Abstract
Uridine diphosphogalactose-4-epimerase (UDP-galactose-4-epimerase, GalE, EC 5.1.3.2) mediates the 4-epimerization of nucleic acid-activated galactose into UDP-glucose. To date, no enzyme is known to mediate 4-epimerization of free monosaccharide substrates. To determine the potential activity of GalE for free monosaccharide, Escherichia coli GalE was expressed and purified using Ni-affinity chromatography, and its ability to mediate 4-epimerization of a variety of free keto- and aldohexoses was assessed. Purified GalE was found to possess 4-epimerization activity for free galactose, glucose, fructose, tagatose, psicose, and sorbose at 0.47, 0.31, 2.82, 9.67, 15.44, and 2.08 nmol/mg protein per minute, respectively. No 4-epimerization activity was found for allose, gulose, altrose, idose, mannose, and talose. The kinetic parameters of 4-epimerization reactions were K m = 26.4 mM and k cat = 0.0155 min−1 for d-galactose and K m = 237 mM and k cat = 0.327 min−1 for d-tagatose. The 4-epimerization of tagatose, a reaction of commercial interest, was enhanced twofold (19.79 nmol/mg protein per minute) when asparagine was exchanged with serine at position 179. The novel activity of GalE for free monosaccharide may be beneficial for the production of rare sugars using cheap natural resources. Potential strategies for developing enhanced GalE with increased 4-epimerization activity are discussed in the context of the above findings and an analysis of a 3D structural model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A large number of studies examining the enzymatic conversion of specific monosaccharides into more valuable isomeric forms have been published in recent years. The isomerization of aldohexoses into ketohexoses is the best known of these reactions. Fructose can be converted from glucose by d-xylose isomerase in the manufacture of high-fructose corn syrup [1]. Tagatose, a rare hexose monosaccharide sweetener, is converted from galactose by l-arabinose isomerase [2]. d-Psicose-3-epimerase mediates 3-epimerization and conversion of fructose into psicose, a non-toxic sugar that can suppress hepatic lipogenic enzyme activity [3] and can be used as a non-caloric sweetener [4]. To date, there have been no reports of an enzyme that can mediate the 4-epimerization of free monosaccharides, although several enzymes have been reported to mediate the 4-epimerization of nucleic acid-activated or phosphate-activated monosaccharides.
Uridine diphosphogalactose 4-epimerase (UDP-galactose-4-epimerase, GalE, EC 5.1.3.2) catalyzes the interconversion of UDP-galactose and UDP-glucose in bacterial and mammalian galactose metabolism [5]. The activity of this enzyme has been characterized in numerous organisms, including Escherichia coli [6], Aeromonas hydrophila [7], Saccharomyces fragile [8], and Lactococcus lactis [9], as well as in bovine [10], human [11], and plant [12] tissues.
A preliminary report suggests that GalE might mediate epimerization between glucose and galactose in mouse brain [13]. As such, we sought to investigate the possibility that GalE might mediate the conversion of non-nucleic acid-activated glucose to galactose because this enzymatic process has potential application in the commercial manufacture of rare saccharides from naturally abundant glucose [14]. Although directed evolution has become a powerful tool to improve enzymes for commercial purposes, the availability of an original template would provide a greater chance of acquiring desirable characteristics [15].
In this study, we report the 4-epimerization activity of GalE from E. coli for free monosaccharide substrates and the associated kinetic parameters. We also show that the activity for tagatose was enhanced when asparagine was exchanged with serine at position 179 by saturation mutation. A further increase in activity is discussed based on an analysis of a 3D model.
Materials and Methods
Strains, Plasmids, and Protein Expression
E. coli ER2566 (New England Biolabs Inc., Hertfordshire, UK) and pET-20b (Novagen, Darmstadt, Germany) were used as the expression host and vector, respectively. The gene encoding GalE (GenBank ACB01960) was PCR-amplified using the oligonucleotides CATATGGTGAGAGTTCTGGTTACC (NdeI restriction site underlined) and CTCGAGTTTATCGGGATATCCCTG (XhoI restriction site underlined), which were derived from the sequences of E. coli W3110 genomic DNA (KCTC 2223; GenBank AC_000091). PCR fragments (1 kb) were cloned into T-vector (T&A Cloning Vector, RBC Co., Taiwan) and sequenced at a DNA sequencing facility (Macrogen Co., Seoul, Korea), then digested with NdeI and XhoI and ligated into similarly digested pET-20b. The resulting vector, pEcGalE, was transformed into ER2566 by electroporation (BTX ECM; Harvard Apparatus, Holliston, MS, USA). This recombinant strain was cultivated by shaking at 200 rpm in a 2,000-mL flask containing 500 mL of Luria–Bertani (LB) medium with 50 μg/mL of ampicillin. When the culture achieved an O.D600 nm of 0.5, 0.1 mM isopropyl-β-d-thiogalctopyranoside was added to induce protein expression.
