Abstract
Marine algae, which make up about 80% of the world’s living organisms, contain many energy sources, such as sugars and lipids. Therefore, the possibility of utilizing structural carbohydrates from marine algae for bioethanol production has been studied. In order to obtain monosaccharides, Undaria pinnatifida, Chlorella vulgaris, and Chlamydomonas reinhardtii were used for the saccharification experiments. The pretreatment was carried out by dilute acid hydrolysis and enzymatic treatment. To find the optimal conditions, experiments were performed at several temperatures, acid concentrations, pH conditions and durations. To test bioethanol production, several ethanolic E. coli W3110 strains, which were developed previously, were used. The maximum yield of bioethanol, 0.4 g ethanol/g biomass, was achieved with pretreated C. vulgaris and E. coli SJL2526, derived from wild-type E. coli W3110 and which includes the adhB, pdc, galP, and glk genes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Global warming is a serious environmental problem affecting all living organisms. Greenhouse gases, such as carbon dioxide, and emissions from using fossil fuels has been discussed as the main contributors to global warming. To solve such issues, developing alternative energy sources has become an international goal. Using bioethanol is an effective way to decrease carbon dioxide emissions [1, 2]. Bioethanol is generated from biomass, which includes plants, microorganisms, and animal excretions. Compared to fossil fuels, bioethanol emits less ozone, benzene, carbon dioxide and other harmful pollutants. It is also easy to use biofuels as transportation fuels, but several hurdles remain before the biofuel technology can be deployed on a commercial scale, such as low productivity and price competitiveness [3]. The use of biofuel has drawn universal attention because of possible global price increases for agricultural products because a large portion of biofuel feedstock is land crops. Many researchers are searching for alternative biomass sources for the production of bioethanol [1–3]. Lately, algae- or cellulose-based bioethanol has been suggested as the next generation biofuel source [4].
The conversion of cellulose or algae biomass into ethanol has been receiving increasing attention in recent years [1, 5, 6]. Lignocellulosic biomass hydrolysis is the most highly developed saccharification method for bioethanol production. Over the past few decades, the overall conversion of cellulosic materials into monosaccharides has been improved by three different methods: physical, chemical and physicochemical. Physical methods were generally basic and simple treatments, for instance, milling, chopping, grinding and gamma irradiation, which can be done using industrial equipment. Chemical methods are the most frequently used saccharification treatments for cellulosic biomass. In particular, acid and alkali methods have been continuously developed for conversion of cellulosic material into fuel ethanol [7, 8]. Physicochemical methods have been developed for lignin degradation. Steam, liquid hot water, ammonia fiber explosion, and ammonia recycle percolation treatments are performed at extreme conditions. Lignocellulosic biomass saccharification techniques have been continually reformed and simplified [9]. Nevertheless, in most cases, conversion of biomass into monosaccharides has been hampered by economic and safety problems.
The ocean makes up 71% of the earth’s surface and accounts for 90% of the entire ecosystem; by contrast, using marine resources are insufficient. The use of photosynthetic marine algae instead of land crops is a promising new possibility for transportation fuel [10–13]. Approximately 20–30% of marine algae biomass is made up of cellulose or starch, and marine algae also contain various monosaccharides such as glucose, xylose, galactose, and others [4]. These sugars can be extracted using two major saccharification methods, namely, dilute acid hydrolysis and enzymatic pretreatment, and these sugars can also be converted into bioethanol [7].
This research presents a comparison of saccharification conditions covering several temperatures, acid concentrations, pH conditions and durations using three different algae species. Additionally, to obtain bioethanol, several ethanolic E. coli W3110 strains, which were developed previously, and sugars derived from algae pretreatment were used in fermentation experiments. By suggesting marine algae as a biomass source and also by improving the pretreatment methods, this study may provide important knowledge to the alternative energy industry field.
Materials and Methods
Materials
Three different algae species, the macro algae Undaria pinnatifida, the marine micro algae Chlorella vulgaris and the freshwater microalgae Chlamydomonas reinhardtii were used in this study. U. pinnatifida was purchased from a local grocery store. It was milled in a common blender for 5 min and was stored at room temperature. C. vulgaris was purchased from Daesang Co., Seoul, Korea. C. reinhardtii was provided by Prof. Kwang-Hwan Jung at Sogang University, Seoul, Korea. It was stored in a chamber at −40 C immediately after it was harvested. Celluclast 1.5 L and Novozyme 188 were purchased from Sigma-Aldrich Co. and were stored in a refrigerator at 4 C.
