Abstract
Glycosylation of flavonoids is mediated by family 1 uridine diphosphate (UDP)-dependent glycosyltransferases (UGTs). Until date, there are few reports on functionally characterized flavonoid glycosyltransferases from Withania somnifera. In this study, we cloned the glycosyltransferase gene from W. somnifera (UGT73A16) showing 85–92 % homology with UGTs from other plants. UGT73A16 was expressed as a His6-tagged fusion protein in Escherichia coli. Several compounds, including flavonoids, were screened as potential substrates for UGT73A16. HPLC analysis and hypsochromic shift indicated that UGT73A16 transfers a glucose molecule to several different flavonoids. Based on kinetic parameters, UGT73A16 shows more catalytic efficiency towards naringenin. Here, we explored UGT73A16 of W. somnifera as whole cell catalyst in E. coli. We used flavonoids (genistein, apigenin, kaempferol, naringenin, biochanin A, and daidzein) as substrates for this study. More than 95 % of the glucoside products were released into the medium, facilitating their isolation. Glycosylation of substrates occurred on the 7- and 3-hydroxyl group of the aglycone. UGT73A16 also displayed regiospecific glucosyl transfer activity towards 3-hydroxy flavone compound, which is the backbone of all flavonols and also for a chemically synthesized compound, not found naturally. The present study generates essential knowledge and molecular as well as biochemical tools that allow the verification of UGT73A16 in glycosylation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among ancient civilizations, India has been known to be rich repository of medicinal plants which produce various types of secondary metabolites including flavonoids, alkaloids, and terpenoids [1]. These compounds undergo modification reactions (methylation, hydroxylation, glycosylation, etc.) and lead to structural diversity of secondary metabolites [2]. Glycosylation is a prominent modification reaction and is often the last step in biosynthesis of natural compounds [3]. It is quantitatively the most significant reaction on earth. Glycosylation of small molecules has key roles in many biological processes. It also plays the role in biosynthesis of bioactive compounds, regulation of hormone activity, and metabolic process of toxins [4, 5]. In addition, glycosylation has been recognized as one of the important mechanism in detoxification of exogenous compounds [6].
Glycosylation of plant secondary products, such as flavonoids, coumarins, terpenoids, and cyanohydrins, is generally catalyzed by PSPGs [7], which belong to family-1 glycosyltransferases, catalyzing glycosyl transfer from nucleoside diphosphate-activated sugars (donor) to aglycon substrate (acceptor) molecules. The activated sugar form is typically uridine diphosphate (UDP)-glucose, but UDP-galactose, UDP-glucuronide, UDP-xylulose, and UDP-rhamnose are also reported. In plants, sugar acceptors contain each and every class of secondary metabolites, for example phenolics, terpenoids, cyanohydrins, etc. [4, 6, 8].
One of the most widely studied classes of plant glycosides is the large and heterogenic group of polyphenols. Until date, a large number of polyphenolic glycosides, including flavonoid glycosides, have been identified. Flavonoids are typical phytochemicals having an impact on human [9, 10] and synthesized via the phenylpropanoid pathway. Flavonoids are associated with different biological activities such as anti-bacterial, anti-thrombotic, anti-inflammatory, anti-carcinogenic, and vasodilatory [11]. Various flavonoid uridine diphosphate-dependent glycosyltransferase (UGT) genes have been cloned and characterized [12–15]. The reported UGTs glycosylate various secondary metabolites (flavonoids and alkaloids), hormones, and xenobiotics [4, 6]. In vitro studies indicate considerable difference in the anti-oxidant potential of different flavonoid subgroups, depending upon their bioavailability, chemical structure, and metabolism [16]. Many studies have been aimed at developing new production platforms for such plant natural products through metabolic engineering in microorganisms and plants [15, 17–21].