Protein Purification
Bacteria were harvested from culture broth via centrifugation (3,000×g, 4 °C) and resuspended in buffer containing 50 mM sodium phosphate (pH 8.0), 10 mM imidazole, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The resuspended cells were disrupted on ice for 1 min using a sonicator (UP200S; Hielscher Ultrasonics GmbH, Teltow, Germany) set at 170 W at 1-s intervals. After removal of cell debris by centrifugation (10,000 rpm for 20 min), the supernatant was applied to a Ni+ affinity chromatography column (Novagen). The active fraction was dialyzed at 4 °C for 24 h against MacIlvain buffer (pH 7.5). The resulting solution contained purified enzyme. All purification steps using columns were carried out in a cold chamber (4 °C). Protein concentration was quantified by the Bradford method. Purified proteins were visualized by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue staining.
Enzymatic Activity Assay
The 4-epimerization reaction (1 mL) was performed in MacIlvain buffer (pH 7.5) containing 50 mM free monosaccharide and 0.5 mg of purified enzyme. After the reaction components were combined, the mixture was incubated at 35 °C for 1 h. The free monosaccharide in the reaction mixture was then subjected to 1-phenyl-3-methyl-5-pyranzolone (PMP) derivatization for HPLC analysis [16–18]. A 0.5 M PMP in methanol (75 μL) and a 1.5 M NaOH (15 μL) were added to the 4-epimerization reaction (60 μL), and the resulting mixture was allowed to derivatize at 70 °C for 3 h. After cooling at room temperature, the mixture was neutralized by adding 50 μL of 0.5 M HCl. Unreacted PMP was dissolved by adding chloroform (0.5 mL), and the organic phase was discarded after vigorous shaking. The removal process was repeated twice and the remaining chloroform was removed by evaporation at 45 °C. The aqueous portion was used for HPLC analysis. PMP-monosaccharides were separated on a Shim-pack CLC-ODS C18 column (6.0 × 150 mm; Shimadzu, Tokyo, Japan) and analyzed with a UV detector at 240 nm (UVD486; Waters, Milford, MA, USA). Eluents A and B were 10% acetonotrile and 25% acetonitrile, respectively, both in 0.1 M sodium phosphate (pH 7.0). The column was equilibrated with eluent A for 20 min and then eluted with eluent B for 20–40 min in a gradient of 0–100% at a flow rate of 1 mL/min.
Glucose 4-epimerization to galactose was verified using a modified glucose dehydrogenase coupling method [19]. The reaction mixture for this assay contained sample, 0.3 mM NAD+ and 0.5 U glucose dehydrogenase (Sigma, St. Louis, MO, USA) in MacIlvain buffer (pH 8.0). The mixture was maintained at 37 °C for 10 min, and then absorbance was measured at 340 nm. Formation of fructose via 4-epimerization of tagatose was confirmed using the fructose dehydrogenase coupling method [20]. A 40 μL reaction mixture was further combined with 0.5 U fructose dehydrogenase (Sigma) and incubated at 37 °C for 10 min. Ten microliters of ferricyanide solution (0.1 M potassium ferricyanide and 0.1% Triton X-100 in MacIlvain buffer, pH 4.5) and 50 μL of ferric sulfate-SDS solution (5 g Fe2[SO4]3∙H2O, 3 g SDS and 95 mL of 85% phosphoric acid in 1,000 mL D.W.) were sequentially added, and the resulting mixture was incubated at 37 °C for 20 min. The absorbance of the mixture was measured at 660 nm.
One unit (U) of enzyme activity was defined as the amount of enzyme required to liberate 1 nmol of product per minute at pH 7.5 and 35 °C.
Effects of pH, Temperature, and [NAD+] on Enzyme Activity
The effects of pH and temperature on GalE were examined in an attempt to optimize the 4-epimerization of galactose. Fifty millimolar galactose was mixed with 0.5 U purified GalE in McIlvaine buffer (1 mL) for 1 h. The pH of the buffer and the temperature of the reaction were varied from 5.0 to 8.0 (at 35 °C) and from 25 to 45 °C (at pH 8.0), respectively. To determine the effect of the NAD+ concentration on the activity of GalE, reactions were supplemented with 0, 10, 50, 100, 200, or 300 μM NAD+. Glucose formation was estimated using the modified glucose dehydrogenase coupling method described above.
