Abstract
A tetrahedral Zn(II) complex with the second generation fluoroquinolone, norfloxacin, was prepared and characterized (NOR–Zn complex, NZC). The antibacterial efficiency of the NZC was tested on two Gram-positive and four Gram-negative bacteria by minimum inhibitory concentration method. The cytotoxic potential of NZC on MDA (human breast adenocarcinoma), Caco-2 (human colon adenocarcinoma), and Hela (human cervix carcinoma) cell lines was studied. The DNA interaction property of the NZC has been investigated using UV–vis, fluorescence, Fourier transform infrared, as well as cyclic voltammetry methods. Intrinsic binding constant (K b ), thermodynamic, and other spectroscopic and voltammetric data indicate that the NZC has more affinity for DNA than for norfloxacin and interacted with DNA via two modes: electrostatic and outside hydrogen binding. The proposed DNA binding mode supports the large enhancement in the cytotoxicity and antibacterial activity of NZC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is well known that the fluoroquinolones are broad-spectrum antibiotics and widely used in the anti-infective chemotherapy field [1]. These microbicidal effects are largely attributed to inhibition of DNA topoisomerases II (DNA gyrase) and IV of prokaryotes, while these influences on mammalian DNA enzymes are several orders of magnitude weaker [2, 3]. However, due to growing evidences of resistant bacterial strains associated to the adverse safety profile of many of these agents [4, 5], many groups have been working on the development of new safer derivates that would induce less side effects and could be used against a broader spectrum of infections [6]. In this way, the researchers found out that transition metal complexes of fluoroquinolones have a remarkable in vitro and in vivo biological activity against Gram-positive and Gram-negative bacteria and cytotoxic effects on mammalian cancer cells [7–10]. In addition, accumulating evidences demonstrated that fluoroquinolones have anti-HIV activity [11], and some of them, especially ciprofloxacin, gatifloxacin, and norfloxacin, are able to modulate lipopolysaccharide-induced proinflammatory cytokine production [12]. Therefore, the evaluation of antimicrobial activity and cytotoxicity, and the introduction of the molecular mechanism of action of metal–fluoroquinolone complexes for the development of antitumor and antibacterial agents are very important [13, 14]. It is well known that the transition metal complexes of fluoroquinolones have more interaction with DNA because of multimodes of interaction such as intercalation, groove bindings, electrostatic forces, and outside hydrogen binding. This effect is a strong motivating force for understanding the basics of these interaction modes and also the development of metal complexes as new anti-inflammatory, antifungal, antibacterial, and anticancer compounds [15–18]. Norfloxacin, (1-ethyl-6-fluoro-1,4-dihydro-4-oxo-7-(1-piperazinyl)-3-quinolinecarboxylic acid (NOR), is a second generation of fluoroquinolones and used in treating bacterial infections of the urinary tract, respiratory tract, and skin. It is also known to be effective in treating diarrhea and can, in addition, treat conjunctivitis when it is administered in the form of eyedrops [19, 20]. Norfloxacin is not, however, effective against infections involving anaerobic bacteria (e.g., yeast, athlete's foot). Like other piperazine-substituted fluoroquinolones (e.g., ciprofloxacin and ofloxacin), norfloxacin is a specific inhibitor of DNA gyrase, a bacterial type II topoisomerase, which unwinds the super coiled DNA helix prior to replication and transcription [21, 22]. It is known that the drug does not inhibit the gyrase but complexes to DNA in a highly cooperative mode of binding [23–25]. It has been suggested that the action of DNA gyrase forms a unique binding pocket on DNA, which can accommodate a dimer of NOR molecules with a sandwich-type arrangement of the chromophores and stabilized by π–π stacking [25, 26]. Alkaysi and et al. reported a spectrophotometric study on the interaction between NOR and Al3+, Mg2+, and Ca2+ ions in pH levels of 3.6 and 8.8. They also reported that the microbiological activity of norfloxacin in the presence of these metal ions was decreased [27]. Refat synthesized metal complexes of NOR with Ag+, Cu2+, and Au3+ ions with molecular structures [Ag2(NOR)2](NO3)2, [Cu(NOR)2(H2O)2]SO4·5H2O, and [Au(NOR)2(H2O)2]Cl3. He shows that NOR is coordinated to Ag+ and Au3+ ions as a neutral monodentate chelating through the N atom of the piperidyl ring, but the Cu2+ is coordinated through the carbonyl oxygen atom (quinolone group) and the oxygen atom of the carboxylic group and has moderate activity against the Gram-positive and Gram-negative bacteria as well as against fungi [28]. Shaikh and coworkers synthesized a Bi–NOR complex with the structural formula of [Bi(NOR)4·2H2O] and reported a remarkable enhancement in the antibacterial activity of the complex compared with NOR [29]. According to our best library knowledge, few reports exist for the synthesis of transition metal ion–NOR complexes [30, 31], and scanty reports exist for the evaluation of norfloxacin–DNA interaction [32, 33]. In the present article, we report the synthesis and evaluation of in vitro cytotoxicity and antibacterial activity of the NOR–Zn complex (NZC). The synthesized complex was checked for its DNA interacting behavior.
Material and Methods
Reagents
Double-strand calf thymus deoxyribonucleic acid (ds-CT-DNA) and norfloxacin were purchased from Sigma (Sigma-Aldrich, Bellefonte, PA) and used as received. Stock solutions of DNA were prepared by dissolving an appropriate amount of DNA in double-distilled water and stored at 4 °C in a refrigerator. The concentration of the stock solution of DNA was determined by ultraviolet absorbance at 260 nm using the molar extinction coefficient = 6,600 M−1 cm−1 [34]. Generally, the stock solution of DNA is fresh for the test; the storage time is not longer than 96 h. A solution of CT-DNA gave a ratio of UV absorbance at 260 and 280 nm more than 1.8, indicating that DNA was sufficiently free from protein. Zn(NO3)2, methanol (MeOH), and other reagents with high analytical grade were purchased from Merck. All spectroscopic and voltammetric measurements were performed in 0.01 M Tris–HCl buffer (pH = 7.3). The cancer cell lines including: MDA (human breast adenocarcinoma), Caco-2 (human colon adenocarcinoma), and Hela (human cervix carcinoma) were purchased from the National Cell Bank, Pasteur Institute of Iran. The antibacterial activities of NZC tested against a panel of bacteria strains including: two Gram-positive bacteria such as: Enterococcus faecalis (ATCC 29122) and Listeria monocytogenes (ATCC 15313), and four Gram-negative bacteria such as: Proteus mirabilis (ATCC 9240), Pseudomonas aeruginosa (ATCC 27853), Shigella dysenteriae (ATCC 29026), and Escherichia coli (ATCC 11522).
