Abstract
To improve the yield of daptomycin by Streptomyces roseosporus, a method of rational screening of He–Ne Laser and N-methyl-N-nitro-N-nitrosoguanidine (NTG)-induced mutants was employed in this work. Under the optimal mutagenesis conditions (NTG of 0.3 mg/L, 40 min; irradiation at 15 mW, 15 min), two steps of combined mutations were conducted. Screening of mutants was done according to the individual resistance to n-decanoic acid and daptomycin. A mutant strain LC-54-16, with the highest daptomycin production ability of 616 mg/L was obtained, which was over 5 times higher than the wild strain LC-52-6. The transcription levels of the main dpt genes in the mutant were approximately five times higher than those in the wild. The superiority of the mutant to the wild strain was further proved by the comparative studies on the kinetics of the mutant and the wild. The decrease of the inhibition of substrate and product was further confirmed, as well. It was concluded that the method of rational screening of He–Ne Laser and NTG-induced mutants could efficiently improve the daptomycin production ability of S. roseosporus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
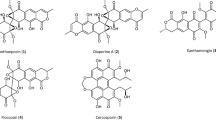
Daptomycin, the first approved lipopeptide antibiotics, is a member of the A21978C family of the cyclic anionic 13-amino acid lipopeptide, produced as a secondary metabolite by Streptomyces roseosporus (Fig. 1a) [1]. Produced by precursor of n-decanoic acid-directed fermentation [2], daptomycin has a novel, calcium-dependent mechanism of action involving binding to bacteria cell membranes, which may account for its efficacy against strains resistant to other antibiotics in clinical use [3]. Daptomycin has been commercialized for the treatment of skin and skin structure infections caused by Gram-positive pathogens [4–6]. Daptomycin is synthesized by a large non-ribosomal peptide synthetase multienzyme (NRPS), encoded by three very large genes, dptA, dptBC, and dptD arranged in tandem [7] (Fig. 1b). The NRPS is composed of three subunits, DptA, DptBC, and DptD, each subunit contains two to six modules to direct the incorporation of the 13 amino acids. The NRPS up stream genes of dptE and dptF show sequence similarities to acyl-CoA ligase and acyl carrier protein genes, respectively. DptE and DptF are predicted to be involved in the acylation of the N-terminal tryptophan (Trp), which initiates the biosynthesis of daptomycin [7]. The group of contiguous genes, including dptG, dptH, dptI, and dptJ, located downstream of the NRPS genes also have important functions in the biosynthesis of daptomycin. Comparative domain analyses suggest involvement of dptG in regulation of expression or export of antibiotics [8]. While dptH may enhance the efficiency of daptomycin production by clearing misincorporated substrates that block the pathway. The dptJ encodes the tryptophan 2, 3-dioxygenase, an enzyme involved in the conversion of Trp to kynurenine [7]. The dptI is involved in the formation of 3-methylglutamic acid in vivo [9].
The market prospect of daptomycin is bright and promising, as the demand of daptomycin has been increasing, since approved by FDA in 2003. Extensive studies of developing new drugs based on the molecular structure of daptomycin through combinatorial biosynthesis have been conducted [7, 8]. However, the yield of daptomycin is not high enough [9], and the cost of industrial production needs to be further reduced. Yet, there is little information available regarding enhancement of daptomycin productivity. Further, few literatures on the precise description of daptomycin fermentation with appropriate kinetic models have been reported.
The modern techniques of gene engineering and metabolic engineering are considered to be effective methods in the microorganism breeding. However, application of these modern techniques frequently encounters serious impediments due to limited insight into the genetics, physiology, and biochemistry of organisms [10]. Thus, traditional mutation is still an effective approach to obtain improved strains for overproduction of both primary and secondary metabolites [11]. Rational screening, on the basis of understanding the metabolism and pathway regulation, allows for a significant improvement in the efficiency of the selection process, by contrast with the time-consuming random selection method. Moreover, precursor-resistant mutants and product-resistant mutants are widely used for screening high producing strains [12].
Low-power laser irradiation technology has brought great interest in the microorganism mutation breeding [13]. Chemical mutation by N-methyl-N-nitro-N-nitrosoguanidine (NTG) has been reported as a successful method for mutation and screening of high-yield strains [14]. However, there was no report about the combined application of low-power He–Ne laser irradiation and NTG in microorganism mutation breeding.