The levels of flavonoid glycosides are very low in plants, and it is necessary to increase the yield of medicinally important compounds. The primary objective of the present study was to isolate and clone a gene which can perform the glycosylation of a wide variety of natural compounds. We chose Withania somnifera as the plant source which is mostly used in Indian ayurvedic medicine, and little is known about functionally characterized flavonoid glycosyltransferases from this plant. Here, we report in vitro characterization of a flavonoid-O-glycosyltransferase and production of milligram quantities of the 7-O-glucoside of iso-flavones (genistein, daidzein, and biochanin A), flavones (apigenin), flavanone (naringenin), and 3-O-glucoside of flavonols (kaempferol) in Escherichia coli cultures expressing UGT73A16.
Materials and Methods
Plant Material, Cloning of WsUGT, and Phylogenetic Analysis of UGTs from Higher Plants
The total RNA was isolated from W. somnifera leaves using TRI Reagent (Sigma, USA) and then reverse-transcribed in a final reaction volume of 20 μL using AMV reverse-transcriptase (Promega, USA) and oligo(dT)18 primer. The first-strand cDNA was synthesized at 42 °C for 1 h and terminated by heating at 70 °C for 10 min. The primers were designed by using conserved domains of glycosyltransferases (GTs) from NCBI (accession no. U32644 and AB360626). Two oligonucleotide primers, SBF1 (5′ATGATGCAAGAACCACTAGA 3′) and SBR1 (5′ GTTGAATTCCAACCACA 3′), were synthesized and used to amplify a partial fragment of glycosyltransferase. This fragment was subsequently used to design the gene-specific primers to clone the full-length cDNA of glycosyltransferase by RACE (rapid amplification of cDNA ends)
The BD SMARTTM RACE cDNA Amplification Kit (Clontech, USA) was used to amplify the 3′ end and 5′ end of UGT73A16 cDNA. The first-strand 3′ RACE-ready and 5′ RACE-ready cDNAs were prepared according to the manufacturer′s protocol and used as templates for 3′ RACE and 5′ RACE, respectively. For 3′ RACE gene-specific primer FLONF (5′AATTGGGCTATTGGCCCGCTTTCCCTGTGCAAC 3′) and UPM (5′ CTAATACGACTCACTATAGGGC 3′, provided by the kit) were used for the primary amplification. For the nested amplification of 3′ RACE, the primers FLONNF (5′ GCTTGACTCAAAGAAACCAAGTTCCATTGT 3′) and NUP (5′ CTAATACGACTCACTATAGGGC 3′, provided by the kit) were used with the products of the first amplification as templates. For 5′ RACE, gene specific primer SACR (5′ TTTGATTAATCGGATCCCCAAATTCT 3′) and UPM were used with 5´ RACE-ready cDNA as the template. For the nested amplification of 5´ RACE, the primers SACNR (5′ GCTTGGCCATGTCTAGTGTAGGAATC 3′) and NUP were used with the products of the first amplification as templates.
A full-length cDNA sequence of UGT73A16 was deduced by assembling the products of the 3′ RACE and 5′ RACE. The nucleotide sequence WSfullF 5′ ATGGGTCAGCTCCATATTTTCTTC 3′ was used as a forward primer, while WSfullR 5′ ATGACCAGTGGAACTATATGTACT 3′ as a reverse primer. The PCR was performed with AccuTaq-LA DNA polymerase (Sigma, USA) under the following conditions: 94 °C for 5 min, followed by 35 cycles of 45 s denaturation at 95 °C, 1 min annealing at 52 °C, and 1.5 min amplification at 72 °C, and a final extension at 72 °C for 8.0 min. The PCR product was purified with Gen EluteTM gel extraction kit (Sigma, USA) and cloned into pGEM-T Easy vector (Promega, USA) and sequenced. The sequence reported in this paper has been deposited in the NCBI GenBank database [GenBank FJ654696]. The translated protein sequence of UGT73A16 was used to construct a phylogenetic tree using bootstrapping MEGA 4.0.2 [22] with the known plant glycosyltransferases (GTs) deposited in NCBI GenBank database by using neighbor-joining method [23].