Bacteria used for Ethanol Fermentation
Escherichia coli (E. coli) SJL25, E. coli SJL2526, E. coli SJL27 and E. coli SJL2627 were used to produce bioethanol. These different strains were designed to enhance the up-take several sugars. E. coli SJL25, a recombinant strain derived from the wild-type E. coli strain W3110, contains the adhB and pdc genes, which are overexpressed. Additionally, E. coli SJL2526, derived from the wild-type E. coli strain W3110, includes the adhB, pdc, galP, and glk genes, which are overexpressed. E. coli SJL27 contains the adhB, pdc, and pgm genes, which are overexpressed. E. coli SJL2627 overexpresses the adhB, pdc, pgm, galP, and glk genes.
Dilute Acid Hydrolysis
The dilute acid hydrolysis process used in this study had several steps. Samples of each of the three different algae species were hydrolyzed in dilute H2SO4 (1–5% on 5% (w/v) dry solid basis (v/v)) and were heated in an autoclave at 100–120 C. The treatment time was 120 min for C. vulgaris and C. reinhardtii and was 24 h for U. pinnatifida. The autoclaved algae samples were cooled using coolant, and then the supernatant was collected by centrifugation (4500 rpm, 10 min, from Vision scientific Co., Korea). The collected marine algae samples were adjusted to pH 6.5–7.0 with CaCO3 before filtration. The filtration process used a vacuum pump with a 0.2-μm cellulose membrane filter.
Enzymatic Treatment
Each of three different algae samples was soaked in citric acid, sodium citrate buffer (5% dry solid basis, w/v) and autoclaved at 121 C for 15 min. Celluclast 1.5 L (4 ml per 100 g of cellulose) and Novozyme 188 (2 ml per 100 g of cellulose) were added to the autoclaved algae. The enzymatic treatment conditions were 35–55 C and pH 3.6–5.6 for 60 min.
Fermentation Experiment
Batch culture was carried out in a 5 L fermentor (Biotron Co., Korea) containing 2 L of LB media with either 20 g/L of glucose or approximately 5 g/L of pretreated C. vulgaris sugar, under the following conditions: pH 7.0, controlled by the addition 1 M NaOH; temperature, 37 C; air flow rate at 1 vvm; and agitation at 150 rpm.
The seed culture was grown in a 10-mL culture tube containing 3 mL of LB media to define the mutants. For the batch cultures, a 500-mL shake flask containing 200 mL of LB media was inoculated with 1 mL of seed culture, which was prepared by inoculation with a single colony and incubation in a shaking incubator for 12 h at 37 C. All of the flask cultures were grown in a rotary shaking incubator (Jeiotech Co., Korea) at 37 C and 170 rpm.
Analysis of Bioethanol and Pretreated Marine Algae Sugar Production
The amount of pretreated marine algae sugar produced was measured by HPLC with an LC-10AT unit (Younglin Co., Korea) with an RI750F monitor (Younglin Co., Korea) and an HPX-87 C carbohydrate analysis column (300 mm × 7.8 mm; Bio-Rad). DDW was used as the mobile phase at 85 C and at a flow rate of 0.6 ml/min.
The supernatant taken from the culture medium of the batch fermentations was analyzed for bioethanol content. The amounts of sugars and bioethanol produced were measured by HPLC with an LC-10AT unit (Younglin Co., Korea) with an RI750F monitor (Younglin Co., Korea) and an Aminex HPX-87H organic acids column (300 mm × 7.8 mm; Bio-Rad). Sulfuric acid (0.01 M) was used as the mobile phase at 55 C and at a flow rate of 0.6 ml/min. All solutions were filtered with a 0.2-μm membrane before use. Supernatants were concentrated 20-fold prior to analysis by the HPLC assay.