Almost all antibiotics have product inhibition effects due to the precise and rigorous regulation and insufficient resistance to self-production antibiotics. Thus, the strains with enhanced resistance to self-production antibiotics often have improved antibiotics productivity. Further, our previous research indicated that the growth of cells would be inhibited by the toxicity of n-decanoic acid by overfeeding of n-decanoic acid during fermentation (data not shown). Therefore, to screen mutants with daptomycin resistance and n-decanoic acid resistance, two steps of selection with He–Ne laser irradiation and NTG induced mutations were developed. Furthermore, a comparative analysis of the NRPS genes and the main accessory genes’ expression at transcription level between the wild strain and the mutant was studied, in order to explore mechanism of mutagenesis. Kinetic models for batch fermentation of daptomycin by both the original strain and the improved mutant including cell growth, substrates consumption and daptomycin production were then established in a 7.5-L fermentor.
Materials and Methods
Microorganism and Spores Suspensions
The wild-type strain of S. roseosporus NRRL11379 was purchased from ARS Culture Collection (NRRL Collection). S. roseosporus LC-52-6 was an early derivative mutant of S. roseosporus NRRL11379, naturally screened by our laboratory which has improved daptomycin production ability, and was used as a wild-type and parent strain.
S. roseosporus LC-52-6 culture stocks were maintained in 15% glycerol at −70 °C. Spores on slant were inoculated into 30 mL of vegetative medium in a 250-mL shake flask and incubated in an orbital shaker at 30 °C and 220 rpm for 48 h. Spores suspension harvested was filtrated and counted microscopically (about 108 spores/mL)
Medium and Cultural Conditions
The slant medium consisted of the following ingredients (g/L): starch, 20; KNO3, 1; MgSO4·7H2O, 0.5; NaCl, 0.5; K2HPO4·3H2O, 0.5 and Fe2 (SO4)3, 0.1, dissolved in 1 L water, with pH adjusted to 7.5.
The seed medium A (g/L): glucose, 5; peptone, 5; yeast extract, 8; dextrin, 15; K2HPO4·3H2O, 0.5; MgSO4·7H2O, 0.5 and CaCO3, 0.2, dissolved in 1 L water, with pH adjusted to 7.5.
Screening agar plate: starch, 20; KNO3, 1; MgSO4·7H2O, 0.5; NaCl, 0.5; K2HPO4·3H2O, 0.5; Fe2(SO4)3, 0.1 and agar, 20, dissolved in 1 L water, with pH adjusted to 7.5 and adding corresponding amount of screening reagents (n-decanoic acid or daptomycin).
The fermentation medium B (g/L): glucose, 60; casein, 1; yeast extract, 5; asparagines, 2; glutamine, 2; NH4SO4, 2; K2HPO4·3H2O, 0.5; MgSO4·7H2O, 0.5 and CaCO3, 0.2, dissolved in 1 L water, with pH adjusted to 7.5.
Fermentations were carried out in 500-mL flasks containing 50 mL medium B, or in a 7.5-L BIOFLO 110 fermentor (NewBrunswick Scientific Company, USA) with 4 L medium B. The inoculum size was 2% (v/v). The flask cultures were incubated at 30 °C and 220 rpm for 7 days. As for the fermentor cultures, the pH was automatically controlled at 7.0 with 8 M NaOH solution and incubated at 30 °C, with agitation of 300∼400 rpm and the aeration speed of 0.8 v/v/m for 7 days. N-decanoic acid was fed at 48 h after inoculation (0.2 g/L). All experiments were done in triplicate.
Analytical Methods
The concentration of the daptomycin was measured by HPLC method (1100 Series, Agilent Company, USA) with a C-18 reversed-phase column (LabAlliance, USA, 250 × 4.6 mm). The mobile phase consisted of 0.1% trifluoroacetic acid in water and acetonitrile (55:45, v/v). The flow rate was 1.0 mL/min. Glucose concentration was determined by a glucose biosensor analyzer (SBA-40C, Biology Institute of Shandong Academy of Sciences, China). The cell weight was determined by filtering the fluid through a pre-weighed filter paper (Whatman GF/C) and dried at 80 °C in a vacuum oven to constant weight. The concentration of decanoic acid in the culture broth was analyzed by Kechuang GC9800 gas chromatography (Shanghai, China) equipped with a flame ionization detector and a 30 m × 0.25 mm PEG-20 M column. The column temperature was 240 °C. The temperatures of the injector and detector were 250 °C and 300 °C, respectively. Nitrogen was used as the carrier gas at a flow rate of 25 mL/min. Each experiment was repeated three times. The data shown in the corresponding tables and figures were the mean values of the experiments and the error bars indicated the standard deviation.
