Abstract
Stem cells derived from synovial lining—synovial lining-derived stem cells or SDSCs—are a promising cell source for cartilage tissue engineering. We hypothesized that negatively selected SDSCs would form cartilage constructs and conventionally passaged SDSCs would be contaminated with macrophages, inhibiting SDSC-based chondrogenesis. We mixed SDSCs with fibrin gel and seeded the cells into polyglycolic acid scaffolds. After 3 days of incubation with a proliferative growth factor cocktail (containing transforming growth factor β1 [TGF-β1], insulin-like growth factor I [IGF-I], and basic fibroblast growth factor [FGF-2]), the cell-fibrin-polyglycolic acid constructs were transferred into rotating bioreactor systems and cultured with a chondrogenic growth factor cocktail (TGF-β1/IGF-I) for up to 4 weeks. Tissue constructs based on negatively selected SDSCs had cartilaginous characteristics; were rich in glycosaminoglycans and collagen II; exhibited high expression of mRNA and protein for collagen II, aggrecan, and Sox 9; exhibited a negligible level of mRNA and protein for collagens I and X; and had an equilibrium modulus in the range of values measured for native human cartilage. Conventional passage yielded SDSCs with contaminating macrophages, which adversely affected the quality of tissue-engineered cartilage. We thus propose functional cartilage constructs could be engineered in vitro through the use of negatively isolated SDSCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Articular cartilage is unique in its limited ability to heal, and thus cartilage lesions often progress to arthritis. Given the well-known limitations of autologous and allogeneic chondrocytes for transplantation procedures, much attention has recently been focused on multipotential cells with chondrogenic potential. Synovial lining might be a useful source of chondrogenic tissue because it is readily available to the surgeon operating on cartilage lesions and it has a propensity to undergo chondrogenesis manifested in the condition of synovial chondromatosis [6, 22, 23, 35]. Furthermore, human chondroprogenitor cells of synovial origin sustain their high proliferative potential and capacity to differentiate into chondrocytes irrespective of the individual’s age [8, 25, 40]. Several studies suggest synovial lining cells as a candidate source of chondrogenic cells. (1) Synovial cells share several properties with chondrocytes, including the production of cartilage oligomeric matrix protein [9, 10, 30], link proteins [12], and sulfated glycosaminoglycans [17]. (2) Studies relating to the ontogenetic development of synovial joints have revealed articular cartilage cells and synovial cells originate from a common precursor pool [26] and exist in a close functional relationship not only during fetal development but also in adult life [2]. (3) Under various pathologic conditions, synovial cells have strong chondrogenic potential [1, 38].
We previously developed a fast and feasible method (“negative isolation”) to remove macrophages (type A synoviocytes) and purify synovial fibroblasts (SFBs, type B synoviocytes) also known as stem cells derived from synovial lining (synovial lining-derived stem cells or SDSCs), believed to be the cell source responsible for cartilage differentiation [27]. In the presence of TGF-β1, SDSCs purified by negative isolation formed a superior cartilage micromass compared to that formed by conventional passage; using a pellet culture system, we also defined the optimum concentration and sequence of growth factors for SDSC proliferation and chondrogenic differentiation [27]. However, the question still remained whether these cells could form functional cartilage tissue constructs.
We asked whether engineered tissue constructs with SDSCs purified by negative isolation had better cartilage properties in molecular, structural, and functional aspects than those by conventional passage. We tested the hypothesis that negatively isolated SDSCs can form cartilage constructs and conventionally passaged SDSCs are contaminated with macrophages, inhibiting SDSC-based chondrogenesis.