Instruments
An HP Agilent (8453) UV–vis spectrophotometer equipped with a peltier (Agilent 89090A) was used for the study of spectrophotometric titrations. All fluorescence measurements were carried out with a Beckman spectrofluorometer (LS45). The maximum excitation wavelength used was 350 nm. The pH values of the solutions were adjusted, employing a Metrohm pH meter model 827 using a combined glass electrode. The cyclic voltammetry measurements were performed by a voltammeter, Metrohm VA 797 computrace (Metrohm), with a three-electrode system: a hanging mercury drop electrode (HMDE) as working electrode, silver–silver chloride (Ag/AgCl) as reference electrode, and a platinum wire as counter electrode. For all types of voltammetric measurements, the supporting electrolyte (0.01 M Tris–HCl buffer solution, pH 7.3) was placed in a voltammetric cell of volume 10 mL and dehydrated via purging with pure N2 gas for 2.0 min. The cyclic voltammetric (CV) measurements were carried out by keeping both concentration of the NZC and the total volume of solution constant, while the ds-DNA concentration varied. The Fourier transform infrared (FT-IR) measurement was carried out using Shimadzu (IR Prestige-21) equipped with a KBr beam splitter. The elemental analysis was performed using Heraeus CHN elemental analyzer. Thermogravimetric studies were performed for the complex by the TG-DTA STA model 503.
Synthesis of the NOR–Zn Complex
The synthesis of the NZC was performed like the work of Patel [10] with some modifications. Briefly, to 5.0 mL of warm methanol, 0.3 mmol (0.01 g) of NOR and 0.3 mmol of Zn(NO3)2·4H2O (0.082 g) were added. The solution was stirred for 24 h and stored overnight under N2 atmosphere. After 24 h, the methanol evaporated, and the resulting solid compound washed with 1.0 mL of very cold methanol and dried at room temperature. The structure of the complex was evaluated by FT-IR, thermogravimetry, and elemental analysis.
The Procedures of Cytotoxicity and Antimicrobial Assessment
Cell Culture
Cell lines were seeded in 75-cm2 tissue culture flasks and maintained in RPMI 1640 and Dulbecco's MEM supplemented with 10 % heat-inactivated fetal bovine serum, 50 U mL−1 penicillin, and 50 μg mL−1 streptomycin. The medium was renewed every 2 days, and the cell cultures were incubated at 37 °C in a humidified atmosphere (95 % air and 5 % CO2).
LDH Cytotoxicity Assay
The cytotoxicity effect of the aqueous extract of NOR and NZC was also examined using lactate dehydrogenase (LDH) assay as described by Linford [35] with some modifications. Briefly, the cell lines were separately plated at a density of 5 × 103 per well in 96-well micro plates with RPMI 1640-DMEM (Dulbecco's modified Eagle medium) containing 10 % FBS and allowed to incubate overnight. After 24 h of early cell culture, the fresh medium with NOR and NZC at concentrations of (0 to 800 μg mL−1) was renewed. At 72 h of treatments, the plates were centrifuged and 100 μL of the media from each plate transferred to a new 96-well micro plate. Thereafter, 100 μL of LDH assay mixture was added to each plate and incubated at 37 °C for 30 min. A group of plates was treated with 1 % Triton X-100 solution for 45 min to maximum LDH release. The LDH release was estimated using a micro plate reader at 492 nm according to the manufacturer's instructions. Tests were performed in triplicate. The mean cell viability was expressed as a percentage of the control.
Antibacterial Screening of NOR and NZC
The susceptibility of certain strains of bacterium towards NOR and NZC was judged by measuring the minimum inhibition concentration (MIC). The compounds were first dissolved in a binary mixture of H2O–DMSO (90:10 v/v %) and then diluted to the highest concentration (100 μg mL−1) to be tested, and then serial dilutions were made in a concentration range from 0 to 64 μg mL−1. The target microorganisms were cultured in Mueller–Hinton broth for 24 h. After that, the suspensions were diluted with 0.5 McFarland standard turbidity and diluted again (1/1,000 ratio) by Mueller–Hinton broth. To 96-well micro plates, 100 μL of Mueller–Hinton broth containing diluted bacteria (1.5 × 105 CFU mL−1) and 100 μL of aliquot from the solutions of the compounds were added. A positive control (containing inoculums but no tested compounds) and negative control (containing tested compounds but no inoculums) were included on each micro plate. The contents of the wells were mixed, and the micro plates were incubated at 37 °C for 24 h under microaerophilic conditions. The MIC was defined as the lowest concentration of the compounds to inhibit the growth of microorganisms.
The Procedures of DNA Binding Assessment
Spectrophotometric Titration of NZC with DNA
The spectrophotometric titrations of NZC with CT-DNA were carried out like our pervious works [36, 37] as follows: the addition of DNA solution (0–5 × 10−5 M) to 2.0 mL solution of NZC (5 × 10−5 M) was carried out using a precalibrated micro pipette. The spectrum of solution was recorded after each addition. The addition of DNA to solution was continued until the desired r i = [DNA]/[NZC] = 1.0 was achieved. At this given r i , no spectrum change was observed. The binding constant (K b ) was measured using Eq. (1):
where ε a is the extinction coefficient of NZC at maximum wavelength after each addition of DNA, ε f is the extinction coefficient of the NZC without any addition of DNA (277 nm), ε b is the extinction coefficient of the NZC when fully bounded to DNA (after that by the addition of DNA, no change in absorbance should be observed). The ε f was measured from the slope of the calibration curve of free NZC, using the Beer–Lambert law. ε b was determined from the absorbance of NZC in maximum wavelength (272 nm), when the addition of DNA did not result in further changes in the absorption spectrum of NZC. Finally, ε a was determined as the ratio between the measured absorbance after each addition of DNA and NZC concentration. The experimental data of [DNA]/(ε a − ε f ) versus [DNA] gives K b as the ratio of the slope to intercept.
Fluorescence Study of Interaction of NZC with DNA
Emission spectra were recorded between 400 and 600 nm with an excitation wavelength of 350 nm, corresponding to the λ max of NZC in the Tris–HCl buffer (0.01 M and pH = 7.3). Increasing amounts of DNA were added directly into the cell containing the NZC solution (1.0 × 10−5 M). The concentration ranged from 0 to 1.6 × 10−5 M for DNA. The solution in the cuvette was thoroughly mixed before each scan. All measurements were performed at 25 °C.