Expression and Purification of WsUGT (UGT73A16) in E. coli
To express (UGT73A16) in E. coli, the full-length cDNA of UGT73A16 was amplified with AccuTaq-LA DNA polymerase by using the primers containing restriction enzyme sites NdeI at forward primer and XhoI at the reverse primer. The amplified fragment was cloned in pGEM-T Easy vector, digested using NdeI and XhoI and digested inserts were cloned into pET-30b(+) (Novagen, USA) using NdeI/XhoI sites. The resulting construct was transformed into E. coli BL21 (DE3) without pLys S or pLys E (Novagen, USA). For the protein induction, the transformant was grown at 37 °C with shaking in 50 ml LB medium containing 50 μg mL−1 kanamycin until absorbance at 600 nm (A 600 nm) reached to 0.5–0.7. Protein expression was induced with 0.08 mM IPTG while incubating for 20 h at 22 °C. The E. coli cells were harvested by centrifugation and resuspended in 10 mL of cell suspension buffer (50 mM Tris–HCl (pH 8), 1 mM EDTA, 0.5 % TritonX-100, 0.1 mM PMSF, 0.7 mM DTT, 100 mM NaCl, and 0.1 % sodium azide) [24] and disrupted with an ultrasonic cell disrupter (ultrasonic liquid processor, MISONIX) and centrifuged ( 8,000×g for 15 min). The His6-tagged soluble protein was purified using the Ni-NTA agarose affinity column (Qiagen).
Kinetic Assays and Analysis of Reaction Product
The reaction mixture for UDP-glycosyltransferase contained 12 μg of purified UGT73A16 protein, 100 mM Tris–Cl (pH 8.0), 100 μM of UDP-glucose, and 18 μM of substrates (Sigma). The reaction mixture was incubated at 37 °C for 3 h. The reaction was terminated and extracted twice by the addition of equal volume of ethyl acetate and evaporated to dryness. The dried reaction product was dissolved in methanol and analyzed by high performance liquid chromatography (HPLC, Perkin Elmer, USA) equipped with a diode array detector and a Waters symmetry C18 column (5 μm particle size, 4.6 mm × 25 cm, Supelco Analytical) except for kaempferol and baicalein (3 μm particle size, 4.6 mm × 15 cm, Supelco Analytical). For generation of an analytical scale, the mobile phase consisted of sterile milliQ water (SMQ) with 0.1 % TFA and programmed as follows: 10 % acetonitrile for 5.0 min, 30 % acetonitrile for 5.0 min, 60 % acetonitrile for 5.0 min, and 90 % acetonitrile for 5.0 min. The flow rate was 1 mL min−1, and UV detection was performed at 250–340 nm.
UGT73A16 as Whole Cell Biocatalyst
E. coli BL21 (DE3) strains carrying the recombinant UGT73A16 construct were grown in 50 mL Luria-Bertani (LB) medium at 37 °C until A 600 nm reached 0.6–0.7. IPTG was then added to the cultures to a final concentration of 0.08 mM. E. coli culture harboring vector pET-30b(+) without insert was used as a control. At the same time, 100 μM of each substrates, genistein (27.02 mg L−1), daidzein (25.4 mg L−1), biochanin A (28.4 mg L−1), apigenin (27.02 mg L−1), naringenin (27.2 mg L−1), and kaempferol (28.6 mg L−1), dissolved in dimethyl sulfoxide were then added to the induced cultures. After an appropriate incubation time (12, 16, 20, 24, and 48 h) at 22 °C, cultures were centrifuged at 5,000×g for 10 min to obtain the supernatant and cell pellet fraction. The cell pellet was resuspended in ethyl acetate to extract aglycone and its glucosides and centrifuged at 10,000×g for 5 min to remove the cell debris. The supernatant fraction was also used for ethyl acetate extraction. Both the supernatant and cell fractions were evaporated to dryness and analyzed on HPLC as given above.