Materials and Methods
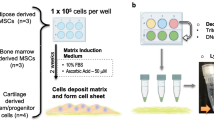
We harvested synovial tissue from both knees of two young pigs. SDSCs purified by either negative isolation or conventional passage were compared to determine the difference in chondrogenic properties utilizing an established cartilage tissue engineering model, which is through bioreactor cultivation of cells on composite scaffolds made of fibrin gel and polyglycolic acid (PGA) mesh (Fig. 1). The proliferation and chondrogenesis of SDSCs were modulated by sequential application of growth factor cocktails, and the physical signaling was provided by the hydrodynamically active environment of rotating bioreactors. The progression of chondrogenesis (chondrogenic gene expression [SRY (sex determining region Y)-box 9 (Sox 9), aggrecan, collagens II and X]) and tissue assembly (compressive functionality) from negatively isolated SDSCs were evaluated over 4 weeks of cultivation by assessing the molecular properties (Western blot was used for evaluation at the protein level and quantitative TaqMan® PCR at the mRNA level with three constructs); structural properties (safranin O staining for GAG expression and immunostaining for collagens I and II, as well as quantitative biochemical analyses for DNA and GAG contents with four constructs); and mechanical properties (equilibrium modulus with four constructs) of tissue constructs. The chondrogenic ability of negatively isolated- and conventionally passaged-SDSCs was compared over 4 weeks of cultivation by assessing the structural (safranin O staining for GAG expression and immunostaining for collagens I, II, and macrophage as well as quantitative biochemical analyses for DNA and GAG contents with four constructs) and mechanical properties (equilibrium modulus) of tissue constructs.
The diagram illustrates the research design. MSCs derived from synovial tissue using two different methods were suspended in fibrin glue and seeded into fibrous biodegradable scaffolds made of PGA for 3 days under static conditions and then cultured for 4 weeks in rotating bioreactors. (A) Synovial tissue harvesting, (B) negative isolation method, (C) purified SDSCs, (D) conventional passage method, (E) PGA disc, (F) fibrin glue, (G) cell-fibrin glue-PGA construct, (H) static culture (3 days), and (I) bioreactor culture (4 weeks) are shown.
Random biopsies of the intimal layer of synovial tissue (Fig. 1A) were obtained aseptically from the knees of two young pigs and pooled together for the study. After temporary storage in culture medium at 4°C, the synovial tissue was finely minced and digested at 37°C on an x-y-z shaker (Clay Adams® Nutator; BD Biosciences, Bedford, MA) for 30 minutes in phosphate-buffered saline (PBS) containing 0.1% trypsin and then for 2 hours in 0.1% solution of collagenase P in Dulbecco’s modified Eagle’s medium (DMEM)/10% fetal bovine serum (FBS). The cell suspension was passed through a 70-μm nylon filter, and the cells were collected from the filtrate by centrifugation. Cells were cultured for 4 days in complete medium (DMEM/10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin). Nonadherent cells were removed by a PBS wash on days 2 and 4, and the remaining adherent cells were processed by two different methods, negative isolation and conventional passaging, as described below.
For negative isolation of SFBs (Fig. 1B) from primary cultures of adherent synovial cells containing macrophages and fibroblasts, cells were detached by trypsinization for 1 minute (0.25% trypsin/0.2% EDTA), washed, and suspended in PBS/2% FBS (107 cell/mL). The suspension was incubated with 5 × 107/mL Dynabeads® M-450 CD14 containing a monoclonal antibody specific for macrophages (Dynal Biotech, Oslo, Norway) for 1 hour at 4°C on an x-y-z shaker. The conjugated cells and the unbound Dynabeads® were collected using the Dynal Magnetic Particle Concentrator® (Dynal Biotech), and the depleted supernatant with SFBs was transferred to a second tube (Fig. 1C). The SFBs isolated by conventional passage, without the negative selection step, served as a control (Fig. 1D).
