Abstract
In this work, the high-level expression of the lipase r27RCL was achieved by optimization of the lipase gene copy number in the host strain Pichia pastoris. The copy number of the lipase gene proRCL from Rhizopus chinensis CCTCC M201021 was quantified by real-time quantitative polymerase chain reaction and a range of Mut+ P. pastoris strains carrying one, three, five, and six copies of proRCL were obtained. The maximum lipase activity was achieved at 12,500 U/mL by the five-copy recombinant strain after 96 h of methanol induction in the 7-L fermenter. However, the enzyme activity of the six-copy recombinant strain decreased remarkably. By transcription analysis of proRCL, ERO1, and PDI, it suggested that unfolded protein response seemed to be triggered in the highest copy recombinant strain after 24 h. Thus, elaborate optimization of foreign gene dosage was very important for the high-level expression of foreign proteins in P. pastoris.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipases (triacylglycerol ester hydrolases EC 3.1.1.3) are well-known hydrolases capable of hydrolyzing the ester bonds of water-insoluble substrates at the interface between substrate and water, which have been widely applied in many industries, such as oleochemical industry (hydrolysis, esterification, or transesterification), pharmaceutical industry, food industry (especially as the additive in the bread baking process), feed industry, and so on [1, 2]. Lipases from Rhizopus sp. exhibit wide applications in industries, such as esterification of docosahexaenoic acid (DHA), biodiesel–fuel production [3, 4], interesterification of olive oil with palmitic acid [5], and synthesis of propylene glycol based nonionic detergents [6]. All these applications in these industries require large quantities of enzymes, which makes the high-level expression of lipases in heterologous systems an important issue.
Over the last few decades, Pichia pastoris expression system has been used successfully for production of various recombinant heterologous proteins. This process offers several advantages, including the alcohol oxidase 1 (AOX1) gene promoter being tightly regulated by methanol, easy growth to high cell densities, high levels of protein expression at the intra- or extracellular level, and the ability to perform eukaryotic protein modifications [7–9]. Many microbial lipases have been successfully expressed in P. pastoris, such as Rhizopus oryzae lipase [10, 11], Candida rugosa lipase [12], Candida parapsilosis lipase/acyltransferase [13], and Candida antarctica lipase B [14]. For improvement of the expression level of foreign proteins in P. pastoris, previous studies mainly focused on the optimization of the fermentation process [15, 16]. However, high-level expression in P. pastoris usually faces some potential bottlenecks, such as limitations in gene dosage, mRNA transcription, protein translation, translocation, or secretion, which could not be solved solely by the fermentation process control. Recently, molecular technology by genetic modification was proved to be one of the most useful and effective method for high-level expression [17]. The lipase production and the enzyme activity of lip2 from Aspergillus niger were increased by 5.3- and 11.6-fold through the codon optimization, respectively [18]. Many researches have shown that gene dosage of foreign protein is one of the key factors for recombinant protein production [19–21]. For instance, increasing the lipase gene copy number of the lip2 from Yarrowia lipolytica increased the protein expression level by twofold [22]. The expression level of GDS(L)-lipase from Streptomyces rimosus was increased by 22-fold compared with that of the original strain by optimization of gene copy number in P. pastoris [23]. Usually, increasing the target gene copy number could dramatically enhance the production of foreign protein in P. pastoris. But in some cases, such as human trypsinogen [24] and Na-ASPI [25], increased gene dosage leads to the reduction of protein expression level. Nevertheless, there was not a sufficient mechanistic investigation about the effects of gene copy number on the protein production in P. pastoris.
In our previous study, R. chinensis CCTCC M201021 screened from Daqu of brewing strong aromatic Chinese spirits showed a high potential for industrial usage, including synthesis of eicosapentaenoic acid and DHA, sorbitan oleate, ethyl esters, and biodiesel [26–30]. And from this strain, the lipase gene proRCL was cloned and successfully expressed in P. pastoris named as r27RCL [31]. In 7-L fermentor, the highest lipase activity reached 2,130 U/mL by optimization of methanol concentration over the production phase [32]. However, the expression level of the lipase r27RCL still could not satisfy the needs of the industrial applications.