FT-IR Studies for Interaction of DNA with NZC
The spectra of the DNA–NZC solutions were acquired using a cell assembled with AgCl windows. Spectra were collected after interaction of different amounts of DNA with NZC solution (5 × 10−3 M) (Ri′ = 1/r i = [NZC]/[DNA] = 50, 5). The spectra were measured over the spectral range 600–4,000 cm−1 with a nominal resolution of 2 cm−1 and a minimum of 50 scans. The blank windows and water were removed by BGK mode of instrument according to our previous work [38]. The spectra were smoothed with Golly procedure.
Voltammetric Measurements
The cyclic voltammetric measurements were carried out by keeping constant both concentrations of the NZC (1 × 10−5 M) and total volume of solution, while the ds-DNA concentration was varied (0 to 2 × 10−4 M). Subsequently, a single-sweep cyclic voltammetric experiment was performed at a given scan rate 50 mV s−1, with an initial potential of 0.0 V and a vertex potential of −1.2 V.
Results and Discussion
The Proposed Structure of NZC
Chemically, NOR is an amphoteric molecule possessing two major ionizable groups, a basic piperazine amine, and acidic carboxyl having pKa = 8 and 6.3, respectively. The structure of NOR with a carbonyl adjacent to the carboxyl group allows for proposing the formation of a complex between transition metal ions and the drug. The possibility of interactions was studied using FT-IR, TG-DTA, and elemental analysis. The FT-IR spectra were recorded on the AgCl window, in the region 4,000–400 cm−1 which was calibrated by polystyrene. There is not any decomposition of the sample due to the effect of AgCl. The comparison between the NOR wave numbers observed in the IR spectra of the complex and free NOR helped in defining the structure of the metal complex. The general mechanism of the interaction between NOR and the transition metal cations is chelation between the metal and the ketonic (4-oxo) and carboxylic groups [27, 39]; therefore, we first focus our attention on these groups' vibrations. In the IR spectra of pure NOR, the absorptions at 1,638 and 1,732 cm−1 in the spectrum are attributed to the absorption of the pyridone stretch ν (C=O)p and carboxylic stretch ν (C=O)carb, respectively. While in NZC, the intensities of these bands were remarkably reduced and appears in the 1,624 and 1,726 cm−1. This shift in band towards lower energy suggests that coordination occurs through the pyridone and carboxylic oxygen atoms [40]. In the case of NOR, strong absorption bands at 1,642 and 1,336 cm−1 could be assigned for ν (COO)assy and ν (COO)sym vibration, respectively, while for NZC, these bands were observed in the range 1,516 and 1,301 cm−1 [40, 41]. The carboxylate group can bind to metal ions in a monodentate, bidentate, or bridging manner. The frequency difference [\( \varDelta \nu =\nu {{\left( {\mathrm{COO}} \right)}_{\mathrm{assy}}}-\nu {{\left( {\mathrm{COO}} \right)}_{\mathrm{sym}}} \)] can be used as an indication of the binding mode of the carboxylate [42]. If Δν is greater than 200 cm−1, this group is probably bound in amono dentate way. The Δ value is greater than 200 cm−1, which indicates the monodentate coordination mode of the carboxylate group of the ligand [43]. The absorption band in the 1,638-cm−1 region attributed to the ketone group in the free ligand spectra is shifted to 1,624 cm−1 in the complex which is good indication that this group is coordinated to the Zn(II) ion [40]. Also, the band in the 1,732 cm−1 shifted to the 1,726 cm−1 and shows that the –COOH bonded to the Zn(II) ion. In the 3,400-cm−1 region, no broad absorption band is observed. Moreover, the infrared spectra of NZC present two weak intensity bands around 2,937 and 2,818 cm−1 assigned to the vibration of the quaternized nitrogen of the piperazinyl group which indicates that the zwitterionic form of norfloxacin is involved in the coordination to the metal ion [7]. The elemental analysis data were as follows: calc. (in percent): C, 54.6; H, 5.1; N, 11.9; Zn, 9.2; found (in percent): C, 54.2; H, 4.98; N, 12; Zn, 9, what suggested a proportion of 1:2 of Zn-(NOR)2 with the molecular formula of [Zn(NOR)2](NO3)2. Already the same structure was reported for other metal complexes of NOR [44, 45]. The water content in the complex was determined by thermal analysis, the apparatus operating at a heating rate of 10 °C min−1 in the range of 30–700 °C in N2 in order to establish their compositional differences as well as to ascertain the nature of associated water molecules. The thermogravimetric results agreed with the proposed formulas. The t.g. curve for the proposed complex shows two weight losses. The first, in the 210–280 °C range, and the second inflections, near 400 and 480 °C, which are attributed to complex thermal decomposition. Based upon analysis of the results obtained, it was possible to identify that in the Zn(II) complex with the NOR, the coordination of the NOR to the metal occurs in the most usual way, i.e., via the ketonic and carboxylic oxygen atoms, and forms a tetrahedral complex containing two stable six-membered ring chelates with a neutral zwitterionic form. Figure 1 shows the proposed structure for the proposed complex.
Cytotoxicity Effect of the NZC on Cancer Cell Lines
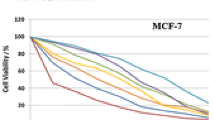
In the cytotoxicity assay, we examined the potential anticancer activity of NZC. For this purpose, we studied the in vitro cytotoxic effect of NZC against three well-known cell lines including Hela, Caco-2, and MDA by LDH assay method. On the basis of obtained results, after 72 h of incubation, NZC significantly reduced the viability of three cell lines at different concentrations compared with the control group (NOR), and this effect was stronger as time increased. On the other hand, the studied cytotoxicity treatments resulted in a dose- and time-dependent manner. Furthermore, the observed results from effective concentrations of NZC were accompanied with differences for various cancer cell lines. In fact, and based on the data in Fig. 2, the cytotoxicity effect of NZC on the Hela cell line was considerably lower than those on the other cell lines. The IC50 (in microgram per milliliter) values of NZC and NOR on cell lines were shown in Table 1.