Selection of the Optimal Mutagenesis Conditions
Effects of NTG concentration and exposure time on the survival rate and positive mutation rate were investigated in this work by varying the NTG concentration (0.1∼0.7 mg/L) and the exposure time of NTG (0.2∼1 h). The effects of exposure time under laser and output power of laser irradiation on the survival rate and positive mutation rate were also investigated in this work by varying the exposure time (5∼25 min) and the output power of laser irradiation (5∼25 mW). In order to avoid the photo-recombination influence, the laser irradiation was carried out in darkness. After each run, the cell suspension was diluted properly and spread over the agar medium in plates. The individual colonies on each screening plate and control plate were separately counted. The survival rate was calculated by Eq. 1. All the individual colonies on each screening plate were inoculated on slants, after 7 days’ cultivation at 30 °C in darkness. Then the performance of each individual colonies was investigated in 500-mL flasks containing 50 mL medium B. The control culture was treated under the same conditions with no laser irradiation and NTG exposure. The positive mutants refer to those strains which have higher daptomycin production ability than that of the wild strain (the control culture) under the same cultivation conditions. The positive mutant rate was calculated by Eq. 2:
Mutagenesis and Rational Screening
Spores of the parent strain were collected by sterile filtration and suspended with sterile water, then diluted with 50 mM Tris–HCl buffer (pH 6.0) to get a suspension of 106∼108 spores/mL.
Every 100 μL spore suspension was treated with 0.1∼0.7 mg/mL NTG in 50 mM Tris–HCl buffer for 0.2∼1 h, collected by centrifugation, washed three times with sterile saline, then plated on corresponding screening agar plates (Slant medium supplemented with 0.6 g/L n-decanoic acid).
Every 100 μL spore suspension was transferred into the glass tubes (16 × 160 mm) containing 2 mL sterile saline water. The obtained suspension was irradiated at optical laser wavelength of 632.8 nm in the darkness, by a He–Ne laser (Guangzhou Institute of Laser Application Technology, P.R.China). The optical fiber was fixed with a delivery arm that permitted precise positioning of the fiber tip 80 mm above the cell suspension. After mutagenesis, the irradiated spore suspension was diluted properly, and 100 μL sample of each diluted suspension was plated on corresponding screening plates (slant medium supplemented with 2,500 mg/L daptomycin).
All the individual colonies on each screening plate were inoculated on slants, for 7 days of cultivation at 30 °C in darkness, then the performance of each individual colonies was investigated in 500-mL flasks containing 50 mL medium B.
After each mutation experiment, the positive mutants were continuously transferred for nine generations from one agar slant medium to the next. Each generation was cultured under the same conditions to determine the daptomycin production ability.
Gene Expression by RT-PCR
Total RNA was isolated from S. roseosporus cells by RNAprep pure Bacteria Kit (TianGen, China), in early stationary phase. The quantity and purity of RNA were determined by measuring the optical density at 260 and 280 nm and further determined by 1% formaldehyde agarose-gel electrophoresis. The very large NRPS genes of dptA, dptBC, and dptD were represented by their specific middle regions (about 600–1,200 bp). The gene specific primers of each gene were shown in supplementary Table 1. Reverse transcription polymerase chain reaction (RT-PCR) reactions were carried out in an iCycler (Bio-Rad, USA) using the QuantiTect SYBRgreen RT-PCR kit (Qiagen, Germany) according to the manufacturer’s procedure. The rpsL gene, housekeeping gene of S. roseosporus, was used as a constitutive reference [15]. RT-PCR was independently performed three times for each gene under identical condition. The relative abundance of each transcript (normalized to internal control gene of rpsL) with respect to the control sample was measured in triplicates and calculated according to the comparative Ct method [16].