Due to the higher specific activity of r27RCL than that of mRCL, the full lipase gene proRCL was expressed in P. pastoris [31]. In this work, the high-level expression of the lipase r27RCL was achieved by optimization of the lipase gene copy number in the host strain P. pastoris. And the transcriptional levels of the target gene proRCL, as well as chaperone genes in the recombinant strains were analyzed, aiming to understand the mechanism of the effects of gene copy number on the lipase expression in P. pastoris.
Materials and Methods
Enzyme and Reagent
Restriction enzyme, T4 DNA ligase, Taq DNA polymerase, polymerase chain reaction (PCR) reagent (TaKaRa Biotechnology (Dalian) Co., Ltd.), primers (SBS Gene Technology (Shanghai) Co., Ltd.), DSTM2000 DNA Ladder Maker, E5000 DNA Ladder Maker, E15000 DNA Ladder Maker (Dongsheng Biotechnology (Guangzhou) Co., Ltd.), Gel Extraction Kit, PCR Purification Kit (Bioflux), Plasmid Mini Kit I (OMEGA BIO-TEK), SsoFast™ EvaGreen® Supermix manual (Bio-Rad), and fluorescence quantification tubes (Bio-Rad), and other analytically pure reagent is purchased from inland or foreign.
Strains, Plasmids, and Medium
P. pastoris GS115 and plasmid pPIC9K from Invitrogen BV was used as the host strain and the vector, respectively. Recombinant plasmid pPIC9K-proRCL was constructed by Yu et al. [31]. Yeast nutrient medium YPD, BMGY, BMMY, and YPD-G418 are prepared by means of “P. pastoris expression Kit” (Pichia Multi-Copy Expression Kit, version A, Invitrogen BV, The Netherlands).
Transformation of P. pastoris and Selection of Recombinants with Different Gene Copy Number
The lipase gene proRCL existed at the downstream of the α-factor section signal from Saccharomyces cerevisiae for the secreted expression. The plasmids pPIC9K-proRCL and pPIC9K linearized by restriction enzyme Sal І were transformed into P. pastoris GS115 by electroporation according to the Invitrogen Protocols (Pichia Multi-Copy Expression Kit, version A, Invitrogen BV, The Netherlands). Competent cells were prepared under the way optimized by Wu et al. [33]. All the transformants were spread on YPD-G418 agar plates with 0.5, 1, 1.5, and 2 mg mL−1 G418, respectively, and incubated at 30 °C for 2–5 days.
Extraction of P. pastoris Genomic DNA
Genomic DNA used for RT-QPCR was prepared by Hoffman et al. reported previously [34].
Design of Primers Set for RT-QPCR
All the primers used in this work were listed in Table 1. Genomic DNA and cDNA were used as the templates in real-time quantitative PCR (RT-QPCR), respectively. The reaction conditions had been established as recommended by SsoFast™ EvaGreen® Supermix manual (Bio-Rad). Each 20 μL reaction contained 10 μL 2× SsoFast EvaGreen Supermix, 0.5 μL 20 μM forward and reverse primers, 1 μL sample genomic DNA or cDNA, and 8 μL sterile deionized water. All RT-QPCR reactions were run in triplicate on MJ chromo4 (MJ, America) using the following program: 98 °C 2 min, 40 cycles of 98 °C for 5 s, and 50 °C for 5 s. The specificity of amplicons was verified by analyzing the melting curve after 40 cycles.
Standard Curve Establishment and Gene Copy Number Quantification
The promoter of alcohol oxidase gene AOX1 (the target gene PAOX1) exists in plasmid pPIC9K and P. pastoris genome simultaneously. The copy number of PAOX1 minus one equals the copy number of the lipase gene proRCL. RT-QPCR data were normalized using GAPDH gene as the endogenous control (reference gene). Genomic DNA of GS115 Strain was employed to establish the standard curve. Briefly, a tenfold serial dilution series of the GS115 genomic DNA ranging from 1 × 100 to 1 × 10−4 dilution was utilized as the templates to establish the standard curves for PAOX1 and GAPDH. The C T value in every sample was measured thrice using a RT-QPCR with the PAOX1 and GAPDH primer sets, and then plotted against the logarithm of the relative template concentration. For analyzing the copy number of the target gene in different recombinant strains, data were treated using the Pfaffl method [35], and the copy number of the target gene and the reference gene in GS115 strain were set as the calibrator to normalize the data (as shown in Table 2 and Formula (1)).