Results
Cloning and Phylogenetic Analysis of WsUGT
The CAZy website (carbohydrate-active enzymes; http://afmb.cnrs-mrs.fr/CAZy) provides classification of GTs from several organisms. We selected UGT genes from Nicotiana tabacum and Lycium barbarum which showed high homology with flavonoid glycosyltransferases. Based on the conserved UGT sequences, a pair of specific primer was designed to obtain a partial fragment of flavonoid glycosyltransferase. This partial fragment was further used for the 3′ RACE and 5′ RACE reaction to get the full-length sequence of glycosyltransferase including UTR region. According to sequence information from 3′ RACE and 5′ RACE, WSfullF and WSfullR were designed to obtain open reading frame. This open reading frame of 1,413 bp encoded 471 amino acid residues with a calculated molecular mass of ∼52 kDa. In addition, a 13-bp 5′ untranslated region and a 101-bp 3′ untranslated region exist in the isolated gene. According to the glycosyltransferase nomenclature guidelines [25], the systematic name of the W. somnifera glycosyltransferase is UGT73A16.
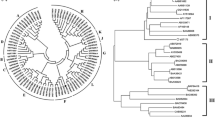
Function and specificity of UGT73A16 cannot be fully predicted based on the sequence information alone; thus, a phylogenetic tree was constructed for the UGT73A16 along with the different UGTs by means of the neighbor-joining method (Fig. 1). The phylogenetic tree of flavonoid UGTs is divided in to three clusters: cluster I contained flavonoid 7-O-glycosyltransferase, salicylic acid-induced glycosyltransferase (accession number AAK28303), and UDP-glucose: flavonoid 3-O-glucoside 7-O-glucosyltransferase (accession number Q9ZQ95); cluster II is composed of UGTs displaying diverse regioselectivity; and cluster III consisted of flavonoid 3-O-glycosyltransferase. Phylogenetic analysis suggests that the UGT73A16 was most similar to cluster I and has the potential to glycosylate a wide range of flavonoids.
Phylogenetic tree analysis. The phylogenetic tree of UDP-dependent glycosyltransferase: 7-O-glycosyltransferase forms cluster I, UGTs showing diverse regioselectivity forms cluster II, and 3-O-Glycosyltransferases forms cluster III. GenBank accession numbers are given in brackets. Following plant UGT sequences were used for constructing phylogenetic tree: L. barbarum UGT73A10, L. barbarum UGT73A12, L. esculentum, N. tabacum TOGT1, S. baicalensis 7GT, Antirhinnum majus UGT73A9, Arabidopsis thaliana 7GT, Dianthus caryophyllus GT, Dorotheanthus bellidiformis GT, A. thaliana AtF3G7GT, Glycine max 7GT, Brassica napus GT, Citrus unshiu GT, Iris hollandica 5GT, Nicotiana tabacum GT, Petunia hybrida 5GT, Torentia hybrida 5GT, Perilla frutescense 5GT, Hordeum vulgare 3GT, Zea mays 3GT, Malus domestica 3GT, A. thaliana 3GT, Vigna mungo 3GaT, P. hybrida 3GT, Gentiana triflora 3GT, Forsythia intermedia 3GT, P. frutescense 3GT
Heterologous Expression of UGT73A16 and Screening of Various Substrates for Glycosylation
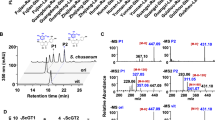
In order to screen various substrates for glycosylation, UGT73A16 was expressed under the control of T7 lac promoter in E. coli BL21 (DE3) cells as His6-tagged fusion protein. The crude extracts of recombinant E. coli having UGT73A16 were subjected to chromatography using a nickel affinity column for purification. The molecular weight of the purified recombinant UGT73A16 was approximately ∼52 kDa (Fig. 2). This purified UGT73A16 was incubated with various acceptors and donors (UDP-glucose and UDP-glucuronic acid). The results showed that UGT73A16 exhibited glycosyltransferase activity towards flavonoids including flavonols (kaempferol and isorhamnetin), flavones (apigenin and baicalein), isoflavones (genistein, biochanin A, and daidzein), flavanone (naringenin) and 3-hydroxy flavone with UDP-glucose. This purified UGT73A16 also showed the flavonoids 7-O-glucuronyltransferase activity only with the baicalein, which transfers glucuronic acid from UDP-glucuronic acid to the 7-OH group of baicalein. HPLC analysis (Fig. 3) suggested that the reaction of UGT73A16 with kaempferol, apigenin, daidzein, genistein, isorhamnetin, baicalein, biochanin A, and 3-hydroxy flavone yielded a single glycosylation product of monoglucosides, whereas the reaction with naringenin yielded two glycosylation products.