Degradable PGA scaffolds (a mesh of 15-μm fibers, void volume of 97; Synthecon, Houston, TX) [28, 29] were punched into eighty 5-mm-diameter × 2-mm-thick discs, sterilized with ethylene oxide, immersed in 100% ethanol, 70% ethanol, and then PBS (without Ca2+ and Mg2+) (Fig. 1E). In a centrifuge tube, 150 μL fibrinogen (100 mg/mL in PBS, from human plasma; Sigma, St. Louis, MO) (Fig. 1F), 140 μL PBS with cells, 5 μL thrombin (0.1 U/μL, from human plasma; Sigma), and 5 μL CaCl2 (50 mM) were sequentially added. Then, 26 μL of the cell-gel mixture was pipetted onto a PGA disc in a Petri dish. This procedure resulted in fibrin-PGA composites containing 2.6 × 106 cells per scaffold (Fig. 1G), corresponding to an initial seeding density of 100 × 106 cells/mL scaffold volume. The whole process was completed in 3 minutes. The dish with constructs was transferred into an incubator at 37°C for 10 minutes. Then complete medium was added to cover the constructs.
After 1 hour, the medium was replaced by chemically defined medium (high-glucose DMEM, 40 μg/mL proline, 100 nmol/L dexamethasone, 0.1 mmol/L ascorbic acid 2-phosphate, 100 U/mL penicillin, 100 mg/L streptomycin, and 1x ITS™ Premix) supplemented with a proliferative growth factor cocktail (10 ng/mL transforming growth factor β1 [TGF-β1], 50 ng/mL basic fibroblast growth factor [FGF-2], and 500 ng/mL insulin-like growth factor I [IGF-I]) for 3 days (Fig. 1H) [27]. The cell-fibrin-PGA constructs were transferred into a rotating bioreactor (RCCS-4; Synthecon, Houston, TX) filled with a chemically defined medium supplemented by a differentiative growth factor cocktail (10 ng/mL TGF-β1 and 500 ng/mL IGF-I) for 4 weeks (Fig. 1I) [27]. Over the course of in vitro cultivation, the bioreactor rotation speed was adjusted so that the growing constructs remained freely suspended in the rotating flow.
For histologic analysis [29], constructs (n = 2) were fixed overnight at 4°C in 4% paraformaldehyde in PBS, paraffin-embedded, and sectioned to 5 μm. Consecutive sections were stained with safranin O/fast green for sulfated glycosaminoglycan (GAG) and were immunostained with monoclonal antibodies against collagen II (II-II6B3; DSHB, Iowa City, IA), collagen I (Abcam, Cambridge, MA), collagen X (Sigma), and macrophage (Spring Bioscience, Fremont, CA). Immunohistochemical sections were hydrated, treated with 1% hydrogen peroxide to inhibit endogenous peroxidase, and incubated for 30 minutes with 2 mg/mL testicular hyaluronidase in PBS (pH 5) at 37°C followed by another 30 minutes with 1.5% normal goat serum and overnight at 4°C with the primary antibody, then stained using a kit (Vectastain ABC, Burlingame, CA), followed by standardized development in diaminobenzidine (DAB, Invitrogen). The sections were counterstained with hematoxylin. The images were recorded by a Provis AX70 light/fluorescence microscope (Olympus, Melville, NY) and SPOT digital microscopy camera (Diagnostic Instruments Inc., Sterling Heights, MI) with software entitled PictureFrame®, version 2.1 (Optronics, Goleta, CA). The stained sections were qualitatively evaluated for stain density and distribution by two of the authors (FH, VLK) who were blinded to treatments. Five sections were used to perform each stain from one single sample. Two samples from the same group were compared to each other to confirm the identity. The positive color for the safranin O stain is red; the positive color for the immunostain is brown.
For biochemical analyses, constructs (n = 4) were digested for 6 hours at 60°C with 125 μg/mL papain in PBE buffer (100 mmol/L phosphate, 10 mmol/L EDTA, pH 6.5) containing 10 mmol/L cysteine, by using 100 μL enzyme per sample. To quantify cell density, the amount of DNA in the papain digests was measured using the Quant-iTTM PicoGreen® dsDNA assay kit (Invitrogen) with a CytoFluor® Series 4000 (Applied Biosystems). GAG was measured by using dimethylmethylene blue dye [11] and a Spectronic™ BioMate™ 3 Spectrophotometer (Thermo Scientific, Milford, MA) with bovine chondroitin sulfate as a standard.
