostatic charge of the system. The systems were subjected to several cycles of energy minimizations and position restraints to equilibrate the protein and the water molecules around the protein. Particle mesh Ewald (PME) algorithm was used for the electrostatic interactions with the cutoff of 1 nm. The time step was 2 fs, and the temperature was kept constant at 300 K using a modified Berendsen thermostat with a time constant of 0.1 ps. The simulation time for each dynamic was 10 ns. GROMACS routines were used to analyze the trajectories in terms of RMSD, RMSF, and gyration radius.
Results and Discussion
Purification and Refolding from Inclusion Bodies
In this work, we present a biochemical characterization of a new alkaliphilic esterase from Rhodococcus sp. The lip3 gene was cloned into a pET-22b expression vector, and the construct was transferred into E. coli BL21(DE3) calcium competent cells. Several expression conditions were investigated, but IPTG induction of E. coli cells resulted in the accumulation of recombinant RhLip as inclusion bodies (IB). The induction at 20 °C was effective in producing the highest amount of RhLip compared to the other contaminant proteins. The IBs were purified and the protein refolded as described in the “Materials and Methods” section. According to the structural analysis, two disulfide bridges were detected, and the refolding protocol was optimized by adding GSSG (oxidized glutathione) and GSH (reduced glutathione) to the refolding solution. Using this protocol, about 8 mg of pure enzyme was obtained from 0.5 g of E. coli cell pellet.
Purity of the protein preparation was evaluated by SDS-PAGE analysis. As shown in Fig. 1, a single band was observed with an apparent mass of about 38 kDa.
RhLip Functional Characterization
The dependence of RhLip activity in the function of pH was estimated using pNP-pentanoate as substrate (Fig. 2a). The absorption of p-nitrophenol changes at different pH values because of variations in equilibrium between p-nitrophenol and p-nitrophenoxide. In this work, we monitored the release of p-nitrophenol at 348 nm, that is, the isosbestic point of p-nitrophenol and p-nitrophenoxide. The maximum activity of RhLip was recorded at pH 11 in 0.1 M CAPS, and the relatively low activity between pH 7 and pH 9 suggests that the enzyme was a highly alkaliphilic esterase. Other alkaliphilic esterases have been identified from genomic and metagenomic sources so far [18] including two other cold-active esterases with optimal pH >10 [19–21]. The relationship between RhLip activity and temperature was evaluated in the range of 10–60 °C using pNP-pentanoate as substrate (Fig. 2b). The apparent maximal activity was recorded at 30 °C, and the activity detected at 10 °C remained approximately the 50 % of the maximum activity.
a Effect of pH on the esterase activity. b Effect of temperature on the esterase activity. c Thermostability of RhLip, at various temperatures, such as 5 °C (black circles), 10 °C (black squares), 20 °C (black up-pointing triangles), 30 °C (black down-pointing triangles), 40 °C (black diamonds), and 50 °C (white circles). The enzyme was incubated in 0.1 M CAPS pH 10.0 at the indicated temperatures and times. The residual activity was measured at 30 °C using pNP-pentanoate as substrate
The RhLip thermal stability was evaluated in the range of 5–50 °C. Enzyme samples were incubated at any given temperature for different lengths of time, and the residual activity was recorded at 30 °C. This study demonstrated that RhLip presented a typical behavior as other psychrophilic enzymes, showing a low kinetic stability at temperature higher than 30 °C [22, 23].
In fact, as shown in Fig. 2c, we observed a very low decrease in activity after 2 h of incubation at 5, 10, and 20 °C, while when the temperature increased up to 30 and 40 °C, we noted a significant decrease in activity. After 2 h of incubation at 50 °C, only 20 % of enzymatic activity was still recorded.
Kinetics Studies
We investigated the activity of RhLip toward different synthetic substrates by using several pNP-esters with different acyl chain lengths. Activity was assessed in the presence of 0.1 M CAPS pH 10 instead of the optimum pH buffer (0.1 M CAPS pH 11) due to the instability of the various substrates at alkaline pH value. All the characterization was also performed at 30 °C and in the presence of 3 % acetonitrile. Concerning the affinity values, we observed that Km values decrease when acyl chain length increases, and this suggests that RhLip possesses a high affinity with longer aliphatic chain substrates. Instead, the Kcat and Kcat/Km values show the opposite behavior: in our standard conditions, the enzyme displays the highest Kcat and Kcat/Km on pNP-butanoate with values of 1.63 s−1 and 2.16 s−1 mM−1, respectively (Table 1). The biochemical characterization of the recombinant enzyme revealed pNP-butanoate (C4) as the preferred substrate, and the hydrolytic activity significantly decreased as the chain length increased above C8, with very little activity toward pNP-tetradecanoate (C14) (data not shown), suggesting that the enzyme was an esterase and not a lipase.