Antibacterial Screening of NZC
The susceptibility of certain strains of bacterium towards NZC and NOR was judged by measuring the MIC. A stock solution of 100 μg mL−1 was prepared by dissolving an appropriate amount of NZC in a binary mixture of H2O–DMSO (90:10 v/v %) solution. Table 2 shows in vitro antibacterial activity data of the NZC and NOR. It is clear from the data that NZC is more active compared to NOR for E. coli, E. faecalis, and S. dysenteriae bacterial species. The observed higher antibacterial activities of the NZC can be explained on the basis of the strong interaction of NZC with DNA. The permeability of NZC to the lipid membrane of bacteria can be explained by Tweedy's chelation theory [46]. The lipid membrane that surrounds the cell favors the passage of only lipid-soluble materials due to which lipophilicity is an important factor which controls the antibacterial activity. On chelation, the polarity of the Zn(II) ion will be reduced to a greater extent due to the overlap of the ligand orbitals and partial sharing of the positive charge of the Zn(II) ion with the donor groups. Further, it increases the delocalization of π electrons over the whole chelate ring and hence enhances the liposolubility of the NZC. This increased liposolubility enhances the penetration of the NZC into the lipid membrane.
The Study Interaction of DNA with NZC
Spectrophotometric Titrations
The absorption spectra are the most common means to examine the interaction between metal complex and DNA. In this work, the absorption spectra of NZC through the interaction exhibit a hyperchromism and weak blue shift due to the interaction between the aromatic chromophore ligand of NZC and DNA. Figure 3 shows the representative absorption spectra of NZC in the presence of increasing amounts of DNA at r i = ([DNA]/[NZC]) = 0.0–1.0. The absorption spectra of NZC in the invisible region were characterized by intraligand π–π* transitions in the NOR chromophore at λ max of 277 and 320 nm. The addition of increasing amounts of DNA to the solution of NZC resulted in the weak hyperchromicity (H) and little blue shift (∼5 nm) of the absorption maxima in the UV–vis spectra of NZC. This increase in absorbance of NZC provides the first indication of the formation of ground-state complex between the NZC and ct-DNA. But these relative small changes in hyperchromicities and blue shift of NZC upon introducing ct-DNA reveal that this complex interacts with ct-DNA maybe by the outside-binding mode [47]. In other words, the NZC interacts with the DNA, maybe through an electrostatic and hydrogen binding between the amine and/or oxygen moieties of the NOR and the nucleotides [48]. In order to obtain the ground-state binding constant K b for the ct-DNA complex with NZC, the following equilibrium can be considered:
K b can be estimated from the changes in absorbance at a fixed wavelength of 277 nm, using Eq. (1). The binding constant was found to be 8.9 × 103 M−1 at 25 °C. We have also checked the effect of the addition of ct-DNA on free NOR. The results show that on addition of ct-DNA, a concomitant decrease in absorbance, opposite to that of the NZC, occurred. The results indicate a different mode of interaction of the NZC compared to the free NOR with ct-DNA. As a result of complex formation occurring between DNA and NZC, the thermodynamic stability and the functional properties of DNA change. Therefore, the thermodynamic studies of drug–DNA interactions are so valuable for the determining of responsible factors of overall binding affinity and specificity of the drug. The first step in most investigations is to experimentally determine the equilibrium binding constant (K b ) and hence the observed Gibbs free energy change, ΔG bind (ΔG b = −RT ln K b ). In this experiment, binding-induced changes in the spectral properties of the NZC were monitored using UV–vis spectroscopy at 278–298 K (Fig. 4a). The ln K values obtained at four different temperatures are plotted against the reciprocal of temperature (Fig. 4b) according to the van’t Hoff equation, and thermodynamic parameters for the interaction are calculated. For NZC, the change in enthalpy due to interaction is −50.0 kJ mol−1 and change in entropy is −93.88 J mol−1 K−1. The negative ΔG value (−27.32 kJ mol−1) for interaction of ct-DNA with the complex indicates the spontaneity of the complexation. The decrease in entropy may be due to the interaction of NZC with DNA, causing stability via electrostatic and outside hydrogen binding interaction. Due to interaction of the complex with the DNA backbone, the structure of the double helix is distorted to a more random one. The thermodynamic parameters like changes in free energy (ΔG), enthalpy (ΔH), and entropy (ΔS) due to ligand binding provide an insight into the binding mode. Hydrogen bonds, van der Waals, hydrophobic, and electrostatic interactions are the major interactions that play a key role in molecular recognition [49]. It is found that the interaction process is enthalpy driven, and the major contribution of ΔG comes from negative ΔH. However, a negative enthalpy change and a negative entropy change indicate that significant immobilization of ct-DNA and the ligand occurs in an initial step involving hydrogen association that results in negative ΔS [49]. In the subsequent interacting complex, the negative ΔH contribution to the overall ΔG may be associated with electrostatic interactions.
The Fluorescence Quenching Studies
Fluorescence quenching may result from a variety of processes such as excited state reactions, energy transfer, ground-state complex formation, and collisional processes. Collisional or dynamic quenching refers to a process where the fluorophore and the quencher come into contact during the lifetime of the excited state, whereas static quenching refers to a fluorophore–quencher complex formation. The fluorescence spectra of NZC in different concentrations of DNA are shown in Fig. 5a. It is obvious that NZC has a strong fluorescence emission peak at 497 nm after being excited with a wavelength of 350 nm. When a fixed concentration of NZC was titrated with increasing amounts of DNA, an intrinsic fluorescence decrease of NZC was observed. Furthermore, there was a slight blue shift at the maximum wavelength of NZC fluorescence emission when the solution of DNA was added. The results indicated that DNA could quench the intrinsic fluorescence of NZC and the binding of NZC to DNA indeed exists. Static and dynamic quenching can be distinguished by their different binding constants' dependence on temperature and viscosity, or preferably by lifetime measurements [50]. In this paper, we have used the lifetime measurements to elucidate the quenching mechanism. The data obtained were analyzed by the Stern–Volmer equation (Eq. 2) [51]:
a Fluorescence spectra of 1 × 10−5 M NZC complex in the absence (dotted line) and presence of the increasing amounts of DNA in Tris–HCl buffer solution at 25 °C. r i = [DNA]/[NZC] = 0.0, 0.3, 0.4, 0.6, 0.8, 0.9, 1.0, 1.2, 1.4, and 1.6; b the Stern–Volmer plots of F 0/F versus [DNA] at 25 °C; c a plot of log(F 0−F)/F versus log [DNA]
whereF 0 and F are the fluorescence intensities of NZC in the absence and presence of the quencher, respectively. K SV is the Stern–Volmer dynamic quenching constant, K q is the quenching rate constant of the biomolecule and the value of the maximum scattering collision quenching constant is 2.0 × 1010 L mol−1s−1, τ 0 is the average lifetime of the biomolecule without quencher, and [Q] is the concentration of the quencher (DNA). Figure 5b shows the Stern–Volmer plots of F 0/F versus [Q] at 298 K; the values of K sυ and K q are 1.9 × 104 M−1 and 1.9 × 1012 M−1 s−1, respectively. The results showed that the value of K q is greater than that of the maximum scattering collision quenching constant, which indicated that the quenching mechanism of NZC by DNA was most likely a static quenching procedure. When small molecules bind independently to a set of equivalent sites on a macromolecule, the binding constant (K) and the numbers of binding sites (n) for the DNA–NZC system can be determined by Eq. (3) [52]:
where K and n are the binding constant and the number of binding sites in base pairs, respectively. Thus, a plot (Fig. 5c) of \( \log \frac{{{F_0}-F}}{F} \) versus log [Q] yielded K = 1.3 × 103 and n = 0.59. The linear correlation coefficient R indicated that the assumptions underlying the derivation of Eq. (3) were convincible. The value of n approximately equal to 0.5 suggested that there were two classes of binding sites for NZC towards DNA. The reported values of the binding constant for some typical intercalators such as ethidium bromide and safranine T were 2.6 × 106 and 4 × 105 L mol−1 [53], respectively. In this work, the binding constant of DNA–NZC was 1.3 × 103 L mol−1 at 298 K, which was smaller than the values of typical intercalators, ethidium bromide and safranine T, and confirmed the nonintercalation binding.