To assess collagen expression [29], constructs (n = 3) were lyophilized, measured for dry weights, incubated using 50-mg dry sample per 15 mL 4 mol/L guanidine hydrochloride (Sigma) for 20 hours at 4°C, homogenized, and digested with 0.6 mg/mL of pepsin (Sigma) per mL in 0.5 mol/L acetic acid (Sigma) at 4°C for 48 hours at a ratio of 10:1 (mg/mg) of dry sample to pepsin. The samples were centrifuged at 48,000 g for 1 hour, and the supernatant was lyophilized and dissolved in radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors. To assess noncollagen protein expression, the samples were homogenized and dissolved in RIPA buffer with protease inhibitors. Total proteins were quantified using BCATM Protein Assay Kit (Pierce, Rockford, IL). The samples were denatured and separated by NuPAGE® Novex® Bis-Tris Mini Gels (Invitrogen) in the XCell SureLockTM Mini-Cell (Invitrogen) at 120 V for 3 hours at 4°C. Bands were transferred onto a nitrocellulose membrane (Invitrogen) using XCell ΙΙTM blot module (Invitrogen) at 15 V overnight at 4°C. Nonspecific binding was blocked with 5% nonfat milk in TBST (100 mmol/L Tris-HCl, 0.9% NaCl, 1% Tween® 20, pH 7.5) for 1 hour. The membrane was incubated with a primary monoclonal antibody in 1% nonfat milk in TBST to collagen II (II-II6B3), collagen X (Sigma), aggrecan (Chemicon, Temecula, CA), and Sox 9 (Abcam) for 1 hour at room temperature (β-actin served as an internal control), followed by the secondary antibody of horseradish peroxidase-conjugated goat antimouse (Pierce) for 40 minutes at room temperature and exposure using SuperSignal® West Femto Maximum Sensitivity Substrate (Pierce) and CL-XPosure™ Film (Pierce). The membrane was then stripped using Restore™ Western Blot Stripping Buffer (Pierce) and incubated with the primary monoclonal and secondary antibody as described above. For semiquantitative analysis, bands were scanned (CanoScan8400F, Canon, Lake Success, NY) and analyzed using NIH Image J software (US National Institutes of Health, Bethesda, MD).
For studies of gene expression, the total RNA was extracted from constructs (n = 3) using an RNase-free pestle in TRIzol® reagent and RNeasy® Mini Kit (Qiagen, Valencia, CA). Sample mRNAs were quantified and 1 μg RNA was used for reverse transcription (RT) with High-Capacity cDNA Archive Kit (Applied Biosystems). Chondrogenic marker genes (collagen II, collagen X, Sox 9, and aggrecan) were customized by Applied Biosystems as part of the Custom TaqMan® gene expression assays (Table 1). Eukaryotic 18S RNA (Assay ID Hs99999901_s1 ABI) was carried out as the endogenous control gene. Real-time PCR was performed with the iCycler iQTM Multi Color RT-PCR Detection System (Bio-Rad Laboratories, Hercules, CA). The cycle parameters were 50°C for 2 minutes, hot start at 95°C for 10 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds, and annealing and extension at 60°C for 1 minute. The cycle threshold (Ct) values for 18S RNA and that of samples was measured and calculated by computer software (Perkin-Elmer, Wellesley, MA). Relative transcript levels were calculated as χ = 2−ΔΔCt, in which ΔΔCt = ΔE − ΔC, ΔE = Ctexp − Ct18s, and ΔC = Ctct1 − Ct18s [21].
Compressive moduli of constructs (n = 4) were determined in uniaxial stress-relaxation using a stepper motor-driven miniature compression device manufactured in-house with a miniature 5-mm DVRT® (Microstrain, Burlington, VT) as in our previous studies [29]. In brief, discs 3 mm in diameter and 1.5 mm thick were harvested from the central region of the construct. Discs were equilibrated in PBS containing protease inhibitors, placed in a cylindrical confining chamber filled with PBS, mounted in a miniature stepper motor-controlled material test machine, and compressed by a porous stainless steel platen by applying a 5% strain followed by four consecutive stress relaxations, each following a 2% strain step. Data were recorded at a sampling rate of 10 points/second over a time increment of 480 seconds. Constructs were considered to fully relax during this increment based on a change in stress of less than 0.006 MPa over the final 180 seconds. The equilibrium modulus was then determined for each sample as the slope of the best linear regression fit (r2 > 0.99) of the measured equilibrium stress versus applied strain.