Results and Discussion
Effects of Parameters for Both Laser Irradiation and NTG Mutagenesis
Two parameters, concentration and exposure time were investigated to get optimal NTG mutagenesis conditions. The NTG concentration increased from 0.1 to 0.7 mg/L and the exposure time varied from 20 to 60 min. The survival rate and positive mutation rate were shown in Fig. 2. The survival rate decreased with the increase of either parameter when the other was fixed. The lowest value approached 7.5% at the condition of 0.7 mg/L NTG, 60 min. Because the concentration of NTG was too high, and the exposure time was too long under this condition, poor survival rate was observed. For the positive mutation rate, low positive mutant rate was observed for both extremes of NTG concentration at 0.1 and 0.7 mg/L. It was speculated that the NTG concentration in these ranges was either too weak to induce mutation or too strong to damage the cells. On the other hand, a high positive mutant rate appeared at a moderate concentration of 0.3 mg/L. This indicated that the positive mutant rate was much more sensitive to the mutagenesis conditions than mortality in middle range of NTG concentrations. The highest positive mutant rate was further obtained (more than 21%) by a combination of NTG concentration of 0.3 mg/L and exposure time of 40 min, respectively. This was then considered as optimal condition of NTG to mutate strains.
Similar phenomena were observed in the irradiation parameters effects. Two irradiation parameters, output power and exposure time, were investigated to get optimal mutagenesis conditions. The output power increased from 5 to 25 mW and the exposure time varied from 5 to 30 min. For survival rate, it decreased with the increase of each parameter when the other was fixed and the lowest value approached 2.7% at the irradiation condition of 25 mW, 25 min, which indicated that the laser irradiation fluence was too high, and the irradiation time was too long under this condition (Fig. 3). Low positive mutant rate was observed for both extremes of output power at 5 and 25 mW. It was proposed that the irradiation in these ranges was either too weak to induce mutation or too strong to damage the cells. Further, a high positive mutant rate appeared at a moderate output of 15 mW. The highest positive mutant rate was further obtained as more than 20%, when the output power was 15 mW and exposure time was 15 min.
Screening Mutants Resistant to n-Decanoic Acid and Daptomycin
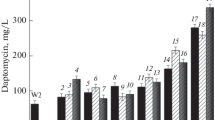
S. roseosporus LC-52-6 was performed by NTG and He–Ne laser under the optimal mutagenesis conditions (NTG of 0.3 mg/L, 40 min; irradiation at 15 mw, 15 min). Two steps of combined mutations were conducted to screen mutants with resistance to n-decanoic acid and daptomycin. After being exposed to NTG and screened by high concentration of n-decanoic acid (0.6 g/L), a total of 62 positive mutants (named form LC-53-1 to LC-53-62) were obtained, which was speculated as the reduced inhibition of n-decanoic acid to the cell growth and daptomycin biosynthesis. The five highest-producing mutants were listed in Table 1. The mutant LC-53-32 with the highest productivity of daptomycin (372 mg/L) and strong resistance to n-decanoic acid was selected for further mutation by He–Ne laser. The mutant S. roseosporus LC-53-32 was exposed to He–Ne laser and screened by high concentration of daptomycin (2,500 mg/L). Then, 53 positive mutants were obtained. According to the performance of the mutants, the five highest-producing mutants were listed in Table 2. The daptomycin titer of mutant LC-54-16 was the highest which approached 616 mg/L.
The three positive mutants (LC-54-16, LC-54-35, and LC-54-21) were transferred nine generations continuously. The results of daptomycin titer and heredity stability for the mutants were shown in Fig. 4. The daptomycin production of mutant LC-54-35 and LC-54-21 decreased dramatically since fourth generation. While daptomycin production of the mutant LC-54-16 was stable and had good heredity stability. Therefore, mutant LC-54-16 was selected in the subsequent studies of the performance comparison with the parent strain.
Performance Comparison Between the Wild and Mutant Strain
Comparison of Tolerance to n-Decanoic Acid and Daptomycin
The tolerance to n-decanoicacid and daptomycin of both the mutant LC-54-16 and the wild LC-52-6 was shown in Fig. 5. The dry cell weight of the wild decreased remarkably with the increase of n-decanoic acid concentration, while the mutant performed high ability of resistance to n-decanoic acid. Although the growth of the wild was basically inhibited, the dry cell weight of the mutant was still over 10 g/L, when the concentration of n-decanoic acid was up to 0.6 mg/L. The results demonstrated that the substrate inhibition of n-decanoic acid to the mutant was greatly reduced. Similar phenomena were observed in the tolerance to daptomycin for both the mutant and the wild. When the concentration of daptomycin increased to 2.5 g/L, the wild could not grow at all, while the dry cell weight of the mutant was over 10.5 g/L. Therefore, all the results above demonstrated that the selected mutant was not only resistant to the substrates of n-decanoic acid, but also to the product of daptomycin in a certain degree, which accounted for improved ability of daptomycin production of the mutant.