HPLC chromatogram: UGT73A16 assay mixture with a naringenin: P1, naringenin 7-O-glucoside; P2, naringenin 4′-O-glucoside; b standard1: naringenin 7-O-glucoside; c kaempferol: P3, kaempferol 3-O-glucoside; d standard2: kaempferol 3-O-glucoside; e genistein: P4, genistein 7-O-glucoside; f standard3: genistein 7-O-glucoside; g biochanin A: P5, biochanin A 7-O-glucoside; h standard4: biochanin A 7-O-glucoside
The glycosylation products obtained from apigenin, naringenin, daidzein, biochanin A, and genistein were co-eluted with their 7-O-glucosides in analytical HPLC. In case of kaempferol, isorhamnetin, and 3-hydroxy flavones, UGT73A16 gives the 3-O-glycosylated reaction product. These results strongly suggested that UGT73A16 belongs to a member of cluster I (Fig. 1). Glycosylation position of kaempferol, biochanin A, daidzein, and naringenin were assigned based on hypsochromic shift data [26] and with HPLC profile, comparison with published data [19, 27, 28] and using authentic reference compounds (Table 1).
Flavonols have two absorption maxima: band I (350–380 nm) and band II (240–280 nm) corresponding to the B- and A-ring, respectively. Conjugation of 3-, 5-, or 4'-hydroxyl groups causes a band I hypsochromic shift, which is larger for a 3-substitution (11–19 nm) than a 4'-conjugation (3–5 nm). For naringenin (peak at 289.72 nm), only the product P1 did not show a hypsochromic shift, suggesting conjugation at the 7-hydroxyl group. P2 (peak at 285 nm) showed a hypsochromic shift of 5 nm, suggesting conjugation at the 4'-hydroxyl group (Fig. 4a). The maximum absorbance of kaempferol was at 266.57 and 366.96 nm, and those of reaction product P3 were at 266.57 and 347.81 nm, signifying conjugation at the 3-hydroxyl group (Fig. 4b). The lack of hypsochromic shift between substrate and reaction product strongly suggests that glycosylation occurred at the hydroxyl group of C-7, which was confirmed by comparison with an authentic reference standard. The maximum absorbance of genistein and biochanin A were at 261.98 and 266.22 nm, respectively, and reaction product P4 and P5 also at 261.98 and 266.22 nm, respectively (Fig. 4c, d).
Hypsochromic shift. Hypsochromic shift of a naringenin: naringenin 7-O-glucoside and reaction product P1 and P2 shown in square dot, round dot, and solid line, respectively; b kaempferol: kaempferol substrate, kaempferol 3-O-glucoside, and reaction product P3 shown in square dot, round dot, and solid line, respectively; c genistein: genistein substrate, genistein 7-O-glucoside, and reaction product P4 shown in square dot, round dot, and solid line, respectively; d biochanin A: biochanin A 7-O-glucoside and reaction product P5 shown in square dot and solid line
The HPLC analysis of reaction product of naringenin P1 had a 9.8-min retention time and UV spectrum similar to those of naringenin 7-O-glucoside, and the reaction product P2 had a 5.5-min retention time and compared by hypsochromic shift. The reaction product of kaempferol, genistein, and biochanin A P3, P4, and P5 had a 9.5-, 11.2-, and 14.5-min retention time, respectively, and UV spectrum similar to those of the kaempferol 3-O-glucoside, genistein 7-O-glucoside, and biochanin A 7-O-glucoside, respectively (Fig. 3). In case of baicalein (data not shown), it gives the reaction product baicalin P7 and P8 and has a 9.75- and 9.6-min retention time similar to baicalein 7-O-glucuronide and baicalein 7-O-glucoside, respectively. Likewise, the reaction products of apigenin daidzein and isorhamnetin were determined by HPLC to be an apigenin 7-O-glucoside, daidzein 7-O-glucoside, and isorhamnetin 3-O-glucoside (data not shown).