7-L Fermenter Fermentation
The 7-L fermentation experiments were performed as reported previously [32]. The culture was incubated at 28 °C with a 2.8 L volume in a 7-L bioreactor (New Brunswick, BioFlo110, Edison, NJ, USA). The inocula were grown for 18 h at 28 °C in shake flasks at 250 rpm with BMGY medium. In the glycerol batch phase, 200 mL of inoculum was directly added into 2.6 L of a fermentation basal salts medium (40 g/L glycerol, 22.7 g/L H3PO4, 0.93 g/L CaSO4, 18.2 g/L K2SO4, 14.9 g/L MgSO4 · 7H2O, 4.13 g/L KOH, and 7.0 g/L K2HPO4) and trace solution (12 mL). Trace solution consisted of 6 g/L CuSO4 · 5H2O, 0.08 g/L NaI, 3.0 g/L MnSO4 · H2O, 0.2 g/L Na2MoO4 · 2H2O, 0.02 g/L H3BO3, 0.5 g/L CoCl2, 20 g/L ZnCl2, 65 g/L FeSO4 · 7H2O, 0.2 g/L biotin, and concentrated sulfuric acid (0.5 % v/v). The medium was sterilized by filtration. The pH of the medium was adjusted and controlled at 5.0 with the addition of 28 % (v/v) ammonium hydroxide. Dissolved oxygen (DO) concentration was constantly maintained between 20 and 60 % saturation and controlled in cascade mode. Aeration was constantly maintained at 1.0 vvm and pure oxygen was supplied as needed. The agitation rate maintained between 300 and 700 rpm.
The fermentation was conducted during the glycerol batch phase at 28 °C until all of the glycerol was consumed, which was indicated by a sharp increase in DO. The process then proceeded to a glycerol fed-batch phase at 28 °C, with feeding with 50 % (v/v) glycerol containing 1.2 % (v/v) trace solution at the average rate of 12.4 g/L h−1. When needed, glycerol feeding rates were adjusted to control DO. After the desired biomasses were reached, the methanol fed-batch phase was initiated at 28 °C, during which the culture was supplied with 100 % (v/v) methanol containing 1.2 % (v/v) trace solution, and the methanol concentration was controlled at 0.1 % by an online methanol analyzer (FC2002, Super-Xinxi, Shanghai, China).
Cell Concentration
Dry cell weight (DCW) of the cell suspension was determined by method previously reported [32]. DCW of the cell suspension was determined by centrifugation of 5 mL cell broth in a pre-weighed centrifuge tube, followed by washing with distilled water and drying to constant weight at 80 °C in an oven.
Lipase Activity Determination
The lipase activity in supernatant was assayed by the method constructed previously [36]. One volume of a 1.08 mM solution of pNPP in 2-propanol was mixed just prior to use with nine volumes of 50 mM Tris–HCl buffer pH 8.0, containing 4 g/L Triton X-100 and 1 g/L arabic gum. The standard reaction was started by pre-equilibration of 2 mL of the above mixture at 37 °C and addition of 0.1 mL of enzyme solution at an appropriate dilution in 50 mM pH 8.0 Tris–HCl buffer. The variation of the absorbance at 410 nm of the assay against a blank without enzyme was monitored for 2–5 min using a UV–vis spectrophotometer (UNICO UV-3102 PC, China). One enzyme unit was defined as the amount of enzyme releasing 1 μmol of p-nitrophenol per minute under the assay conditions. One unit of enzymatic activity was defined as the amount of enzyme releasing 1 μmol of fatty acid per minute under the assay conditions. All the assays were conducted in triplicate and significant differences (p < 0.05) were measured.
RNA Extraction and cDNA Synthesis
During the fermentation of recombinant strains, 1 mL fermentation broth was taken and centrifuged to get the cells every 24 h. Total RNA was extracted by Yeast RNAiso Kit (TaKaRa Bio Co., Ltd). RNA integrity was tested in 1.2 % agarose gels and its concentration measured by densitometry and by 260/280 nm absorbance ratio. Five hundred nanograms of total RNA was subjected to reverse transcription using PrimeScript® RT reagent Kit (TaKaRa Bio Co., Ltd). The reaction was terminated by heating at 85 °C for 5 s.