Comparison of Gene Expression by RT-PCR
A comparative analysis of the NRPS genes and the main accessory genes’ expression at transcription level between the wild and the mutant was studied by RT-PCR, in order to explore mechanism of mutagenesis. As shown in Fig. 6, transcription levels of NRPS genes dptA, dptBC, and dptD and the main accessory genes (including dptE, dptF, dptG, dptH, dptI, and dptJ) in the mutant were approximately five times higher than those in the wild. While, the S. roseosporus housekeeping gene of rpsL exhibited similar gene expression levels in both the wild strain LC-52-6 and the mutant strain LC-54-16. Thus, the enhancement of the dpt genes expression could explain the improvement of daptomycin biosynthesis ability of the mutant strain. Similar phenomena were found by Rhee and Julian [15]. They compared the expression levels of the dpt genes between the S. roseosporus wide-type UA031 and high-producing mutant strain UA343 and found that the majority of the dpt genes were up-regulated in the UA343. It was suggested that dpt genes might be transcribed on a polycistronic transcript or on overlapping large transcripts and the promoter probably resided in the upstream of dpt genes (Coeffet-Le Gal et al.) [17]. The mutation may exist in the promoter and regulation region, upstream of the dpt genes, which probably resulted in the up-regulation of the dpt genes at the approximate extent.
Batch Fermentation in the Fermenter and Kinetics Modeling
Model Development
The specific growth rate was modeled in the form of Monod kinetic model with glucose as the sole rate-limiting substrate. The above studies demonstrated that the substrate n-decanoic acid and the product daptomycin may inhibit cell growth. Further, inhibition of glucose was also taken into account. Therefore, the Monod model was modified to account for the inhibitions of substrates and product by incorporating two additional parameters, n and Smax, for each substrate and product as proposed by Luong [18] and Maiti et al. [19], respectively. The Leudeking–Piret model was used to investigate the product formation for both the wild and mutant strains of S. roseosporus. The model equations for cell growth, substrate consumption, and product formation kinetics were:
The meaning of the variables and constants of the model was explained in the nomenclature part.
Estimation of the Model Parameters
The first-order ordinary differential equations of the model were integrated by the ode23t subroutine in MATLAB (Mathworks, Natick, NA, USA). The values of the kinetic parameters for the wild and mutant strains were regressed by lsqnonlin function in MATLAB and presented in Table 3. The μ max of the mutant was obviously higher than that of the wild and the values of saturation coefficient K S and n s of the mutant were all lower than those of the wild. The value of S max was higher than that of the wild, which indicated that the inhibition of glucose decreased after the mutagenesis. Further, the values of \( {S_{\rm{de}}}^{{ \max }}\,{\hbox{and}}\,{S_{\rm{da}}}^{{ \max }} \) of the mutant were higher than that of the wild, while the values of n de and n da were much lower than that of the wild. The results showed that the inhibition of n-decanoic acid and daptomycin decreased after the mutagenesis by He–Ne irradiation NTG exposure. Kinetic constants α and β of the mutant were higher than that of the wild, especially β was over five times higher than that of the wild, which indicated that the catalytic ability of daptomycin biosynthsis improved after mutageesis. This was constant with the up-regulation of the dpt genes, reflected by RT-PCR. Moreover, Y P/S value of the mutant was much higher than that of the wild, but there was little difference on Y X/S values of the mutant and wild strain, which also indicated that the catalytic capability of daptomycin biosynthsis had been improved after the mutagenesis. Y P/de of the mutant was obviously higher than that of the wild, which demonstrated that the mutant could utilize n-decanoic acid more efficiently for daptomycin biosynthesis. However, both the value of Y P/de for the mutant and the wild was much lower than the theoretical value (9.42 g/g), which indicated that the n-decanoic acid was partially into the biomass biosynthesis.
The curves of experimental data of the fermentation process and the fitted value of the model for both the mutant and wild strains were shown in Fig. 7. The theoretical curve fitted well with the experimental data for the wild strain. The proposed model appeared to provide an adequate representation of fermentation kinetics of S. roseosporus. The mutant strain showed faster consumption of glucose and n-decanoic acid than that of the wild strain. Much faster daptomycin biosynthesis rate was observed for the mutant strain, in spite of similar overall growth profiles for both the wild and the mutant. The similar result was found by Rhee and Julian [15]. Moreover, the experimental data exhibited faster glucose consumption at stationary phase (about 80∼130 h), than the fitted values of the model for the mutant, which indicated more glucose was utilized for the daptomycin biosynthesis and the inhibition of glucose was decreased.