FT-IR Measurements
We used FT-IR to determine NZC binding sites, sequence preference, and the structural variations of the NZC–DNA complex in aqueous solution. The IR spectral features for NZC–DNA interaction are presented in panels a–c of Fig. 6. Figure 6a represented the FT-IR spectrum of pure NZC. The vibrational bands of NZC at 2,917, 2,837, 1,726, 1,624, 1,516, 1,301, and 1,255 cm−1 are assigned to the vibration of the quaternized nitrogen of the piperazinyl group, carboxylic stretch ν (C=O)carb, pyridone stretch ν (C=O) p , ν (COO)assy, and ν (COO)sym, respectively. In the presence of DNA at r i = [DNA]/[ZNC] = 1/50, shifting for the bands at 1,726 (carboxylic stretch) to 1,732, and 1,516 (ν (COO)assy) to 1,518 were observed. The observed shifting was accompanied by an increase in intensities of all vibration bands. A minor increase of intensity of the NZC in plane vibrations can be attributed to a weak NZC–DNA complexation. As DNA concentration increased (r i = 1/5), shifting for the bands at 1,624 (pyridone stretch ν (C=O) p ) to 1,618 and 1,516 (ν (COO)assy) to 1,512, were observed. Also, a major decrease in the intensities of carboxylic and ketonic bands was observed. However, these results indicate that the major interaction of DNA with carboxylic and ketonic groups occurred.
Electrochemical Behavior of NZC
In order to have a valuable judgment of voltammetric titration data of NZC with DNA, first of all, the electrochemical behavior of NZC in pH = 7.3 at the surface of HMDE was studied. A typical cyclic voltammogram of 1 × 10−5 M of NZC is shown in Fig. 7a. In forward sweep potentials (−0.4 to −1.2 V with scan rate of 10 mV S−1), one irreversible cathodic peak in the potential of −0.89 V was obtained, and in backward sweep potentials (−1.2 to −0.4 V), no peak was observed. The cathodic peak at −0.89 V corresponds to the reduction of the –C=O group of NOR to the –CHOH group [54]. The dependence of the cathodic peak current of NZC at the HMDE to the scan rate (υ) in the absence and presence of an excess of DNA was examined. Under selected experimental conditions, by increasing the scan rate, the cathodic peak currents (i pc) of NZC in the absence of DNA considerably increased, while in the presence of 5 × 10−5 M of DNA, the peak current slowly increased on the HMDE electrode (Fig. 7a, b). The negative shift of peak potential (E pc) with increasing scan rate υ demonstrated an irreversible electrode process for NZC. The plots of i pc versus υ 1/2 and υ for NZC in the absence and presence of DNA are shown in Fig. 8a, b. As it is observed, the cathodic peak current (i pc) of NZC varies linearly with the square root of the scan rate, υ 1/2, rather than with ν, with a linear equation of: \( {i_{\mathrm{pc}}}\left( {\mathrm{nA}} \right)=-4.77{\nu^{{{1 \left/ {2} \right.}}}}{{\left( {\mathrm{mV}\,{{\mathrm{S}}^{-1 }}} \right)}^{{{1 \left/ {2} \right.}}}}+1.97 \) and regression coefficient R = 0.982. The results indicate that the mass transport of NZC to the HMDE surface is a diffusion-controlled process. In the presence of DNA (R = [DNA]/[NZC] = 5), the cathodic peak of NZC sharply decreased and slightly shifted towards the negative potentials. These findings suggested there were almost no free NZC in the solution when R = 5. The lines i pc versus υ and ν 1/2 of the complex in the presence of DNA were linear and revealed that the mass transport of the DNA–NZC to the HMDE surface was carried out by both adsorption and diffusion mechanisms. The potential peak (E p ) versus the lnυ for NZC and DNA–NZC has been plotted (Fig. 9), and the corresponding fitting lines are (E p = −0.036 lnν − 0.808) for NZC and (E p = −0.039 lnν − 0.791) for DNA–NZC. To measure the symmetry of the energy barrier and all electrons, Eq. (4) was used.
The cathodic peak current (i pc) variation of NZC (diamonds) and NZC-DNA complex (triangles) with a scan rate (ν) and b square root of the scan rate (ν 1/2), the concentrations are the same as those of Fig. 7
where E p/2 is the potential, where i = i p/2 in cyclic voltammograms. In cyclic voltammograms, the average value of |E p−E p/2| was 62.07 mV. The value of αn f calculated from Eq. (1), was 1.302. Generally, α in the totally irreversible electrode process, is assumed as 0.5. Hence, two electrons are involved in the oxidation process of NZC and the value of α was 0.651.