Transcription Levels of Intracellular Relevant Genes
The transcription levels of proRCL, ERO1, and protein disulfide isomerase (PDI) in all constructed recombinant strains with different gene copy number were normalized using GAPDH gene as the endogenous control (housekeeping gene). In this case, the transcription levels of lipase gene and chaperone genes in XY RCL-0 strain were set as control to normalize the data. RT-QPCRs were run in triplicate with biological replicates to allow for statistical confidence in differential gene expression.
Results and Discussion
Establishment of RT-QPCR Approach for Copy Number Determination
The traditional methods of gene copy number determination in P. pastoris system are mainly Southern blotting and dot blotting, which are laborious and time consuming [25, 37]. RT-QPCR has been developed into an important and widely used analytical tool [38], which could be divided into two kinds of quantification method, absolute and relative quantification method. Genomic DNA concentration of each sample is not necessarily known accurately in the relative quantification method, and the gene copy number is determined by the ratio of copy number of target gene and reference gene [39]. Previous methods on RT-QPCR mainly focused on the expression analysis in animal, bacterial, and plant cells, while only a few reports were concerned with gene copy number determination in P. pastoris [19, 40–44].
In our study, a modified relative RT-QPCR method was established to quantify the copy number in all constructed strains. Genomic DNA of GS115 strain and the housekeeping gene GAPDH are used as an external standard and the reference gene, respectively. The standard curves for PAOX1 and GAPDH (Fig. 1) were established using 1 × 100 to 1 × 10−4 dilutions of the genomic DNA of GS115 strain. Figure 1 showed that the standard curves of target gene and reference gene were linear in the tested range and the amplification efficiencies were 102 % and 95 %, respectively. A single peak melting curve suggested that the PCR products were single (data not shown). When amplification efficiencies of target gene and reference gene are equal, 2−ΔΔCT is the best relevant quantification method. But if the amplification efficiencies are different between them, the Pfaffl method was selected to determine the copy number of the target gene in the recombinant strains [35]. The PAOX1 copy number was then determined by measuring the copy number ratio of PAOX1 to GAPDH, which has only one copy in the genome of P. pastoris GS115. Zhu et al. [19] also reported a method of RT-QPCR for determining the copy number of PIP gene (the foreign protein gene) in P. pastoris transformants. Different from the above RT-QPCR method, the copy number of lipase gene (the foreign protein gene) was measured according to the copy number of the promoter sequence of gene AOX1 in this study. Thus, it is a more convenient and more widely applicable method for determination of copy number in any system using a promoter sequence PAOX1.
Effect of Gene Dosage on Lipase r27RCL Production
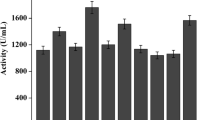
Using RT-QPCR, we selected a series of recombinant strains (each copy of recombinant strains were set as duplicate samples) with one, three, five, and six copy number of lipase gene from the G418 agar plates to evaluate the effects of proRCL dosage on the lipase expression level and designated them as XY RCL-1 (one copy), XY RCL-3 (three copies), XY RCL-5 (five copies), and XY RCL-6 (six copies), respectively. XY RCL-0 was used as the control which was inserted by the linearized plasmid pPIC9K. As shown in Fig. 2a, the DCW of XY RCL-6 was lower than that of XY RCL-1, XY RCL-3 and XY RCL-5 after 48 h. For strains with less than five lipase gene copies, the enzyme activities were positively correlated with the gene dosage. The lipase enzyme activity reached the maximum value of 12,500 U/mL at 96 h in XY RCL-5, which had an increase of 6.2- and 1.8-fold over those of the recombinant strains XY RCL-1 and XY RCL-3, respectively. But the enzyme activity decreased in XY RCL-6 (Fig. 2b). Extracellular protein concentration of XY RCL-3 and XY RCL-5 showed an increase of approximately 1.22 and 2.56 times over that of the single copy strain XY RCL-1 at 96 h, respectively. Nevertheless, the protein expression level of XY RCL-6 was very similar to that of XY RCL-5 (Fig. 2c). The specific activity of the supernatant of the recombinant strain XY RCL-5 reached 1,562,500 U g−1, which was much higher than that of the recombinant strain XY RCL-6. The sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of the supernatant of these recombinant strains was shown in Fig. 3. Furthermore, the lipase gene copy number in all these recombinant strains was genetically stable during the fermentation process, which were identified by the RT-QPCR. In brief, the enzyme activity, the protein quality and the protein concentration were extremely improved by the optimization of the gene dosage.