Model Validation
The adequacy of the model was checked by the method described by Mani et al. [19], i.e., checking the error between measured and predicted values for experiments that were not used during the parameter estimation. For both the mutant and the wild, profiles of concentrations of biomass, product, and substrates predicted by the model were compared with those obtained experimentally for six different media combinations (three for the wild strain and the other three for the mutant), which were not used while developing the model. The quality of model fit was analyzed by computing coefficient of determination R 2 and square root of sum of squares of residual errors. The fit with these parameters was quite good. The standard deviation of fit errors averaged across the measured variables was 1.36 which indicated that the model error was in the acceptable range. The average R 2 was up to 0.95 (Table 4), thus, the model was significant.
Further, the model sensitivity to its parameters and significance of the key parameters were also tested. Each parameter was perturbed, one at a time, by ±20% around its estimated value for sensitivity calculation via the following expression [20]:
In Eq. 8, σ i t , k, βp is the sensitivity of k th response for i th experiment at time t to a parameter β p and \( \widehat{y}_{t,k}^i\left( \beta \right) \) represents the simulated values at time t of k th variable for i th experiment with estimated parameter vector β and \( \widehat{y}_{t,k}^i\left( {\beta + \delta {\beta_p}} \right) \) represents the simulated values at time t of k th variable for i th experiment when only parameter β p is perturbed. The model response was found to be sensitive to four parameters: Y P/S, Y P/de, n de, and n da, which indicted that these four parameters were the key parameters of the model. Moreover, the confidence intervals (CI) of the four sensitive parameters are estimated using bootstrapping-based percentile method [21].The values for 95% CI was presented in Table 5. It could be concluded that all the sensitive parameters were statistically significant thereby indicating model adequacy, since the widths of the confidence intervals were reasonably small.
Conclusion
A method of rational screening of He–Ne Laser and NTG corporately induced mutants was employed in this work to improve the production ability of daptomycin by S. roseosporus. A positive mutant (LC-54-16) with resistance to n-decanoic acid and daptomycin was obtained under the optimal mutagenesis conditions (NTG of 0.3 mg/L, 40 min; irradiation at 15 mw, 15 min). The daptomycin titer of the mutant increased to 616 mg/L in a 7.5-L fermentor, which was over fivefold higher than that of the wild strain LC-52-6. Transcription levels of NRPS genes of dptA, dptBC, and dptD and the main accessory genes (including dptE, dptF, dptG, dptH, dptI, and dptJ) in the mutant were approximately five times higher than those in the wild. Further kinetic models for batch fermentation of daptomycin including cell growth, substrates consumption, and daptomycin production were established in a 7.5-L fermentor. The theoretical curve fitted well with the experimental data for the wild strain. Comparative studies on kinetics of the mutant and the wild further confirmed improved performance of the mutant. It was concluded that the method of rational screening of He–Ne Laser and NTG induced mutants could efficiently improve the daptomycin production ability of S. roseosporus, and might provide an alternative approach to enhance the metabolites productivity in other Streptomyces.
Abbreviations
- Xmax (gL−1):
-
Maximum cell dry weight
- X0 (gL−1):
-
Initial cell dry weight
- μmax (h−1):
-
Maximum specific growth rate
- K S (gL−1):
-
Saturation coefficient of glucose
- Pmax (g L−1):
-
Maximum daptomycin titer
- α:
-
Growth associated product formation rate constant
- β (h−1):
-
Non-growth-associated product formation rate constant
- YX/S (gg−1):
-
Cell yield coefficient of glucose
- YP/S (gg−1):
-
Product yield coefficient of glucose
- m (gg−1):
-
Cell maintenance coefficient
- Smax (gL−1):
-
Maximum glucose concentration at which specific growth rate is zero
- n S :
-
Exponent of glucose repression
- \( S_{de}^{{ \max }}\left( {{\hbox{g}}{{\hbox{L}}^{ - {1}}}} \right) \) :
-
Maximum n-decanoic acid concentration at which specific growth rate is zero
- n de :
-
Exponent of n-decanoic acid repression
- \( S_{da}^{{ \max }}\left( {{\hbox{g}}{{\hbox{L}}^{ - {1}}}} \right) \) :
-
Maximum n-decanoic acid concentration at which specific growth rate is zero
- n da :
-
Exponent of daptomycin repression
- \( {Y_{P/{S_{de}}}} \) :
-
Product yield coefficient of n-decanoic acid
- X (gL−1):
-
Biomass concentration
- S (gL−1):
-
Glucose concentration
- S de (gL−1):
-
n-decanoic acid concentration
- P (gL−1):
-
Daptomycin concentration
References
Debono, M., Barnhart, M., Carrell, C. B., Hoffmann, J. A., Occolowitz, J. L., Abbott, B. J., et al. (1987). The Journal of Antibiotics, 40, 761–777.