Cyclic Voltammetric Titration of NZC with DNA
The electrochemical titration is more valuable to quantify the interaction parameters of an electro active molecule with DNA than other methods [55]. By addition of different amounts of DNA to the voltammetric cell containing 1.0 × 10−5 M of NZC, the cathodic peak currents of NZC begin to decrease, and the formal potential shifts to more positive values which suggest the interaction of the NZC with DNA (Fig. 10). When R = [DNA]/[DM] = 20.0, CV currents of NZC decreased and were stable. The decrease of all anodic and cathodic peak currents is due to the decrease of diffusion coefficients of the NZC–DNA species and equilibrium concentration of NZC in the solution. Under the experimental conditions and in the potential range of −0.4 to −1.2 V at the HMDE, the pure DNA is not electro active. This was mainly due to the merging of cathodic peaks at low DNA concentrations with the background discharge. According to these observations, it seems that the decrease of peak currents of NZC after an addition of excess DNA is caused by the interaction of NZC to the bulky, slowly diffusing DNA, which results in a considerable decrease in the apparent diffusion coefficient. For measurement of binding constant (K b ) and binding site size (s) of the NZC–DNA, diffusion coefficients for free D f , and bound molecules D b , we used the following measurements.
If no electro active molecule (E) specifically reacts with a DNA duplex at a binding site, which is composed of base pairs (S), a DNA-electro active molecule complex (E–S) is produced as follows:
As the NZC nonspecifically binds to the DNA duplex and covers S consecutive base pairs (i.e., one binding site), the binding constant K can be given by Eq. (6):
where C b , C f , and C s represent the equilibrium concentrations of the DNA–NZC complex, free NZC, and free binding site, respectively. The total concentration of NZC, C t , is:
The average number of binding sites (x) along a DNA duplex molecule with an average total number of base pairs L can be described by the following form: x = L/s, where s is the binding site size of the electro active molecule interacting with DNA. It means that the number of DNA base pairs is occupied (or covered) by a binding molecule. Thus, the total concentration of binding sites (xC DNA) can be expressed as follows:
C NP represents the concentration of nucleotide phosphate, which is determined by the UV absorption at 260 nm. The total concentration of binding sites can also be expressed as follows:
The ratio of the NP concentration and the total concentration of electro active molecules can be defined as R:
For an irreversible reaction in CV, the total cathodic current (I pc) under the fixed potential with any R can be calculated:
In fact, B represents the appropriate, concentration-independent terms in the voltammetric expression. A Nernstain reaction in CV at 25 °C is shown as follows:
where n is the number of electron transferred, A is the electrode area, v is the scan rate, D f and D b are the diffusion coefficients for free and bound molecules, α f and α b are the electron transfer coefficients for free and bound molecules, and C f and C b are the bulk concentrations of the free and bound irreversibly electro active species. The binding constant K can be expressed as the following form:
where s is the size of binding site in terms of base pairs. Making appropriate substitutions and eliminating C b and C f from Eq. (12), a new equation was obtained:
where \( b=1+K{C_t}+\frac{{KR{C_t}}}{2s } \).
Since i pc, C t , and [NP] are experimentally measurable and (αn) f , (αn) b , have already been acquired as mentioned above, the binding constant (K) and binding site size (s) of the NZC–DNA, D f , and D b can be obtained from a nonlinear regression analysis of the experimental data (i pc versus [R] plot) according to Eq. (15). The nonlinear fit analysis yielded K = 9.9 × 103 and s = 0.64, D f = 2.1 × 10−4 cm2 s−1, and D b = 5.5 × 10−5 cm2 s−1.
Conclusion
A tetrahedral zinc (II) complex with the second generation fluoroquinolone, NZC, was synthesized, and its cytotoxicity effect, antibacterial activity, and binding affinity to the ct-DNA were evaluated. Different instrumental methods were used to investigate the interaction mechanism. The following results support that the NZC can bind to DNA via two nonintercalation modes. In voltammetric studies, in the presence of DNA, the cyclic voltammograms of the NZC exhibited a small negative shift in cathodic peak potentials followed by decrease in peak current, indicating the interaction existing between the NZC and ct-DNA. The nonlinear fit analysis of cyclic voltammetric titration yielded K b = 9.9 × 103 and binding site size of s = 0.64. Thermodynamic studies indicate that the binding action of the NZC and ct-DNA is an exothermal reaction. However, a negative entropy change indicates that significant immobilization of NZC and ct-DNA occurs via hydrogen binding. In the subsequent interacting complex, the negative ΔH contribution to the overall ΔG may be associated with electrostatic interactions. The study of the fluorescence quenching of the NZC by ct-DNA shows that the probable quenching mechanisms are static quenching procedure. The FT-IR study revealed that DNA interacts more with the ketonic oxygen atoms. The in vitro cytotoxic effects of the NOR and NZC against three well-known cell lines, including Hela, Caco-2 and MDA, were tested using the LDH assay method. Results revealed that the NZC exhibited better cytotoxic activities than the NOR. The NZC was found to be more potent than NOR against E. faecalis, S. dysenteriae, and E. coli. The strong antibacterial and cytotoxicity activities of NZC against tested cell lines and organisms suggest further investigation on metal ion complexes of fluoroquinolones. Hence, it seems worthy to generate a database from the proposed work to develop new effective useful DNA probes.
References
Emami, S., Shafiee, A., & Foroumadi, A. (2006). Structural features of new quinolones and relationship to anti bacterial activity against gram-positive bacteria. Mini-Reviews in Medical Chemistry, 6, 375–386.
Hooper, D. C., & Wolfson, J. S. (1985). The fluoroquinolones: pharmacology, clinical uses, and toxicities in human. Antimicrobial Agents and Chemotherapy, 28, 716–721.
Hussy, P., Maass, G., Tümmler, B., Grosse, F., & Schomburg, U. (1986). Effect of 4-quinolones and novobiocin on calf thymus DNA polymerase a primase complex, topoisomerases I and II, and growth of mammalian lymphoblasts. Antimicrobial Agents and Chemotherapy, 29, 1073–1078.
Sharma, P. C., Jain, A., & Jain, S. (2009). Fluoroquinolone anti bacterials: a review on chemistry, microbiology and therapeutic prospects. Acta Poloniae Pharmaceutica, 66, 587–604.
Sepcić, K., Perković, O., Turel, I., Sepcić, J., & Vjesn, L. (2009). Adverse effects and interactions of fluoroquinolones. Lijec̆nic̆ki vjesnik, 131, 74–80.