We used a one-way ANOVA F test to compare the differences in biochemical analyses (n = 4), real time PCR (n = 3), and compressive modulus (n = 4 for negative isolation and 5 for conventional passage) between the groups. Statistical analysis was performed with SPSS 13.0 statistical software (SPSS Inc., Chicago, IL). Post-hoc power analyses were performed using a significance level of 0.05 and a minimum power equal to 0.80, assuming a two-sided alternate hypothesis. Estimates of standard deviation were obtained from the observed samples.
Results
SDSCs purified by negative isolation could be engineered into cartilage tissue constructs. The qualities were reflected by a number of key parameters, including mesenchymal stem cell (MSC) and early chondrogenic marker (collagen I), chondrogenic markers (Sox 9, collagen II and aggrecan), and a hypertrophy marker (collagen X) at morphology (histology), protein level (biochemistry and Western blot), mRNA level (real-time RT-PCR), and biomechanical property (equilibrium modulus). Our goal was to engineer cartilage tissue constructs containing a low level of collagen I, high levels of Sox 9, collagen II and aggrecan, nondetectable collagen X, and functionality comparable to native cartilage. Our data suggested the stain from two samples in each group was consistent. Sulfated GAG accumulated with time of culture from undetectable levels at Day 3 to high levels at 1 month (Fig. 2A/a–C/c). The concentration of GAG was particularly high in the outer ∼400-μm-thick region (Fig. 2C/c), which also contained high levels of Type II collagen (Fig. 2D/d), low levels of Type I collagen (Fig. 2E/e), and no Type X collagen. The biochemistry data suggested SDSCs purified by negative isolation proliferated over the first 3 days of culture and then maintained cell numbers at relatively steady levels (Fig. 2F) over the 4-week bioreactor culture with the treatment of a differentiative growth factor cocktail. The amounts of GAG increased rapidly between days 3 and 30 (Fig. 2G). The mRNA expression of collagen X was generally much lower than the other chondrogenic marker genes (Fig. 2H). The mRNA expression of collagen II, aggrecan, and Sox 9 increased between days 3 and 15 and then decreased slightly between days 15 and 30. Collagen II, aggrecan, and Sox 9 proteins, hardly detectable at Day 3, were upregulated throughout the duration of the bioreactor culture (Fig. 2I). We did not detect collagen X protein at any time. One-month tissue constructs had equilibrium moduli of 0.39 ± 0.14 MPa (Fig. 2J), which are within the range of data for native articular cartilage [5] and consistent with the presence of GAG (Fig. 2C/c, 2G) and Type II collagen (Fig. 2D/d).
Negatively isolated SDSCs could be engineered in vitro into cartilage tissue constructs. (A–E) Histologic cross-sections and (a–e) face sections are shown for constructs sampled at (A/a) 3 days, (B/b) 15 days, and (C/c–E/e) 1 month of cultivation. The tissue constructs (A/a–C/c; stain, Safranin O; magnification, ×100) shows the accumulation of sulfated GAG. The tissue constructs (D/d–E/e; stain, immunostain; magnification, ×100) also show the presence of (D/d) Type II collagen (Col II) and (E/e) Type I collagen (Col I) after 1 month of cultivation. All scale bars are 200 μm. (F) DNA content and (G) GAG content are expressed as total amounts per construct. (H) Relative expression levels of cartilage marker gene mRNAs for Sox 9, aggrecan (AG), collagen II (Col II), and collagen X (Col X) were evaluated by TaqMan® PCR for cartilage constructs cultured for 3 days, 15 days, and 1 month. (I) Protein expressions of Sox 9, aggrecan, collagen II, and collagen X were evaluated using Western blots of constructs cultured for 3 days (Lane 1), 15 days (Lane 2), and 1 month (Lane 3). (J) Equilibrium modulus was measured from four randomly selected constructs after 1 month of cultivation. The dotted line shows average ± SD for all constructs. Differences between the groups are indicated as follows: * = p < 0.05; ** = p < 0.01; and *** = p < 0.001. Data are shown as average ± SD for n = 4 (biochemical analyses) (F,G) and n = 3 (TaqMan® PCR) (H) constructs per group and time point.