Kinetic Analysis
The concentrations of d-galactose and d-tagatose were varied from 10 to 500 mM in the above activity assays to determine the kinetic parameters of GalE-mediated 4-epimerization of each substrate. Reactions were performed in McIlvaine buffer (pH 7.5) at 35 °C for 1 h. Kinetic parameters such as V max (mM/min), K m (mM), and k cat (min−1) were determined by fitting data to the Michaelis–Menten equation.
3D Structural Modeling
Reference protein coordinates used for docking were taken from the X-ray structure of E. coli GalE in complex with UDP-glucose (Protein Data Bank [PDB] access code: 1XEL) [21]. The structure of the bound ligand UDP-glucose was extracted from the complex using Pymol software (www.pymol.org) and served as the reference for docking. The PDB structure of d-glucose was obtained from www.nyu.edu/pages/mathmol/library/sugars and subsequently input as a potential complex to be sorted by shape complementarity criteria into PatchDock software (bioinfo3d.cs.tau.ac.il/PatchDock) [22].
Saturation Mutagenesis
To introduce random mutation at positions 179, 231, and 299 of GalE, site-directed mutagenesis was performed using a QuickChange kit (Stratagene, Beverly, MA, USA) according to the manufacturer’s protocol. Plasmid pEcGalE was used as the template and the oligonucleotides were as follows: GCGCTACTTCNNNCCGGTTGGCG and CGCCAACCGGNNNGAAGTAGCGC for mutation at position 179; TACTGGCGTANNNGATTACATCC and GGATGTAATCNNNTACGCCAGTA for position 232; CTTCCGGCCNNNTGGGCGGAC and GTCCGCCCANNNGGCCGGAAG for position 299. The resulting plasmids were transformed into E. coli ER2566.
For the primary screening, mutants were selected based on the formation of fructose from tagatose and analyzed by the fructose dehydrogenase coupling method. Cells were grown on agar plates containing LB medium with ampicillin (50 μg/mL) at 37 °C. One thousand colonies were transferred to 96-well plates containing 200 μL of LB-ampicillin as well as a master plate. The 96-well plates were incubated at 37 °C and 900 rpm overnight to allow growth of cells. Ten-microliter samples of culture were transferred to another 96-well plate containing 200 μL of fresh induction medium (LB-ampicillin–2 mM lactose) and incubated at 37 °C and 900 rpm for 6 h to induce protein expression.
Cell growth was measured on a multiplate reader (Microplate Reader 550; Bio-Rad, Richmond, VA, USA) at 600 nm. Eighty microliters of induced cells was mixed with 20 μL of lysis buffer (0.1% Triton X-100 and 1 mM PMSF in MacIlvain buffer, pH 7.5) and incubated at 37 °C and 400 rpm for 30 min to lyse cells. Cell extracts were used to assay tagatose 4-epimerization activity, and fructose formation was estimated by the fructose dehydrogenase coupling method. The enzymatic activity of the selected mutant enzyme in terms of tagatose-into-fructose conversion and the reverse reaction was further characterized by measuring 4-epimerization product formation via HPLC.
Results
Vmax/Km Values of GalE for Free Monosaccharides
To determine if UDP-galactose-4-epimerase (GalE) harbors 4-epimerization activity for galactose, purified GalE was mixed with galactose and the formation of the 4-epimerization product, glucose, was detected by the modified glucose dehydrogenase coupling method. Glucose was formed at a rate of 0.5 nmol/mg protein per minute. Since the optimal reaction conditions for the conversion of free monosaccharide were unknown, the optimal pH, temperature, and concentration of NAD+ were determined (Electronic Supplementary Material (ESM) Fig. S1). The optimal temperature and pH were found to be 35 °C and 7.5, respectively, which are similar to those for the 4-epimerization of UDP-galactose [6]. GalE enzymatic activity was not significantly increased by varying the NAD+ concentration in the reaction buffer. Thus, subsequent reactions with free monosaccharides were performed under these conditions.
After obtaining the above results, 4-epimerization by GalE of various free aldohexoses (glucose, galactose, allose, gulose, altrose, idose, mannose, and talose) and ketohexoses (fructose, tagatose, psicose, and sorbose) was investigated and verified by HPLC (Table 1 and ESM Fig. S2). GalE was able to mediate 4-epimerization of glucose, galactose, fructose, tagatose, psicose, and sorbose into galactose, glucose, tagatose, fructose, sorbose, and psicose at K m/V max values of 0.03, 0.04, 0.21, 0.84, 1.14, and 0.17 min−1, respectively. No 4-epimerization activity was detected for allose, gulose, altrose, idose, mannose, and talose.