The fermentation properties of the recombinant strains with increased gene dosage in the 7-L fermenter. a The growth curves. b The enzyme activity curves. c The extracellular protein concentration curves. Filled square XY RCL-0, filled circle XY RCL-1, filled triangle XY RCL-3, filled inverted triangle XY RCL-5, filled diamond XY RCL-6
Several Rhizopus lipases have been successfully expressed in the heterogenous hosts. The ROL (R. oryzae lipase) was functionally expressed and secreted in the P. pastoris, the enzyme activity of which reached 1,334 U/mL [45, 46]. The RNL (Rhizopus niveus lipase) was expressed in the yeast S. cerevisiae, the specific activity of which reached 870,000 U g−1 [47]. R. arrhizus lipase production reached a maximum lipase activity of 315 U/mL by Tan et al. [48, 49]. In our study, the activity of the lipase r27RCL was dramatically improved by optimization of the gene dosage. The highest lipase activity and protein concentration reached 12,500 U/mL and 8 g/L by the recombinant strain XY RCL-5, respectively. In addition, the DCW and the enzyme activity of XY RCL-6 were both lower than those of XY RCL-5, and the protein concentration of XY RCL-6 was similar to that of XY RCL-5. Li et al. also reported that increasing gene copy number lead to lower biological activity of foreign protein in P. pastoris [50]. One possible explanation is due to the metabolism stress and the misfolded protein in XY RCL-6 which harbors high gene copy number. Therefore, we further explored the relationship between the gene copy number and the transcription levels of intracellular relevant genes.
Transcription Levels of the Lipase Gene proRCL during the Fermentation Process
As shown in Fig. 4, the copy number of the lipase gene proRCL affected the transcription level of proRCL significantly. The transcription level of proRCL increased with the increase of gene dosage at 24 h. The transcription level of proRCL in strains XY RCL-1, XY RCL-3, and XY RCL-5 reached the highest value at 48 h (Fig. 4). However, the transcription level in XY RCL-6 reached the maximum, then decreased gradually and was lower than that in XY RCL-5 after 24 h. The transcription level of lipase gene in XY RCL-6 was lower than that in XY RCL-5 after 24 h (Fig. 4), which may be due to the increased metabolism stress in XY RCL-6. Consistent with the former, the enzyme activity and the specific activity in XY RCL-6 were also lower than that in XY RCL-5 (Figs. 2b and 3), which may be also due to the limitation of the transcription level of proRCL or other limitations in protein synthesis, such as translation, folding, translocation, secretion, and so on. Thus, it is vitally necessary to investigate the transcription levels of relevant chaperones genes related to the protein synthesis in vivo.
Transcription Levels of Chaperone Genes ERO1 and PDI during the Fermentation Process
Through the homologous modeling, there are three disulfide bonds in the structure of lipase r27RCL (data not shown). As a multifunctional protein, PDI is critical to the protein-folding process, especially to the disulfide bond formation in the endoplasmic reticulum (ER). Furthermore, Ero1p (ER oxidoreduction) oxidizes PDI and then transfers disulfide bonds to substrate proteins [51–53]. Figure 5 showed the relative transcription levels of the two important chaperones located in the ER, ERO1p, and PDI in these recombinant strains. The transcription level of ERO1 was similar in XY RCL-0, XY RCL-1, XY RCL-3, and XY RCL-5 during the whole fermentation process, but it was increased gradually in XY RCL-6 after 24 h and was increased by 67 % at 96 h (Fig. 5). The transcription level of PDI showed a similar trend in XY RCL-0, XY RCL-1, XY RCL-3, and XY RCL-5, but it was even higher (increased by 3.7-fold) in XY RCL-6 at 96 h (Fig. 5).