In case of 3-hydroxy flavone (3-HF) which has the single OH group at the C-3 position of B-ring, the reaction product P9 had a 6.3-min retention time (chromatogram not shown) and determined by LC-MS (Fig. 5), which shows the mass of unused substrate with the majority of hydrogen ion (m/z 239.198 [M + 1] +) and the reaction product with majority of sodium ion (m/z 423.386 [M + 23]+). In addition, the K m and V max values (Table 2) of UGT73A16 for naringenin and genistein were determined at pH 8.0 and 37 °C. According to k cat/K m ratio that reflects the enzyme catalytic efficiency, UGT73A16 used naringenin most efficiently although a high enzymatic affinity towards genistein was observed. The relative activity of UGT73A16 was examined with six different flavonoids. The best substrates were naringenin and genistein followed by others (Table 2).
Biotransformation Analysis
To investigate the production of glucosides by recombinant E. coli, we incubated induced cell cultures of E. coli BL21 (DE3) carrying the recombinant UGT73A16 with equal concentration of each substrates for different times. The medium and cell pellet were extracted with ethyl acetate, and products were analyzed by HPLC. More than 95 % of glucoside products were released into the medium extract than the cell pellet extract. The expected glucosides were produced and could be detected in the culture medium after 12 h (Fig. 6, Table 3). Throughout a time course of 24 h, the bulk of aglycone was converted to its specific glucoside and the glucoside product yield was about 7–21 mg L−1 culture medium with conversion rates from 28 to 80 % (Table 3). Most of the conversion occurs during first 24 h (Fig. 6).
Discussion
In this study, a UGT73A16 gene from W. somnifera was isolated and cloned, which exhibited strong sequence similarity with other plant GTs. Phylogenetic tree analysis of UGT73A16 (Fig. 1) showed that it is highly similar to the glycosyltransferases of Lycopersicum esculentum and Scutellaria baicalensis which accept only UDP-glucose as preferred sugar donor and flavonoids as acceptor molecules. The functional characterization of UGT73A16 was confirmed and exhibited a wide range of substrate specificity towards flavonoids. Generally, the position of glycosylation depends upon the presence of double bond between C2 and C3 and the presence of 3-OH group, in aglycone ring. UGT73A16 transferred a glucose molecule to the hydroxyl group of C-3, C-7, or C-4'. When both C-7 and C-4' were present as seen in apigenin and naringenin, the main compound was determined by the presence of C-7 hydroxyl group. In case of flavonols (kaempferol, isorhamnetin, and 3-HF), only C-3 glycosylated product was observed; on the other hand, flavones, flavonones, and iso-flavones gave the C-7 glycosylated product. In summary, UGT73A16 produced 7-O-glucoside product with flavones, flavonones, and iso-flavones and 3-O-glucoside with flavonols. Phylogenetic tree revealed that UGT73A16 is a member of cluster I, suggesting that this enzyme might be a flavonoid 7-O-glucosyltransferase with broad acceptor specificity. HPLC analysis showed that the reactions of UGT73A16 with flavonoids yielded a single transfer product except naringenin, which yielded two transfer products (naringenin 7-O-glycoside and naringenin 4′-O-glycoside).
All major parts of W. somnifera such as the roots, fruits, and leaves provide high content of polyphenols including flavonoids and antioxidant activities with the leaves containing the highest amounts of polyphenols, especially catechin with strong antioxidant properties [29]. In contrast to catechin, the endogenous levels of flavonoid aglycone have also been reported by Alam et al. [29]. However, flavonoid glycosides have not been reported, so far, from W. somnifera. In case of potato, tomato, and tobacco, flavonoid glycosides have been reported [30–32]. Similar compounds may also exist in W. Somnifera, and it will be worthwhile to decipher the pattern of endogenous substrates for UGT73A16.