Effect on Enzymatic Activity by Organic Solvents, Detergents, Metals, and NaCl
The effect of the presence of water-miscible solvents on RhLip enzymatic activity on pNP-pentanoate at 30 °C in 0.1 M CAPS pH 10.0 was investigated. For all the solvents tested, except DMFA, we observed a similar behavior as shown in Fig. 3a. The increasing concentration of organic solvent in the assay mixture enlarged RhLip catalytic activity up to a critical concentration, and further addition of solvents led to a gradual protein inactivation. These results are coherent to what were observed for other esterases belonging to hormone-sensitive lipase protein family, as demonstrated by Mandrich and coworkers [24].
a Effect of organic solvents on the esterase activity. Enzyme activity was evaluated in the presence of increasing concentration of acetonitrile (black squares), diethyl ether (black up-pointing triangles), DMSO (black down-pointing triangles), and DMFA (black diamonds). The relative activity was measured at 30 °C in 0.1 M CAPS pH 10.0 using pNP-pentanoate as substrate. b Effect of NaCl on the esterase activity. Enzyme activity was evaluated in the presence of increasing concentrations of NaCl. The relative activity was measured at 30 °C in 0.1 M CAPS pH 10.0 with pNP-pentanoate as substrate
We reported the best enzymatic activation (more than 200 % of the relative activity) in the presence of 5 % diethyl ether. This behavior has been explained in literature as the ability of the organic solvents to stabilize ionic intermediates in the case of aprotic solvents [25]. RhLip was incubated in the presence of various denaturants or metal ions for 1 h, and the residual activity was measured using pNP-pentanoate as substrate at 30 °C. The resulting values are summarized in Table 2, which demonstrated that few tested compounds had an inhibitory effect on RhLip activity, although at various extents. The strongest inhibitory effect was observed in the presence of Ca2+ ions suggesting the absence of a Ca2+-binding motif sequence. A similar effect of enzymatic inactivation was detected in the presence of Tween-80. On the contrary, a strong activation was observed by incubating the RhLip in the presence of Tween-20 and EDTA. Tween 20 was more easily hydrolysed than Tween-80, indicating that the chain length may play an important role on substrate specificity [26].
The effect of the presence of NaCl on RhLip enzymatic activity was evaluated on pNP-pentanoate in 0.1 M CAPS pH 10.0 at 30 °C. We observed an improved activity with the highest concentration of 1 M NaCl (Fig. 3b). A similar behavior may be explained as the ability of salt to enhance the hydrophobic interaction between enzyme and substrate [27].
RhLip Model
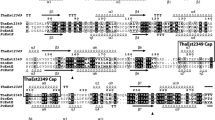
The three-dimensional modeling of RhLip was performed by a homology modeling approach using the C. antarctica lipase A (PDB ID: 3GUU) structure as template. The CAL A structure was chosen due to sequence identity of 30 % between the RhLip sequence and the template. The multiple alignments between RhLip and the best scoring templates are shown in Fig. 4. Starting from the alignment of RhLip sequence with the reference structure, a set of 100 all-atom models was generated. The best model (Fig. 5a) was selected in terms of energetic and stereochemical quality. In details, it had 89.2 % of residues in most favored regions of the Ramachandran plot according to PROCHECK program and a WhatIf Z score of 0.050. These values, compared with those of the template structure, i.e., Z score = −1.181 and 88.6 % of residues in most favored regions of the Ramachandran plot, indicated that the quality of the model was really high. The RhLip model showed an alpha-beta structure characterized by 13 α-helices (H3, H5–H16), three 310 helices (H1, H2, and H4), and nine beta-strands (B1–B9), corresponding to 39.6 and 14.5 % of the sequence, respectively. RhLip secondary structures are represented in Fig. 6. RhLip was stabilized by two disulfide bonds as well as the template structure (Cys47–Cys224, Cys299–Cys351). The SASAs of RhLip model and CAL A structure calculated by POPS algorithm showed a hydrophobic area of 61.23 % and a hydrophilic area of 38.77 % for RhLip, while a hydrophobic area of 49.83 % and a hydrophilic area of 50.17 % for CAL A. Hydrophobicity of the surface exposed to the solvent is an important feature of lipases and esterases because these classes of enzymes work at the interface between a polar and an apolar phase. Structural analysis was carried out by analyzing the most conserved residues of RhLip multiple alignment (Fig. 7). Ser137, Asp283, and His323 form the catalytic triad of RhLip (Fig. 5b). These three amino acids are located in three different loops connecting three strands to three α-helices. In particular, Ser137 is within a conserved motif (GTSXGG), and the presence of the glycines gives some flexibility at this area. The side chain atoms NE2 and ND1 of H323 make two h-bonds (2.69 and 3.19 Å) with the side chain atom OG of S137 and the side chain atom OD2 of D283. These bonds hold together the members of the catalytic triad. Moreover, carbonyl oxygen of His323 makes a hydrogen bond (2.58 Å) with the hydroxyl group of Tyr136, another conserved residue located in a strand of beta-sheet B. This bond further helps this β-strand to keep close to the catalytic triad.