In eukaryotic cells, most secreted and trans-membrane proteins fold and mature in the lumen of the ER. Proteins enter the ER as unfolded polypeptide chains. Overexpression of foreign proteins in P. pastoris will trigger unfolded protein response (UPR). The induction of UPR is disadvantageous to cell survival. An imbalance (called ER stress) between the load of unfolded proteins that enter the ER and the capacity of the cellular machinery that handles this load sets three main responses in motion, which include a reduction in the protein load that enters the ER, an increase in the capacity of the ER to handle unfolded proteins which means transcriptional activation of chaperone genes (such as ERO1, PDI, and so on), and cell death triggered [54, 55]. David et al. also predicted that when ROL was overexpressed, UPR seemed to be triggered in P. pastoris by the analysis of transcriptional levels of the genes Kar2 and PDI involved in protein synthesis and secretion [56]. In our study, the transcription level of proRCL in XY RCL-6 was lower than that in XY RCL-5 after 24 h induction and the transcription levels of chaperone genes in XY RCL-6 were much higher than that in XY RCL-1, XY RCL-3, and XY RCL-5 at 48, 72, and 96 h (Fig. 5), indicating that the cell physiology and metabolism were changed and the so-called UPR was probably triggered (The transcription level of chaperone genes will not be dramatically increased under the normal status). The lower extracellular lipase enzyme activity and the decreased specific activity in XY RCL-6 after 24 h further confirmed that the yeast cells probably suffered from remarkable oxidative stress resulted from the folding of a large flux of r27RCL and large amounts of unfolded or misfolded r27RCL accumulated in the ER. At first, we wanted to detect the unfolded or misfolded r27RCL in vivo. However, the enzyme activity was extremely low in vivo and there was no obvious band of lipase r27RCL by SDS-PAGE analysis. Misfolded or aggregated proteins in the ER are recognized by the strict quality control (QC) system, which leads to binding of the proteins by the BiP complex and eventual redirection to the cytosol for degradation, namely ER-associated protein degradation [57]. Thus, the misfolded lipase r27RCL in the ER would probably be degraded quickly under the strict QC in the six-copy recombinant strain and the very low amount of lipase r27RCL in vivo could not be detected by SDS-PAGE. Through the transcription analysis of chaperones (Fig. 5), it suggested that UPR would not be triggered until reaching some extent (more than five copies in our case) when expressing the foreign genes in P. pastoris. Therefore, the foreign gene copy number in P. pastoris has a great influence on foreign protein production and intracellular metabolism status. And so as not to trigger UPR, elaborate optimization of foreign gene dosage is greatly important for the high level expression of foreign protein in P. pastoris.
Conclusions
In our study, a modified method for quantification of gene copy number in P. pastoris was established by RT-QPCR, which could be applied to determine the gene copy number in any system using the AOX1 promoter sequence in P. pastoris. A range of Mut+ recombinant strains containing proRCL gene dosage of one, three, five, and six copies were generated. The enzyme activity reached 12,500 U/mL by the five-copy recombinant strain XY RCL-5. Then, the investigation of the relationship between gene dosage and the transcription levels of the relative genes (lipase gene proRCL and the UPR-related genes (PDI and ERO1)) showed that the foreign gene dosage in P. pastoris had a great influence on the protein expression quality and the intracellular metabolism status. The UPR was also possibly triggered when the lipase gene copy number increased to six. The high level expression of the lipase r27RCL would offer a greater value in the industries and the analysis of the transcription level of these relative genes provided a possible strategy for further improved lipase production through co-expression with relative chaperone genes.
References
Bornscheuer, U. T., Bessler, C., Srinivas, R., & Hari Krishna, S. (2002). Trends in Biotechnology, 20, 433–437.
Reetz, M. T. (2002). Current Opinion in Chemical Biology, 6, 145–150.
Hama, S., Yamaji, H., Fukumizu, T., Numata, T., Tamalampudi, S., Kondo, A., Noda, H., & Fukuda, H. (2007). Biochemical Engineering Journal, 34, 273–278.
Kaieda, M., Samukawa, T., Matsumoto, T., Ban, K., Kondo, A., Shimada, Y., Noda, H., Nomoto, F., Ohtsuka, K., & Izumoto, E. (1999). Journal of Bioscience and Bioengineering, 88, 627–631.
Komatsu, T., Nagayama, K., & Imai, M. (2005). Journal of Chemical Engineering of Japan, 38, 450–454.