SDSCs derived from conventional passage were contaminated with macrophages, which may inhibit SDSC-based chondrogenesis. Compared to negatively isolated SDSC-based constructs (Fig. 3A–E), conventional passage purified SDSC-based constructs (Fig. 3A–E) showed weak and sparse distribution of GAG (Fig. 3A) and Type II collagen (Fig. 3B) but intense strong expression of Type I collagen (Fig. 3C) and macrophage antigen (Fig. 3D-E). Negatively isolated SDSC-based constructs densely and broadly expressed GAG (Fig. 3A) and Type II collagen (Fig. 3B) but weakly expressed Type I collagen (Fig. 3C). There was no macrophage antigen detectable in the constructs (Fig. 3D–E). The DNA content of conventional passage purified SDSC-based constructs dramatically decreased with time, whereas the DNA content of negatively isolated SDSC-based constructs was maintained at relatively steady levels (Fig. 3F). The negatively isolated SDSC-based constructs contained almost twice as much GAG as the conventional passage purified SDSC-based constructs at 15 days (125 ± 10 μg versus 68 ± 3 μg, p = 0.00058) and 1 month (158 ± 22 μg versus 92 ± 4 μg, p = 0.0074) of culture (Fig. 3G), consistent with histologic data (Fig. 3A/a–E/e). For GAG, we assume that the smallest meaningful difference in means between the two groups that we would like to detect is 57 units. Then the standard deviation of the GAG scores is approximately 7.0 units with a total sample size of eight constructs (four in each group). The observed power is greater than 0.999. Biomechanical evaluation corroborated the histologic and biochemical findings (Fig. 3F–G) and showed that negatively isolated SDSC-based constructs had compressive moduli that were twice as high as those of conventional passage purified SDSC-based constructs (0.39 ± 0.14 MPa versus 0.19 ± 0.08 MPa, respectively) (Fig. 3H). For the compressive moduli variable, we assume that the smallest meaningful difference between the two groups that we would like to detect is 0.2 units. Therefore, the standard deviation of the difference in mean values is approximately 0.11 units with a total sample size of nine constructs (four constructs in the experimental group and five constructs in the control group). That means that the observed power is approximately 0.650.
SDSCs derived from conventional passage were contaminated with macrophages, inhibiting SDSC-based chondrogenesis. Histologic sections of 1-month constructs were stained: sulfated GAG (A/a; stain, Safranin O; magnification, ×100), collagen II (B/b; stain, immunostain; magnification, ×100), collagen I (C/c; stain, immunostain; magnification, ×100), and macrophage (stain, immunostain; magnification, D/d, ×100 and E/e, ×400). Sections A–E are for constructs engineered using SDSCs isolated by conventional passaging; sections a–e are for constructs engineered using SDSCs purified by negative isolation. Scale bars are 200 μm for A/a to D/d and 50 μm for E/e. (F) DNA content (μg/constructs), (G) GAG content (μg/constructs), and (H) equilibrium moduli of 1-month constructs are shown. Differences are indicated as follows: * = p < 0.05, ** = p < 0.01, and *** = p < 0.001. Data are shown as average ± SD (n = 4 constructs per group and time point).