Kinetics of 4-Epimerization of Galactose and Tagatose
To estimate the kinetic parameters of GalE 4-epimerization of galactose and tagatose, substrate concentrations were varied and the 4-epimers of these sugars (glucose and fructose) were measured by the modified glucose and fructose dehydrogenase coupling methods, respectively (Table 2). The K m and k cat values for galactose were 26.4 mM and 0.0155 min−1, respectively, and those for tagatose were 237 mM and 0.327 min−1, respectively. The catalytic efficiencies of GalE for galactose (5.8 × 10−4 mM−1 min−1) and tagatose (1.4 × 10−3 mM−1 min−1) were much lower than that previously reported for UDP-galactose (2.0 × 105 mM−1 min−1) [27].
Saturation Mutation at Active Site of GalE
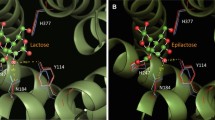
To choose targets for the saturation mutation, the X-ray structure of E. coli GalE in complex with UDP-glucose was compared with a model structure combined with glucose (Fig. 1). This structure was obtained by replacement of UDP-glucose with free glucose in ligand docking and sorted by shape complementarity criteria using PatchDock software. Residues in the active pocket of GalE were categorized as those near the UDP moiety, the NAD+ moiety, and the glucose moiety. The residues near the UDP moiety (Asp179, Arg231, Tyr299) were chosen as the saturation mutation targets for 4-epimerization of free monosaccharide substrates.
Random mutations were introduced into the target positions, and mutated GalE showing the greatest increase in 4-epimerization activity for tagatose was selected by the fructose dehydrogenase coupling assay from among 240 mutants of each target. In the selected clone, Asp179 was mutated to Ser, and its V max/K m values for tagatose and fructose substrates were determined by HPLC (Table 3). The V max/K m values for tagatose and fructose 4-epimerization of N179S were 1.76 ± 0.21 and 0.31 ± 0.04 min−1, respectively, which were 2 and 1.5 times higher than those of the wild type. The protein expression was not significantly varied by the mutation at 179 (ESM Fig. S3). No mutations at 231 and 299 showing greater 4-epimerization activity were found.
Discussion
UDP-galactose-4-epimerase was able to mediate 4-epimerization of free monosaccharides (Table 1 and ESM Fig. S2), as well as it does that of nucleic acid-activated sugars. Among the free monosaccharide substrates, ketohexoses such as psicose and tagatose were more efficiently converted by GalE compared to aldohexoses. The range of free monosaccharide substrates was wider than that previously reported for nucleic acid-activated sugars; E. coli GalE activity has only been reported for UDP-galactose and UDP-glucose [23, 24].
Ligand docking model enabled us to better understand GalE-mediated 4-epimerization of free monosaccharides and to define potential means of improving GalE activity (Fig. 1). The crucial catalytic residues of the enzyme are known to be Ser124 and Try149, with the NAD+ cofactor also being required for catalysis [25–28]. The exact positioning of the 4-OH of a sugar moiety between Ser124, Try149, and the NAD+ cofactor is essential for 4-epimerization. The docking model in Fig. 1 shows that the 4-OH group of glucose is located within 3.7 and 3.9 Å of Ser124 and Try149, respectively; as such, hydrogen bond formation could initiate electron transfer. In the case of a UDP-sugar, the bulky UDP moiety contributes to the correct positioning of the sugar moiety (Fig. 1a). Free monosaccharides, however, have no C1 branch to facilitate the correct positioning of a 4-OH group (Fig. 1b); this difference in structure is thought to account for the significantly lower catalytic efficiency of GalE for free monosaccharide compared with that observed for a UDP-sugar substrate (Table 2). The catalytic efficiency (k cat/K m) for galactose was eight orders of magnitude less than that previously reported for UDP-galactose; this was mainly due to slower turnover (k cat; six orders of magnitude lower) rather than lower affinity (K m; two orders of magnitude lower). This finding implies that the positioning of the 4-OH group between crucial catalytic residues is not stable with a free monosaccharide substrate compared with a UDP-sugar substrate; thus, slower turnover in the 4-epimerization reaction is the result. If the binding capacity of free monosaccharide increases with more stable positioning, specific alteration of GalE might enhance turnover and subsequently enhance catalytic efficiency.