Rodrigues, R. C., & Fernandez-Lafuente, R. (2010). Journal of Molecular Catalysis B: Enzymatic, 64, 1–22.
Cos, O., Ramon, R., Montesinos, J.L., & Valero, F. (2006). Microbial Cell Factories, 5.
Valero, F. (2012). Methods in Molecular Biology (Clifton, NJ), 861, 161–178.
Macauley-Patrick, S., Fazenda, M. L., McNeil, B., & Harvey, L. M. (2005). Yeast, 22, 249–270.
Minning, S., Serrano, A., Ferrer, P., Sola, C., Schmid, R. D., & Valero, F. (2001). Journal of Biotechnology, 86, 59–70.
Resina, D., Serrano, A., Valero, F., & Ferrer, P. (2004). Journal of Biotechnology, 109, 103–113.
Brocca, S., Schmidt-Dannert, C., Lotti, M., Alberghina, L., & Schmid, R. D. (1998). Protein Science, 7, 1415–1422.
Brunel, L., Neugnot, V., Landucci, L., Boze, W. N., Moulin, G., Bigey, F., & Dubreucq, E. (2004). Journal of Biotechnology, 111, 41–50.
Rotticci-Mulder, J. C., Gustavsson, M., Holmquist, M., Hult, K., & Martinelle, M. (2001). Protein Expression and Purification, 21, 386–392.
He, Y. Q., & Tan, T. W. (2006). Journal of Molecular Catalysis B: Enzymatic, 43, 9–14.
Surribas, A., Stahn, R., Montesinos, J., Enfors, S., Valero, F., & Jahic, M. (2007). Journal of Biotechnology, 130, 291–299.
Idiris, A., Tohda, H., Kumagai, H., & Takegawa, K. (2010). Applied Microbiology and Biotechnology, 86, 403–417.
Yang, J., & Liu, L. (2010). Journal of Molecular Catalysis B: Enzymatic, 63, 164–169.
Zhu, T., Guo, M., Tang, Z., Zhang, M., Zhuang, Y., Chu, J., & Zhang, S. (2009). Journal of Applied Microbiology, 107, 954–963.
Karakas, B., Inan, M., & Certel, M. (2010). Journal of Molecular Catalysis B: Enzymatic, 64, 129–134.
Yu, H., Yan, X., Shen, W., Hong, Q., Zhang, J., Shen, Y., & Li, S. (2009). Current Microbiology, 59, 573–578.
Yu, M., Wen, S., & Tan, T. (2010). Engineering in Life Sciences, 10, 458–464.
Vujaklija, D., Schroeder, W., Abramic, M., Zou, P., Lescic, I., Franke, P., & Pigac, J. (2002). Archives of Microbiology, 178, 124–130.
Hohenblum, H., Gasser, B., Maurer, M., Borth, N., & Mattanovich, D. (2004). Biotechnology and Bioengineering, 85, 367–375.
Inan, M., Aryasomayajula, D., Sinha, J., & Meagher, M. M. (2006). Biotechnology and Bioengineering, 93, 771–778.
Sun, S. Y., Xu, Y., & Wang, D. (2009). Bioresource Technology, 100, 2607–2612.
Sun, S. Y., Xu, Y., & Wang, D. (2009). Journal of Chemical Technology and Biotechnology, 84, 435–441.
Xu, Y., Wang, D., Mu, X. Q., & Ni, Y. Q. (2003). Journal of the American Oil Chemists Society, 80, 647–651.
Xu, Y., Wang, D., Mu, X. Q., Zhao, G. A., & Zhang, K. C. (2002). Journal of Molecular Catalysis B: Enzymatic, 18, 29–37.
He, Q., Xu, Y., Teng, Y., & Wang, D. (2008). Chinese Journal of Catalysis, 29, 41–46.
Yu, X. W., Wang, L. L., & Xu, Y. (2009). Journal of Molecular Catalysis B: Enzymatic, 57, 304–311.
Wu, D., Yu, X. W., Wang, T. C., Wang, R., & Xu, Y. (2011). Biotechnology and Bioprocess Engineering, 16, 305–311.
Wu, S., & Letchworth, G. J. (2004). Biotechniques, 36, 152–155.
Hoffman, C. S., & Winston, F. (1987). Gene, 57, 267–272.