Discussion
We believe synovial tissue is a promising cell source for cartilage tissue engineering. Our previous studies developed a quick and feasible method to purify stem cells derived from synovial lining and also characterized the sequence and combination of key growth factors for optimization of SDSC proliferation and chondrogenic differentiation in a micromass culture system. In the presence of TGF-β1, SDSCs purified by negative isolation formed a superior cartilage micromass than that by conventional passage [27]. However, the question remained whether these cells can form functional cartilage tissue constructs. We also questioned whether the engineered tissue constructs with SDSCs purified by negative isolation had better cartilage properties in molecular, structural, and functional aspects compared to conventional passage.
Our study design still presents some limitations. First, the samples designed and collected for the in vitro analyses were not sufficient for equilibrium modulus (n = 4) analyses. Therefore, our data suggested negatively isolated SDSC-based constructs had compressive moduli that were twice as high but statistically similar to those of conventional passage-derived SDSC-based constructs (0.39 ± 0.14 MPa and 0.19 ± 0.08 MPa, respectively). Second, the SDSCs used in this study were harvested and isolated from both knees of two pigs. This is a weakness of the research design. In the future, synovial tissue from many pigs will be collected and pooled together for the SDSC-based chondrogenesis study. Third, our preliminary data has demonstrated negative isolation could remove macrophages from the synovial cell population (data not shown). However, if we could use flow cytometry to further quantify the percentage of macrophage-like cells in the preparations (conventional versus selected), it would be more convincing for the efficacy of the negative isolation technique. Finally, compared to conventional passage, SDSCs purified by negative isolation were devoid of macrophages and these SDSC-based tissue constructs were more cartilage-like. However, the underlying mechanisms need further elucidation.
We demonstrated cartilage constructs can be engineered in vitro using negatively isolated SDSCs and hybrid scaffolds followed by culturing in a rotating bioreactor system and treating with sequential growth factor cocktails. Our previous study demonstrated that, in chondrogenesis of SDSCs, TGF-β1 plays a key role, either on cell proliferation when applied in combination with FGF-2 and IGF-I or on chondrogenic cell differentiation when applied with IGF-I [27]. In this study, after we mixed SDSCs with fibrin glue and seeded the cells into PGA scaffolds, we incubated the constructs in the presence of TGF-β1/IGF-I/FGF-2 for 3 days, followed by 4 weeks with TGF-β1/IGF-I in a rotating bioreactor. The engineered tissue constructs exhibited type-specific protein expression high for collagen II, aggrecan, and Sox 9, low for collagen I , and negligible for collagen X. One-month constructs appeared rich in GAG and Type II collagen and had a measured equilibrium modulus in the range of values for native cartilage [5]. Data collected in this study and in our previous work suggest supplementation with TGF-β1/FGF-2/IGF-I enhances SDSC proliferation and TGF-β1/IGF-I enhances chondrogenic differentiation and can form a basis for SDSC-based cartilage tissue engineering.
Recently, there have been many reports comparing SDSCs with other sources of MSCs, such as bone marrow, periosteum, muscle, and adipose tissue, for their proliferation capacity and multilineage differentiation potential [24, 31, 34, 41]. The consensus from all these investigations is the superiority of synovial lining as a source of MSCs in cartilage tissue engineering and regeneration. The number of colony-forming unit fibroblasts (CFU-Fs) identified from synovial lining was reportedly 1 in 12.5 to 80 nucleated cells plated, which was much greater than that of bone marrow, 1 in 104 to 105 cells [7, 16, 37]. When appropriately stimulated in vivo, synovial cells can be induced to migrate from the synovial lining into partial-thickness articular cartilage defects and therein to differentiate into chondrocytes [19, 20]. With regard to the availability of SDSCs for clinical use, synovial lining can be obtained arthroscopically with a low degree of invasiveness and without causing complications at the donor site due to its high regenerative capacity [14, 36]. There is a report from Sakaguchi et al. [31] showing that an average of 21,000 cells per milligram of synovial lining collected were obtained after the nucleated cells were plated at optimal density and cultured for 14 days. A small sample of synovial lining harvested with a punch biopsy would be sufficient to obtain SDSCs for future treatments. From the standpoint of super chondrogenic differentiation capacity and the ready clinical availability, the synovial lining is an excellent source of MSCs for articular cartilage repair.