Molecular Dynamics
Since the three-dimensional model accurately described the structural organization of this protein, we tried to have a more dynamic view of its structure. We have subjected the best RhLip model to molecular dynamics simulations in order to evaluate its stability. These studies were made at neutral as well as at alkaline pH because the sequence of this protein shows many positively and negatively charged residues sensitive to pH changes. The RhLip model reached a stable equilibrated state after 6 ns simulation at both pH levels; in fact, the related RMSD values (Fig. 7a), computed by superposing the various structures obtained during the simulations and the initial structure at time zero, were almost constant in the remaining simulation time. However, we noted that the fluctuations observed at alkaline pH levels in the first 3–4 ns were slightly more severe in comparison with the simulation computed at neutral pH (Fig. 7b). This behavior highlighted a different way for the protein to reach an equilibrium state compared to the results at neutral pH. Then, in order to compare the overall size of the two systems at different pH levels, we computed the gyration radius concerning all atoms of 346 residues (Fig. 8). The gyration radius trend showed a quite similar evolution in the simulations, and practically, no variation in molecule compactness was observed.
Finally, the superposition of the RMS fluctuations at neutral and alkaline pH revealed that the residues with a high degree of flexibility fell in the loop region. This was due to the high presence of charged residues in these regions that, at neutral and alkaline pH, behaved differently. This suggests the important functional role played by these flexible loops in RhLip at structural as well as at functional levels.
Conclusion
The present study shed light on a new lypolitic cold-adapted enzyme, which was demonstrated to possess interesting features and various potential applications, as additive for detergent production and biocatalyst for regio- and stereoselective reactions in chemical synthesis [6]. The recombinant enzyme was successfully purified from the inclusion body and characterized. RhLip was revealed to be an extremely alkaliphilic and cold-adapted esterase. As other psychrophilic enzymes, RhLip showed a low thermostability at temperature higher than 30 °C, and this heat lability can be exploited in several applications, especially in the food industry. In this case, an industrial process catalyzed by an esterase can be easily stopped by a little increase in temperature, preserving food integrity and flavor.
The enhanced catalytic activity in tested organic solvents could make it useful for some industrial purposes such as production of chemicals, biopolymers, and fuels [28]. RhLip also showed an improved activity in the presence of 1 M NaCl. This evidence could be useful for industrial application of food processing in the presence of a high concentration of salt. The analysis of the RhLip model revealed some structural features of the catalytic site of this enzyme, showing the three-dimensional arrangement and the interactions between residues of the catalytic triad. Moreover, results of MD simulations highlighted that the protein seems to be quite stable at neutral as well as at alkaline pH. Further developments of this study could be directed to identify potential targets for site-directed mutagenesis to improve protein stability and catalytic activity.
References
Rhee, S. K., Liu, X., Wu, L., Chong, S. C., Wan, X., & Zhou, J. (2004). Applied and Environmental Microbiology, 70, 4303–4317.
Gupta, R., Gupta, N., & Rathi, P. (2004). Applied Microbiology and Biotechnology, 64, 763–781.