Huber, F. M., Pieper, R. L., & Tietz, A. J. (1988). Journal of Biotechnology, 7, 283–292.
Zhou, P. F., Wang, Y. W., Guo, M., Wang, H. H., & Xie, J. P. (2009). Review of Medical Microbiology, 20, 12–18.
Chamberlain, R. S., Culshaw, D. L., Donovan, B. J., & Lamp, K. C. (2009). Surgery, 14, 6316–6324.
Arbeit, R. D., Maki, D., Tally, F. P., Campanaro, E., & Eisenstein, B. I. (2004). Clinical Infectious Diseases, 38, 1673–1681.
Kirkpatrick, P., Raja, A., LaBonte, J., & Lebbos, J. (2003). Nature Reviews. Drug Discovery, 2, 943–944.
Miao, V., Coeffet-LeGal, M. F., Brian, P., Brost, R., Penn, J., Whiting, A., et al. (2005). Microbiology-Sgm, 151, 1507–1523.
Yeats, C., Bentley, S., & Bateman, A. (2003). BMC. Microbiology. 2003. Available from: http://www.biomedcentral.com/1471-2180/3/3. Accessed February 6, 2003.
Nguyen, K. T., Kau, D., Gu, J. Q., Brian, P., Wrigley, S. K., Baltz, R. H., et al. (2006). Molecular Microbiology, 61, 1294–1307.
Olano, C., Lombo, F., Mendez, C., & Salas, J. A. (2008). Metabolic Engineering, 10, 281–292.
Khaliq, S., Akhtar, K., Ghauri, M. A., Iqbal, R., Khalid, A. M., & Muddassar, M. (2009). Microbiological Research, 164, 469–477.
Wu, Z. F., Bajpai, R. K., & Yan, W. (2008). Biocatalysis and Biotransformation, 26, 444–449.
Kohli, R., Bose, B., & Gupta, P. K. (2001). Journal of Photochemistry and Photobiology. B: Biology, 60, 136–142.
Kumar, K. S., Manimaran, A., Permaul, K., & Singh, S. J. (2009). Bioscience and Bioengineering, 107, 494–498.
Rhee, K. H., & Julian, D. (2006). Journal of Microbiology and Biotechnology, 16, 1841–1848.
Livak, K. J., & Schmittgen, T. D. (2001). Methods, 25, 402–408.
Coeffet-Le Gal, M. F., Thurston, L., Rich, P., Miao, V., & Baltz, R. H. (2006). Microbiology-Sgm, 152, 2993–3001.
Luong, J. H. T. (1987). Biotechnology and Bioengineering, 29, 242–248.
Maiti, S. K., Singh, K. P., Lantz, A. E., Bhushan, M., & Wangikar, P. P. (2010). Biotechnology and Bioengineering, 105, 109–120.
Fouchard, S., Pruvost, J., Degrenne, B., Titica, M., & Legrand, J. (2009). Biotechnology and Bioengineering, 102, 232–245.
Dogan, G. (2007). System Dynamics Review, 23, 415–436.
Acknowledgments
The authors wish to acknowledge the financial support provided by the National High Technology Research, Development Program of China (2007AA02Z200), the National Natural Science Foundation of China (No. 20706042, No. 20906070), and the Program of Introducing Talents of Discipline to Universities (No. B06006).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Primers used for RT-PCR (DOC 42 kb)
Rights and permissions
About this article
Cite this article
Yu, G., Jia, X., Wen, J. et al. Strain Improvement of Streptomyces roseosporus for Daptomycin Production by Rational Screening of He–Ne Laser and NTG Induced Mutants and Kinetic Modeling. Appl Biochem Biotechnol 163, 729–743 (2011). https://doi.org/10.1007/s12010-010-9078-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-010-9078-x