Pfaffl, M. W. (2001). Nucleic Acids Research, 29, 2002–2007.
Kordel, M., Hofmann, B., Schomburg, D., & Schmid, R. (1991). Journal of Bacteriology, 173, 4836–4841.
Pushnova, E. A., Geier, M., & Zhu, Y. S. (2000). Analytical Biochemistry, 284, 70–76.
D’Haene, B., Vandesompele, J., & Hellemans, J. (2010). Methods, 50, 262–270.
Abad, S., Kitz, K., Hormann, A., Schreiner, U., Hartner, F. S., & Glieder, A. (2010). Biotechnology Journal, 5, 413–420.
Hartner, F. S., Ruth, C., Langenegger, D., Johnson, S. N., Hyka, P., Lin-Cereghino, G. P., Lin-Cereghino, J., Kovar, K., Cregg, J. M., & Glieder, A. (2008). Nucleic Acids Research, 36, 1–15.
Schroer, K., Peter Luef, K., Stefan Hartner, F., Glieder, A., & Pscheidt, B. (2010). Metabolic Engineering, 12, 8–17.
Marx, H., Mecklenbrauker, A., Gasser, B., Sauer, M., & Mattanovich, D. (2009). Fems Yeast Research, 9, 1260–1270.
Williams, K. E., Jiang, J., Ju, J., & Olsen, D. R. (2008). Enzyme and Microbial Technology, 43, 31–34.
Norden, K., Agemark, M., Danielson, J. A., Alexandersson, E., Kjellbom, P., & Johanson, U. (2011). BMC Biotechnology, 11, 47.
Minning, S., Schmidt-Dannert, C., & Schmid, R. D. (1998). Journal of Biotechnology, 66, 147–156.
Minning, S., Serrano, A., Ferrer, P., Solá, C., Schmid, R. D., & Valero, F. (2001). Journal of Biotechnology, 86, 59–70.
Kohno, M., Enatsu, M., Yoshiizumi, M., & Kugimiya, W. (1999). Protein Expression and Purification, 15, 327–335.
Yang, X., Wang, B., Cui, F., & Tan, T. (2005). Process Biochemistry, 40, 2095–2103.
Li, D., Wang, B., & Tan, T. (2006). Journal of Molecular Catalysis B: Enzymatic, 43, 40–43.
Li, Z. G., Moy, A., Gomez, S. R., Franz, A. H., Lin-Cereghino, J., & Lin-Cereghino, G. P. (2010). Biochemical and Biophysical Research Communications, 402, 519–524.
Pollard, M. G., Travers, K. J., & Weissman, J. S. (1998). Molecular Cell, 1, 171–182.
Frand, A. R., & Kaiser, C. A. (1998). Molecular Cell, 1, 161–170.
Tu, B. P., Ho-Schleyer, S. C., Travers, K. J., & Weissman, J. S. (2000). Science, 290, 1571–1574.
Ron, D., & Walter, P. (2007). Nature Reviews Molecular Cell Biology, 8, 519–529.
Guerfal, M., Ryckaert, S., Jacobs, P.P., Ameloot, P., Van Craenenbroeck, K., Derycke, R., & Callewaert, N. (2010). Microbial Cell Factories, 9.
Resina, D., Bollók, M., Khatri, N. K., Valero, F., Neubauer, P., & Ferrer, P. (2007). Microbial Cell Factories, 6, 21.
Römisch, K. (2005). Annual Review of Cell and Developmental Biology, 21, 435–456.
Acknowledgments
Financial support from the National Key Basic Research and Development Program of China (973 Program; no. 2011CB710800), the National High Technology Research and Development Program of China (863 Program; no. 2012AA022207, 2011AA02A209, and 2011AA02A210), the Fundamental Research Funds for the Central Universities (JUSRP11014), the Programme of Introducing Talents of Discipline to Universities (111 Project; 111-2-06 ), and the Ministry of Education, China, and from NSFC (20802027) are greatly appreciated.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Sha, C., Yu, XW., Li, F. et al. Impact of Gene Dosage on the Production of Lipase from Rhizopus chinensis CCTCC M201021 in Pichia pastoris . Appl Biochem Biotechnol 169, 1160–1172 (2013). https://doi.org/10.1007/s12010-012-0050-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-012-0050-9