Al-Trawneh, S. A., Zahra, J. A., Kamal, M. R., El-Abadelah, M. M., Zani, F., Incerti, M., et al. (2010). Synthesis and biological evaluation of tetracyclic fluoroquinolonesas anti bacterial and anticancer agents. Bioorganic & Medicinal Chemistry, 18, 5873–5884.
da G, D., Batista, J., da Silva, P. B., Stivanin, L., Lachter, D. R., Silva, R. S., et al. (2011). Co(II), Mn(II) and Cu(II) complexes of fluoroquinolones: synthesis, spectroscopical studies and biological evaluation against Trypanosoma cruzi. Polyhedron, 30, 1718–1725.
Sousa, I., Claro, V., Pereira, J. L., Amaral, A. L., Cunha-Silva, L., de Castro, B., et al. (2012). Synthesis, characterization and anti bacterial studies of a copper(II) levofloxacin ternary complex. Journal of Inorganic Biochemistry, 110, 64–71.
Patel, M., & Chhasatia, M. (2010). Anti bacterial and DNA interaction studies of zinc (II) complexes with quinolone family member, ciprofloxacin. Journal of Medicinal Chemistry, 45, 439–446.
Patel, M. N., Joshi, H. N., & Patel, C. R. (2012). DNA interaction, in vitro antimicrobial and SOD-like activity of copper(II) complexes with norfloxacin and terpyridines. Journal of Organic Chemistry, 701, 8–16.
Ohmine, T., Katsube, T., Tsuzaki, Y., Kazui, M., Kobayashi, N., Komai, T., et al. (2002). Inhibition of human immunodeficiency virus type 1 Tat-trans-activation-responsive region interaction by an antiviral quinolone derivative. Bioorganic & Medicinal Chemistry Letters, 12, 739–743.
Ogino, H., Fujii, M., Ono, M., Maezawa, K., Hori, S., & Kizu, J. (2009). In vivo and in vitro effects of fluoroquinolones on lipopolysaccharide-induced pro-inflammatory cytokine production. Journal of Infection and Chemotherapy, 15, 168–173.
Chollet, C., Baliani, A., Wong, P. E., Barrett, M. P., & Gilbert, I. H. (2009). Quinol derivatives as potential trypanocidal agents. Bioorganic & Medicinal Chemistry, 17, 2512–2523.
Nenortas, E., Burri, C., & Shapiro, T. A. (1999). Antitrypanosomal activities of fluoroquinolones with pyrrolidinyl substitutions. Antimicrobial Agents and Chemotherapy, 43, 2066–2068.
Sandstrom, K., Warmlander, S., Leijon, M., & Graslund, A. (2003). H NMR studies of selective interactions of norfloxacin with double-stranded DNA. Biochemical and Biophysical Research Communications, 304, 55–59.
Wiles, J. A., Wang, Q., Lucien, E., Hashimoto, A., Song, Y., Cheng, J., et al. (2006). Isothiazoloquinolones containing functionalized aromatic hydrocarbons at the 7-position: synthesis and in vitro activity of a series of potent anti bacterial agents with diminished cytotoxicity in human cells. Bioorganic & Medicinal Chemistry, 16, 1272–1276.
Pfeiffer, E. S., & Hiasa, H. (2004). Replacement of ParC α4 helix with that of GyrA increases the stability and cytotoxicity of topoisomerase IV-quinolone-DNA ternary complexes. Antimicrobial Agents and Chemotherapy, 48, 608–611.
Pansuriya, P. B., & Patel, M. N. (2007). Synthesis, spectral, thermal, DNA interaction and antimicrobial properties of novel Cu (II) heterochelates. Applied Organometallic Chemistry, 21, 739–749.
Lee, C., & Ronald, A. R. (1987). Norfloxacin: its potential in clinical practice. American Journal of Medicine, 82, 27–34.
Robert, C., & Moellering, J. (1987). Norfloxacin: a fluoroquinolone carboxylic acid antimicrobial agent. American Journal of Medicine, 82, 1–2.
Traganos, F., Kaminska-Eddy, B., & Darzynkiewicz, Z. (1991). Caffeine reverses the cytotoxic and cell kinetic effects of novantrone (mitoxantrone). Cell Proliferation, 24, 305–319.
Reece, R. J., & Maxwell, A. (1991). DNA gyrase: structure and function. CRC Critical Reviews in Biochemistry and Molecular Biology, 26, 335–375.
Shen, L. L., & Pernet, A. G. (1985). Mechanism of inhibition of DNA gyrase by analogues of nalidixic acid: the target of the drugs is DNA. Proceedings of the National Academy of Sciences of the United States of America, 82, 307–311.
Shen, L. L., Kohlbrenner, W. E., Weigl, D., & Baranowski, J. (1989). Mechanism of quinolone inhibition of DNA gyrase. Journal of Biological Chemistry, 264, 2973–2978.
Shen, L. L. (1994). Molecular mechanisms of DNA gyrase inhibition by quinolone anti bacterials. Advances in Pharmacology, 29, 285–304.
Shen, L. L., Mitscher, L. A., Sharma, P. N., O'Donnell, T. J., Chu, D. W. T., Cooper, C. S., et al. (1989). Mechanism of inhibition of DNA gyrase by quinolone anti bacterials. A cooperative drug-DNA binding model. Biochemistry, 28, 2886–2894.
Alkaysi, H. N., Abdel-Hay, M. H., Sheikh Salem, M., Gharaibeh, A. M., & Na'was, T. E. (1992). Chemical and microbiological investigations of metal ion interaction with norfloxacin. International Journal of Pharmaceutics, 87, 73–77.
Refat, M. S. (2007). Synthesis and characterization of norfloxacin-transition metal complexes (group 11, IB): spectroscopic, thermal, kinetic measurements and biological activity. Spectrochimica Acta Part A, 68, 1393–1405.
Shaikh, A. R., Giridhar, R., & Yadav, M. R. (2007). Bismuth-norfloxacin complex: synthesis, physicochemical and antimicrobial evaluation. International Journal of Pharmaceutics, 332, 24–30.
Chen, Z. F., Xiong, R. G., Zhang, J., Chen, X. T., Xue, Z. L., & You, X. Z. (2001). Mixed-valence Cu (II)/Cu (I) complex of quinolone ciprofloxacin isolated by a hydrothermal reaction in the presence of l-histidine: comparison of biological activities of various copper–ciprofloxacin compounds. Inorganic Chemistry, 40, 4075–4085.
Turel, I., Gruber, K., Leban, I., & Bukovec, N. (1996). Crystal structure and characterization of the bismuth(III) compound with quinolone family member (ciprofloxacin). Anti bacterial study. Journal of Inorganic Biochemistry, 61, 197–212.