Results
Marine Algae Pretreatment
Dilute acid hydrolysis was carried out at various temperatures, at acid concentrations and for different lengths of time. As shown in Table 1, only 0.075 g glucose/g biomass was extracted from C. reinhardtii at 100 C, 5% sulfuric acid and 120 min. However, 0.12 g glucose/g biomass, 0.033 g xylose/g biomass, 0.047 g galactose/g biomass and 0.007 g fructose/g biomass were extracted from C. vulgaris at 110 C, 5% sulfuric acid and 120 min. Additionally, 0.065 g glucose/g biomass, 0.002 g xylose/g biomass and 0.004 g fructose/g biomass were extracted from U. pinnatifida at 120 C, 5% sulfuric acid, and 24 h. The three marine algae species had different optimal conditions. Monosaccharides could not be extracted from C. reinhardtii under high temperature conditions, that is, at temperatures greater than 110 C. The optimum conditions for C. vulgaris were comparatively mild at 3% sulfuric acid and 105 min at 110 C, but U. pinnatifida only produced monosaccharides after more sulfuric acid was added at 120 C (see Fig. 1).
Enzymatic treatment was performed at several temperatures and pH conditions and for different lengths of time to find the optimal conditions for enzyme degradation activity. Table 2 summarizes the results for the different enzymatic conditions. At 45 C, pH 4.6, and 60 min, the enzyme activity was higher than for any other condition; under these conditions, 0.037 g glucose/g biomass was extracted from C. reinhardtii, and 0.013 g glucose/g biomass was extracted from U. pinnatifida. C. vulgaris had the highest enzymatic degradation yield, with 0.13 g glucose/g biomass and 0.006 g fructose/g biomass. In contrast to the dilute acid hydrolysis experiments, the enzymatic degradation experiments revealed that the three different algae species had same optimal conditions, but quantities of extracted monosaccharides were less than those for dilute acid hydrolysis.
Bioethanol Fermentation
Batch culturing, which was done in 5-L fermentors containing 2 L of LB media with 20 g/L glucose, was carried out using four different ethanolic E. coli W3110 strains. Other than glucose, the fermentation medium did not contain any monosaccharides, and the four different ethanolic E. coli W3110 strains consumed only glucose as the carbon source. Generally, the acetate and lactate production rates were higher than ethanol production rate (see Fig. 2).
The maximum bioethanol concentration was achieved using E. coli SJL25, with a concentration of 2.88 g/L. The lactate concentration of E. coli SJL25 was increased and the acetate concentration was decreased compared to concentrations of the other strains. In contrast, the acetate concentration of E. coli SJL2627 was increased and the lactate concentration was decreased compared to the concentrations of the other strains (see Table 3).
Batch culturing, which was done in 5-L fermentors containing 2 L of LB media with 5 g/L pretreated C. vulgaris sugar, was carried out using four different ethanolic E. coli W3110 strains. In contrast to the results obtained using 20 g/L glucose as the carbon source for fermentation, E. coli SJL2526 and E. coli SJL2627 were able to consume glucose, galactose and xylose as carbon sources when using pretreated C. vulgaris sugar, which contained several monosaccharides, such as glucose, galactose, xylose and fructose, as the carbon source. Generally, the acetate production rate was higher than the ethanol and lactate production rates when using pretreated C. vulgaris sugar (see Fig. 3).
LB-pretreated chlorella sugar fermentation experiments: (A) E. coli SJL25, (B) E. coli SJL2526, (C) E. coli SJL27, and (D) E. coli SJL2627. Filled square cell growth; filled circle glucose; filled upright triangle galactose; open inverse triangle xylose; open diamond ethanol; open left-pointing triangle acetate, filled right-pointing triangle lactate
The maximum concentration of bioethanol obtained from pretreated C. vulgaris sugar batch fermentation was achieved using the 0.6% pretreated C. vulgaris sugar batch fermentation by E. coli SJL2526, with a concentration of 1.67 g/L. Additionally, the lactate concentration of E. coli SJL2526 was increased and the acetate concentration was decreased compared to the concentrations of other strains (see Table 3). Consequently, the maximum yield of bioethanol when using pretreated C. vulgaris sugar batch fermentation was achieved by E. coli SJL2526, with a yield of 0.4 g ethanol/g biomass. Compared to 2-L fermentation with glucose as the carbon source, fermentation with C. vulgaris sugar resulted in an ethanol yield that was slightly higher, about 0.01–0.17 g ethanol/g biomass; the acetate yield was substantially higher, and the lactate yield was lower.