Plant-derived glucosides have attracted great attention due to their widespread applications. The isolation and purification of significant amount of specific flavonoid glucosides from plant source is time consuming since plants contain wide and variable spectra of glucosides with different types of sugar attachment [33]. On the other hand, chemical synthesis of specific glucoside is notoriously complicated. The flavonols aglycone has five potential glycosylation sites, and to synthesize any single monoglucoside, four other hydroxyl groups must be blocked [34, 35]. Thus, the sequential blocking and deblocking of the hydroxyl groups for regioselective glycosylation is a time-consuming and complicated process, and the presence of some contamination is also found [34]. The purpose of this study was to explore the utility of plant UGTs as regioselective biocatalyst.
Nucleotide-activated sugar is the essential component in small molecule glycosylation. One of the major problems for large-scale application of glycosylation is the provision of UDP-glucose to the in vitro system. Although different methods have been reported such as chemical method, enzymatic process [34, 35] and synthesis of nucleoside diphosphate sugars using UDP-glucose pyrophosphorylase and pyrophosphatase. All of these approaches are very laborious, require sugar phosphate (phosphoenolpyruvate and 5′ triphosphate), and thus involve high cost. As usual, it is well known that UDP-glucose participate in bacterial cell wall synthesis and thus glycosyltransferase-engineered bacteria should be an efficient system to overcome the complexity associated with the synthesis of sugars. On basis of genetically engineered E. coli, many studies have been reported for the production of flavonoid glucoside [15, 19, 36].
Due to its broad range of substrate and low regioselectivity, UGT73A16 may be an attractive enzyme for engineering flavonoid diversity. It will also be interesting to determine if UGT73A16 can glycosylate other structurally related and chemically synthesized compounds because it glycosylated the 3-hydroxy flavone, which is a chemically synthesized compounds. To the best of our knowledge, UGT73A16 is a first ever isolated glycosyltransferase gene from W. somnifera which transfer the glucose molecules towards chemically synthesized compounds.
Conclusion
In conclusion, this study provides evidence that UGT73A16 is a typical UDP-dependent glycosyltransferase and shows broad range substrate specificity towards flavonoids. It uses flavonoids as a glucose acceptor and preferentially transfers a glucose group to the 7 - and 3-hydroxyl group of flavonoids, and this enzyme produces a single glycosylation product except in case of naringenin, where two glycosylation products were seen. It also transfers glucose molecule towards 3-hydroxy flavone which is a chemically synthesized compound. The recombinant UGT73A16 used as whole cell biocatalyst in E. coli for enhanced production of glucosides. These results indicate that UGT73A16 can glycosylate the other valuable, natural, and chemically synthesized compounds by the way of either enzymatic modification or biotransformation.
Abbreviations
- IPTG:
-
Isopropyl-β-d-1-thiogalactopyranoside
- UGT:
-
Uridine diphosphate-dependent glycosyltransferase
- WsUGT:
-
Withania somnifera uridine diphosphate glycosyltransferase
References
Wink, M. (1999). Annual Plant Reviews, 2, 358.
Schwab, W. (2003). Phytochemistry, 62, 837–849.
Heller, W., & Forkmann, G. (1994). In The Flavonoids. Advances in Research since 1986, 499–535
Bowles, D., Isayenkova, J., Lim, E. K., & Poppenberger, B. (2005). Current Opinion in Plant Biology, 8, 254–263.
Bowles, D., Lim, E. K., Poppenberger, B., & Vaistij, F. E. (2006). Annual Review of Plant Biology, 57, 567–597.
Jones, P., & Vogt, T. (2001). Planta, 213, 164–174.
Noguchi, A., Saito, A., Homma, Y., Nakao, M., Sasaki, N., Nishino, T., et al. (2007). Journal of Biological Chemistry, 282, 23581–23590.
Vogt, T., & Jones, P. (2000). Trends in Plant Science, 5, 380–386.
Cornwell, T., Cohick, W., & Raskin, I. (2004). Phytochemistry, 65, 995–1016.
Usha, S., Johnson, I. M., & Malathi, R. (2005). Journal of Biochemistry and Molecular Biology, 38, 198–205.
Middleton, E. J., Kandaswami, C., & Theoharides, T. C. (2000). Pharmacological Reviews, 52, 673–751.