Aurilia, V., Parracino, A., & D’Auria, S. (2008). Microbial carbohydrate esterases in cold adapted environments. Gene, 410, 234–240.
Paredes, D. I., Watters, K., Pitman, D. J., Bystroff, C., & Dordick, J. S. (2011). BMC Structural Biology, 11, 42.
Cavicchioli, R., Charlton, T., Ertan, H., Mohd Omar, S., Siddiqui, K. S., & Williams, T. J. (2011). Microbial Biotechnology, 4, 449–460.
Jeon, J. H., Kim, J. T., Kang, S. G., Lee, J. H., & Kim, S. J. (2009). Marine Biotechnology (NY), 11, 307–316.
de Pascale, D., De Santi, C., Fu, J., & Landfald, B. (2012). Marine Genomics, 8, 15–22.
Hjerde, E., Pierechod, M. M., Williamson, A. K., et al. (2013). Genome Announcements, 7(1). e0005513.
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Bioinformatics, 16, 944–945.
Laemmli, U. K. (1970). Nature, 227, 680–685.
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). Nucleic Acids Research, 22, 4673–4680.
Sali, A., & Blundell, T. L. (1993). Journal of Molecular Biology, 234, 779–815.
Laskowski, R. A. (2001). Nucleic Acids Research, 29, 221–222.
Laskowski, R. A., MacArthur, M., Moss, D. S., & Thornton, J. M. (1993). Journal Applied Crystallography, 26, 283–291.
Vriend, G. (1990). Journal of Molecular Graphics, 8(29), 52–56.
Cavallo, L., Kleinjung, J., & Fraternali, F. (2003). Nucleic Acids Research, 31, 3364–3366.
Hess, B., Kutzner, C., van der Spoel, D., & Lindhal, E. (2008). Journal of Chemical Theory and Computation, 4, 435–447.
Kademi, A., Ait-Abdelkader, N., Fakhreddine, L., & Baratti, J. (2000). Applied Microbiology and Biotechnology, 54, 173–179.
Park, H. J., Jeon, J. H., Kang, S. G., Lee, J. H., Lee, S. A., & Kim, H. K. (2007). Protein Expression and Purification, 52, 340–347.
Hu, X. P., Heath, C., Taylor, M. P., Tuffin, M., & Cowan, D. (2012). Extremophiles : Life Under Extreme Conditions, 16, 79–86.
Kim, Y. H., Kwon, E. J., Kim, S. K., Jeong, Y. S., Kim, J., Yun, H. D., et al. (2010). Biochemical and Biophysical Research Communications, 393, 45–49.
De Santi, C., Tutino, M. L., Mandrich, L., Giuliani, M., Parrilli, E., Del Vecchio, P., et al. (2010). Biochimie, 92, 949–957.
Wang, Q., Hou, Y., Ding, Y., & Yan, P. (2012). Molecular Biology Reports, 39, 9233–9238.
Mandrich, L., De Santi, C., de Pascale, D., & Manco, G. (2012). Journal of Molecular Catalysis B Enzymatic, 83, 46–52.
Peters, K. S. (2007). Chemical Reviews, 107, 859–873.
Gandolfi, R., Marinelli, F., Lazzarini, A., & Molinari, F. (2000). Journal of Applied Microbiology, 89, 870–875.
Morimoto, K., Furuta, E., Hashimoto, H., & Inouye, K. (2006). Journal of Biochemistry, 139, 1065–1071.
Egorova, K., & Antranikian, G. (2005). Current Opinion in Microbiology, 8, 649–655.
Acknowledgments
This work was supported by P.N.R.A. (Italian National Antarctic Research Programme) 2009–2011. We also thank the Yggdrasil application grants 2011–2012 and 2012–2013, funded by the National Research Council of Norway, for supporting the research activities of Dr. Concetta De Santi.
Author information
Authors and Affiliations
Corresponding author
Additional information
Concetta De Santi and Pietro Tedesco contributed equally to this work.
Rights and permissions
About this article
Cite this article
De Santi, C., Tedesco, P., Ambrosino, L. et al. A New Alkaliphilic Cold-Active Esterase from the Psychrophilic Marine Bacterium Rhodococcus sp.: Functional and Structural Studies and Biotechnological Potential. Appl Biochem Biotechnol 172, 3054–3068 (2014). https://doi.org/10.1007/s12010-013-0713-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0713-1