Evstigneev, M. P., Rybakova, K. A., & Davies, D. B. (2006). Complexation of norfloxacin with DNA in the presence of caffeine. Biophysical Chemistry, 121, 84–95.
Son, G.-S., Yeo, J.-A., Kim, J.-M., Kim, S. K., Hyung Rang Moon, U., & Nam, W. (1998). Base specific complex formation of norfloxacin with DNA. Biophysical Chemistry, 74, 225–236.
Ahmadi, F., Alizadeh, A. A., Bakhshandeh-Saraskanrood, F., Jafari, B., & Khodadadian, M. (2010). Experimental and computational approach to the rational monitoring of hydrogen-bonding interaction of 2-imidazolidinethione with DNA and guanine. Food and Chemical Toxicology, 48, 29–36.
Linford, N. J., & Dorsa, D. M. (2002). 17 β-Estradiol and the phytoestrogen genistein attenuate neuronal apoptosis induced by the endoplasmic reticulum calcium-ATPase inhibitor thapsigargin. Journal of Steroids, 67, 1029–1040.
Kashanian, S., Gholivand, M. B., Ahmadi, F., Taravati, A., & HosseinzadehColagar, A. (2007). DNA interaction with Al–N, N-bis(salicylidene) 2,2-phenylendiamine complex. Spectrochimica Acta Part A, 67, 472–478.
Kashanian, S., Askari, S., Ahmadi, F., Omidfar, K., Ghobadi, S., & AbasiTarighat, F. (2008). In vitro study of DNA interaction with clodinafop-propargyl herbicide. DNA and Cell Biology, 27, 581–586.
Ahmadi, F., Jamali, N., Moradian, R., & Astinchap, B. (2012). Binding studies of pyriproxyfen to DNA by multispectroscopic atomic force microscopy and molecular modeling methods. DNA and Cell Biology, 31, 259–268.
Efthimiadou, E. K., Karaliota, A., & Psomas, G. (2008). Mononuclear dioxomolybdenum(VI) complexes with the quinolone senrofloxacin and sparfloxacin: synthesis, structure, anti bacterial activity and interaction with DNA. Polyhedron, 27, 349–356.
Macias, B., Villa, M. V., Sastre, M., Castiñeiras, A., & Borras, J. (2002). Complexes of Co(II) and Zn(II) with ofloxacin. Crystal structure of [Co (oflo) 2(MeOH) 2] · 4MeOH. Journal of Pharmaceutical Sciences, 91, 2416–2423.
Sadeek, A. S. (2005). Synthesis, spectroscopic, thermal and biological activity investigation of new Y (ΙΙΙ) and Pd (ΙΙ) norfloxacin complexes. Journal of Molecular Structure, 753, 1–12.
Nakamoto, K. (1986). Infrared and Raman spectra of inorganic and coordination compounds (pp. 191–196). New York: Wiley. 29.
Deacon, G. B., & Phillips, R. J. (1980). Synthesis, structures, thermal and magnetic properties of a series of lanthanide [Ln = Sm, Gd, Er, Yb] complexes with 4-quinolineacarboxylate. Coordination Chemistry Reviews, 33, 227–250.
Sagdinca, S., & Bayarı, S. (2004). Spectroscopic studies on the interaction of ofloxacin with metals. Journal of Molecular Structure, 691, 107–113.
Lıgia, M. M., Vieira, A., Mauro, V., de Almeida, A., Cristina, S. M., Lourenço, B., et al. (2009). Synthesis and antitubercular activity of palladium and platinum complexes with fluoroquinolones. European Journal of Medicinal Chemistry, 44, 4107–4111.
Tweedy, B. G. (1964). Microwave synthesis, spectroscopic, thermal and biological significance of some transition metal complexes containing heterocyclic ligands. Phytopathology, 55, 910–914.
Nakabayashi, Y., Watanabe, Y., Nakao, T., & Yamauchi, O. (2004). Interactions of mixed ligand ruthenium (II) complexes containing an amino acid and 1, 10-phenanthroline with DNA. Inorganica Chimica Acta, 357, 2553–2560.
Mudasir, E., Wijaya, K., Tri Wahyuni, E., Inoue, H., & Yoshioka, N. (2007). Base-specific and enantioselective studies for the DNA binding of iron(II) mixed-ligand complexes containing 1,10-phenanthroline and dipyrido[3,2-a:2′,3′-c]phenazine. Spectrochimica Acta Part A, 66, 163–170.
Ross, P. D., & Subramanian, S. (1981). Thermodynamic studies with acetylthiocholine on nicotinic receptors of mammalian skeletal muscle in vitro. Biochemistry, 20, 3096–3102.
Lakowicz, J. R. (1999). Principles of fluorescence spectroscopy (2nd ed., pp. 237–265). New York: Plenum Press.
Zhang, Y. Z., Zhou, B., Zhang, X. P., Huang, P., Li, C. H., & Liu, Y. (2009). Interaction of malachite green with bovine serum albumin: determination of the binding mechanism and binding site by spectroscopic methods. Journal of Hazardous Materials, 163, 1345–1352.
Shakir, M., Azam, M., Parveen, S., Khan, A. U., & Firdaus, F. (2009). Synthesis and spectroscopic studies on complexes of N, N0-bis-(2-pyridinecarboxaldimine)- 1, 8-diaminonaphthalene (L); DNA binding studies on Cu(II) complex. Spectrochimica Acta, 71, 1851–1856.
Cao, Y., & He, X. W. (1998). Studies of interaction between safranine T and double helix DNA by spectral methods. Spectrochimica Acta, 54, 883–892.
Gholivand, M. B., & Ahmadi, F. (2008). Simultaneous determination of trans-cinnamaldehyde and benzaldehyde in different real samples by differential pulse polarography and study of heat stability of trans-cinnamaldehyde. Analytical Letters, 41, 3324–3341.
Ahmadi, F., & Jafari, B. (2011). Voltammetry and spectroscopy study of in vitro interaction of fenitrothion with DNA. Electroanalytical, 23, 675–682.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ahmadi, F., Saberkari, M., Abiri, R. et al. In Vitro Evaluation of Zn–Norfloxacin Complex as a Potent Cytotoxic and Antibacterial Agent, Proposed Model for DNA Binding. Appl Biochem Biotechnol 170, 988–1009 (2013). https://doi.org/10.1007/s12010-013-0255-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0255-6