Discussion
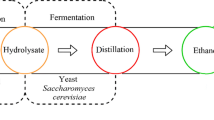
In this study, saccharification of three different algae species was performed. Additionally, bioethanol was produced using pretreated C. vulgaris sugar as the main carbon source and mutated ethanolic E. coli as the fermentor.
The marine algae pretreatment in this study did show the same high level of sugar production observed with other previously reported cellulosic biomass saccharification methods [7, 8]. Dilute acid hydrolysis and enzymatic saccharification of cellulosic biomass has been shown to have a maximum yield of about 0.5 g total sugar/g biomass [7]. In contrast, the maximum yield obtained by marine algae saccharification was approximately 0.25 g total sugar/g biomass, half of the cellulosic biomass saccharification results. Marine algae contain a large amount of water, and pretreated marine biomass had fewer monosaccharides than other lignocellulosic biomass sources. Nevertheless, marine algae do not contain lignin, which has a complex and rigid structure; thus, the saccharification method could be simpler for marine algae than for lignocellulosic biomass. In this study, we found that the optimal conditions for C. vulgaris pretreatment were 3% sulfuric acid (v/v) at 110 C for 105 min, as shown in Table 1, but the pretreatment conditions for lignocellulosic biomass were complex and included extreme steps, such as strong acid concentrations, long residence times and high temperatures [7, 9]. In order to obtain a high yield of algal monosaccharides, researchers interested in algal biomass saccharification methods should try to develop economical and simple steps so that it can be turned into an industrial process. Most importantly, dilute acid hydrolysis, which was carried out using sulfuric acid, should be changed so that the reagents are reusable. In saccharification using the enzymatic method, the price of the enzyme was high, and the efficiency was below expectations, but enzymes are reusable and represent a simple method for industrial applications. As a result, enzymatic treatment should be improved instead of dilute acid hydrolysis, and new, more efficient enzymes must be developed.
Bioethanol is generated from biomass, which is agricultural residue such as corn stover, sugar cane, wheat straw, rice straw and other crops [8, 14]. In many previous studies, the number of fermentation microorganisms was very limited, and the bioethanol production rate was not high enough for commercialization [8, 9, 14–16]. For example, bioethanol fermentation from wheat straw hydrolysate by the recombinant E. coli strain FBR5 resulted in a maximum ethanol yield of only 0.24 g ethanol/g biomass [8]. Therefore, in this study, we presented a new bioethanol production method. By utilizing non-edible marine biomass and effective sugar up-taking ethanolic E. coli, a new method for bioethanol production was developed. Consequently, in this study, bioethanol fermentation from marine algae hydrolysate by E. coli SJL2526 resulted in a maximum ethanol yield of 0.4 g ethanol/g biomass, which was about 0.15 g ethanol/g biomass higher than that found in previous studies. The pretreated C. vulgaris sugar in this study contained various sugars, such as glucose, galactose, xylose, etc. Additionally, the ethanol production yield of E. coli SJL2526 was higher for the pretreated C. vulgaris sugar than for glucose added by nearly twofold, considering that glucose was added at a concentration of 20 g/L and the pretreated C. vulgaris sugar was added a concentration of approximately 5 g/L to a 2-L fermentation batch culture. Nevertheless, industrial bioethanol processes using algal biomass still have a long way to go because of the current inefficient nature of the ethanol fermentation process. Bioethanol fermentation scale-up research should be designed and completed. Our experiments on bioethanol fermentation using marine algae experiments, which achieved a yield of 0.4 g ethanol/g biomass, demonstrate the possibility of extended application to commercial production processes.