Hirotani, Kuroda, M., Suzuki, R. H., & Yoshikawa, T. (2000). Planta, 210, 1006–1013.
Kim, J. H., Kim, B. G., Ko, J. H., Lee, Y., Hur, H. G., Lim, Y., et al. (2006). Plant Science, 170, 897–903.
Kramer, C. M., Prata, R. T., Willits, M. G., Luca, D. V., Steffens, J. C., & Graser, G. (2003). Phytochemistry, 64, 1069–1076.
Willits, M. G., Giovanni, M., Prata, R. T., Kramer, C. M., Luca, D. V., Steffens, J. C., et al. (2004). Phytochemistry, 65, 31–41.
Rice-Evans, C. A., Miller, N. J., & Paganga, G. (1996). Free Radical Biology & Medicine, 20, 933–956.
Deavours, B. E., & Dixon, R. A. (2005). Plant Physiology, 138, 2245–2259.
Leonard, E., Chemer, J., Lim, K. H., & Koffase, M. A. G. (2005). Applied Microbiology and Biotechnology, 15, 1–7.
Lim, E. K., Ashford, D. A., Hou, B. K., Jackson, R. G., & Bowles, D. J. (2004). Biotechnology and Bioengineering, 87, 623–631.
Xian-Zhi, H., Xiaoqiang, W., & Richard, A. D. (2006). Journal of Biological Chemistry, 281, 34441–34447.
Yu, O., Shi, J., Hession, A. O., Maxwell, C. A., McGonigle, B., & Odell, J. T. (2003). Phytochemistry, 63, 73–76.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). Molecular Biology and Evolution, 24, 1596–1599.
Saitou, N., & Nei, M. (1987). Molecular Biology and Evolution, 4, 406–425.
Qing-hu, M. (2007). Journal of Experimental Botany, 58, 2011–2021.
Mackenzie, P. I., Bock, K. W., Burchell, B., Guillemette, C., Ikushiro, S., Iyanagi, T., et al. (2005). Pharmacogenetics and Genomics, 15, 677–685.
Mabry TJ, M. K., Thomas MB. (1970). New York: Springer-Verlag, 354
Quiel, J. A., & Bender, J. (2003). Journal of Biological Chemistry, 278, 6275–6281.
Vogt, T., Grimm, R., & Strack, D. (1999). The Plant Journal, 19, 509–519.
Alam, N., Hossain, M., Khalil, M. I., Moniruzzaman, M., Sulaiman, S. A., & Siew, H. G. (2011). BMC Complementary and Alternative Medicine, 11, 65.
Harborne, J. B. (1962). Biochemical Journal, 84, 100–106.
Gweanae, I., Susan Dupont, M., Fred, A. M., Adrienne, L. D., Geoff, J. C., Martine, E. V., et al. (2003). Journal of Agricultural and Food Chemistry, 51, 2438–2446.
Watanabe, R., & Wender, S. H. (1965). Archives of Biochemistry and Biophysics, 112.
Harborne, J. B., & Baxter, H. (1999). Wiley, New Yark
Bouktaib, M., Atmani, A., & Rolando, C. (2002). Tetrahedron Letters, 43, 6263–6266.
Li, M., Han, X., & Yu, B. (2002). Tetrahedron Letters, 43, 9467–9470.
Xian, Z. H., Wen, S. L., Jack, W. B., & Richard, A. D. (2008). Applied Microbiology and Biotechnology, 80, 253–260.
Acknowledgments
This work was supported by grants from Council of Scientific and Industrial Research—Network Project (CSIR-NWP0009), India. We are grateful to Dr. Shrinivas Hotha for providing LC-MS facility and to his student Ashif Shaikh for the help in LC-MS experiment and analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, S., Vishwakarma, R.K., Kumar, R.J.S. et al. Functional Characterization of a Flavonoid Glycosyltransferase Gene from Withania somnifera (Ashwagandha). Appl Biochem Biotechnol 170, 729–741 (2013). https://doi.org/10.1007/s12010-013-0230-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-013-0230-2