Saturation mutations near the active pocket provided enhanced 4-epimerization activity for tagatose and fructose, an outcome that might be interesting from the viewpoint of rare sugar production (Table 3). This result enabled us to infer that GalE can be used as a template for the directed evolution to develop enzymes mediating the 4-epimerization of other free monosaccharides at an enhanced rate. The 3D structure of GalE in complex with free glucose described herein will be helpful when such an attempt is made. Production of evolved GalE exhibiting enhanced conversion of glucose into galactose and fructose into tagatose may be of commercial interest because it might enable the manufacture of rare sugars from naturally abundant sources.
References
Bhosale, S. H., Rao, M. B., & Deshpande, V. V. (1996). Microbiological Reviews, 60(2), 280–300.
Kim, P. (2004). Applied Microbiology and Biotechnology, 65(3), 243–249.
Matsuo, T., et al. (2001). Asia Pacific Journal of Clinical Nutrition, 10(3), 233–237.
Matsuo, T., et al. (2002). Journal of Nutritional Science and Vitaminology (Tokyo), 48(1), 77–80.
Leloir, L. F. (1951). Archives of Biochemistry, 33(2), 186–190.
Vanhooke, J. L., & Frey, P. A. (1994). Journal of Biological Chemistry, 269(50), 31496–31504.
Agarwal, S., et al. (2007). Biochimica et Biophysica Acta, 1774(7), 828–837.
Samanta, A. K., & Bhaduri, A. (1983). Journal of Biological Chemistry, 258(18), 11118–11122.
Grossiord, B. P., et al. (2003). Journal of Bacteriology, 185(3), 870–878.
Geren, C. R., & Ebner, K. E. (1977). Journal of Biological Chemistry, 252(6), 2082–2088.
Daude, N., et al. (1995). Biochemical and Molecular Medicine, 56(1), 1–7.
Kotake, T., et al. (2009). Biochemical Journal, 424(2), 169–177.
Shneour, E. A., & Hansen, I. M. (1969). Brain Research, 16(2), 501–510.
Granstrom, T. B., et al. (2004). Journal of Bioscience and Bioengineering, 97(2), 89–94.
Kim, H. J., et al. (2010). Journal of Microbiology and Biotechnology, 20(6), 1018–1021.
Shen, X., & Perreault, H. (1998). Journal of Chromatography A, 811(1–2), 47–59.
Honda, S., et al. (1989). Analytical Biochemistry, 180(2), 351–357.
Suzuki, S., Kakehi, K., & Honda, S. (1996). Analytical Chemistry, 68(13), 2073–2083.
Ray, M., & Bhaduri, A. (1975). Journal of Biological Chemistry, 250(10), 3595–3601.
Ameyama, M., et al. (1981). Journal of Bacteriology, 145(2), 814–823.
Thoden, J. B., Frey, P. A., & Holden, H. M. (1996). Biochemistry, 35(16), 5137–5144.
Schneidman-Duhovny, D., et al. (2003). Proteins, 52(1), 107–112.
Darrow, R. A., & Rodstrom, R. (1968). Biochemistry, 7(5), 1645–1654.
Thoden, J. B., et al. (2001). Journal of Biological Chemistry, 276(18), 15131–15136.
Thoden, J. B., Frey, P. A., & Holden, H. M. (1996). Biochemistry, 35(8), 2557–2566.
Thoden, J. B., et al. (1997). Biochemistry, 36(21), 6294–6304.
Liu, Y., et al. (1997). Biochemistry, 36(35), 10675–10684.
Thoden, J. B., Frey, P. A., & Holden, H. M. (1996). Protein Science, 5(11), 2149–2161.
Acknowledgments
This work was supported by the Korean Ministry of Education, Science, and Technology (R01-2009-0070677, R1-2007-000-202310). P. Kim was further supported by a 2009 Research Grant from the Catholic University of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Figure S1
Optimal conditions (pH, temperature, and [NAD+]) for 4-epimerization of glucose (PDF 23 kb)
Figure S2
SDS-PAGE of GalE wild-type and variant N179S (soluble fraction) (PDF 17 kb)
Figure S3
Chromatograms of GalE-mediated conversion of glucose into galactose (a), fructose into tagatose (b), and psicose into sorbose (c). Chromatograms obtained after 4 h were superimposed on chromatograms obtained before the reaction (PDF 99 kb)
Rights and permissions
About this article
Cite this article
Kim, HJ., Kang, S.Y., Park, J.J. et al. Novel Activity of UDP-Galactose-4-Epimerase for Free Monosaccharide and Activity Improvement by Active Site-Saturation Mutagenesis. Appl Biochem Biotechnol 163, 444–451 (2011). https://doi.org/10.1007/s12010-010-9052-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-010-9052-7