Conclusion
This research compared saccharification conditions including several temperatures, acid concentrations, pH conditions and different periods of time using three different algae species, C. reinhardtii, C. vulgaris, and U. pinnatifida. Pretreatment was carried using dilute acid hydrolysis or enzymatic treatment. Additionally, to obtain bioethanol, several ethanolic E. coli W3110 strains, were E. coli SJL25, E. coli SJL2526, E. coli SJL27 and E. coli SJL2627, were used, and algae pretreatment sugars were used for fermentation experiments. The maximum yield of bioethanol when using pretreated C. vulgaris sugar batch fermentation was achieved with E. coli SJL2526, with a yield of 0.4 g ethanol/g biomass.
The results of this study demonstrate the potential of marine biomass and also demonstrate the ethanol productivity of pretreated C. vulgaris sugar using an efficient marine algae saccharification method. This study suggests new possibilities in the bio-energy-producing field using microorganisms.
References
Regalbuto, J. R. (2009). Cellulosic biofuels—Got gasoline? Science, 325, 822–824.
Gressel, J. (2008). Transgenics are imperative for biofuel crops. Plant Sci, 174, 246–263.
Hoekman, S. K. (2009). Biofuels in the U.S.—Challenges and opportunities. Renewable Energy, 34, 14–22.
Anders S Carlsson, Jan B van Beilen, Ralf Möller and David Clayton (2007) Micro-and macro-algae: Utility for industrial applications. CPL Press. 1-82
Patil, V., Tran, K.-Q., & Giselrod, H. R. (2008). Towards sustainable production of biofuels from microalgae. Int J Mol Sci, 9, 1188–1195.
Valderrama, L. T., Del Campo, C. M., Rodriguez, C. M., de-Bashan, L. E., & Bashan, Yoav. (2002). Treatment of recalcitrant wastewater from ethanol and citric acid production using the microalga Chlorella vulgaris and the macrophyte Lemna minuscule. Water Res, 36, 4185–4192.
Saha, B. C., Iten, L. B., Cotta, M. A., & Victor Wu, Y. (2005). Dilute acid pretreatment, enzymatic saccharification and fermentation of wheat straw to ethanol. Process Biochem, 40, 3693–3700.
Zhisheng, Yu, & Zhang, H. (2003). Pretreatments of cellulose pyrolysate for ethanol production by Saccharomyces cerevisiae, Pichia sp. YZ-1 and Zymomonas mobilis. Biomass Bioenergy, 24, 257–262.
Michael S Kent and Katherine M Andrews (2007) Biological research survey for the efficient conversion of biomass to biofuels. Sandia Report. 1-24
Sialve, B., Bernet, N., & Bernard, O. (2009). Anaerobic digestion of microalgae as a necessary step to make microalgal biodiesel sustainable. Biotechnol Adv, 27, 409–416.
Svein Jarle Horn (2000) Bioenergy from brown seaweeds. Ph.D. Thesis. Department of Biotechnology Norwegian University of Science and Technology NTNU Trondheim, Norway. 1-69
Rioux, L. E., Turgeon, S. L., & Beaulieu, M. (2007). Characterization of polysaccharides extracted from brown seaweeds. Carbohydr Polym, 69, 530–537.
Aikaterini Papazi and Kiriakos Kotzabasis. (2007). Bioenergetic strategy of microalgae for the biodegradation of phenolic compounds—Exogenously supplied energy and carbon sources adjust the level of biodegradation. J Biotechnol, 129, 706–716.
Mielenz, J. R. (2001). Ethanol production from biomass: Technology and commercialization status. Ecology and industrial microbiology, 4, 324–329.
Hirano, A., Ueda, R., Hirayama, S., & Ogushi, Y. (1997). CO2 fixation and ethanol production with microalgal photosysthesis and intracellular anaerobic fermentation. Energy, 22, 137–142.
Balat, M., Balat, H., & Oz, C. (2008). Progress in bioethanol processing. Prog Energy Combust Sci, 34, 551–573.
Acknowledgments
This research was supported by a grant from Development of Marine-Bioenergy Program Funded by Ministry of Land, Transport and Maritime Affairs of Korean Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, S., Oh, Y., Kim, D. et al. Converting Carbohydrates Extracted from Marine Algae into Ethanol Using Various Ethanolic Escherichia coli Strains. Appl Biochem Biotechnol 164, 878–888 (2011). https://doi.org/10.1007/s12010-011-9181-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-011-9181-7