However, there are currently few studies focusing on SDSC-based cartilage tissue engineering. Yokoyama et al. [40] mixed SDSCs with collagen gel and incubated the constructs in a serum-free defined medium supplemented with 10 ng/mL TGF-β1 and 500 ng/mL bone morphogenetic protein 2 (BMP-2). Composites with higher cell densities (5 × 107 and 108 cells/mL) after 21 days were richer in proteoglycans than those with lower cell densities, a finding consistent with our data (high cell seeding density, 108 cells/mL). They also reported that, after 1 day, MSC/gel composites contracted and the diameter decreased by 30% even though they were stable thereafter [40]. This is not consistent with our data. Our data demonstrated that the size of tissue constructs increased about 34.13 ± 3.07% after 4-week incubation in a bioreactor (data not shown), presumably due to our use of complementary fibrin gel/PGA hybrid scaffolds in which fibrin gel allows homogeneous cell distribution and chondrogenic differentiation [13, 18] and biodegradable PGA scaffolds provide sufficient load-bearing capacity and structural and mechanical integrity [29]. Both TGF-β1 and BMP-2 were reported to enhance chondrogenic differentiation of MSCs; however, TGF-β1 inhibited chondrocyte hypertrophy [39] and BMP-2 promoted chondrocyte hypertrophy [15, 33]. Therefore, TGF-β1 (10 ng/mL) was chosen in our study combined with a high concentration of IGF-I (500 ng/mL), which produced the highest chondrogenic differentiation of SDSCs [27]. Our preliminary data suggested that if the ratio of IGF-I to TGF-β1 (10 ng/mL) was less than 10, the chondrogenic index (GAG/DNA) of SDSC-based pellets would decrease in a 14-day serum-free incubation compared to the use of TGF-β1 (10 ng/mL) alone. Our study corroborated the findings of Sakimura et al. [32]. They seeded SDSCs into PGA scaffolds and treated with a growth factor combination of TGF-β1 (10 ng/mL) and IGF-I (100 ng/mL) in a serum-free defined medium. After 4 weeks, they found the chondrogenic index of the combination group was less than that of TGF-β1 alone; however, after 8 weeks, the index increase rate of the combination group passed that of TGF-β1 alone. The underlying mechanism is still unknown.
Another important finding in this study is that conventionally passaged SDSCs are contaminated with macrophages, inhibiting SDSC-based chondrogenesis. Our data suggest the negative isolation approach yielded more purified SDSCs without contamination by macrophages and that these cells yielded more mature cartilage constructs. In contrast, the tissue constructs engineered with SDSCs isolated from conventional passage exhibited about half of the DNA and GAG content as well as equilibrium modulus. Histologic immunostaining also suggested there were macrophage antigens detectable in the sections from conventionally passaged SDSC-based constructs. In osteoarthritis, synovial macrophages exhibit an activated phenotype and produce degradative enzymes resulting in the destruction of cartilage [3, 4]. Cartilage tissue engineering mimics some aspects of cartilage development and remodeling in vivo. Therefore, we postulated that macrophages would become active and inhibit chondrogenic differentiation when conventional passage was used to isolate SDSCs from synovial tissue. The mechanism for the inhibition of macrophages in SDSC chondrogenic differentiation needs further research.
Taken together, our data suggest MSCs isolated from synovial tissue can be used to engineer cartilaginous tissue constructs with well-differentiated structural, molecular, and biomechanical properties and the method of cell isolation (negative selection versus conventional passaging) is critical for chondrogenic properties of the cells cultured on scaffolds in the bioreactor containing differentiation growth factors.
References
Allard SA, Maini RN, Muirden KD. Cells and matrix expressing cartilage components in fibroblastic tissue in rheumatoid pannus. Scand J Rheumatol Suppl. 1988;76:125–129.
Bandara G, Georgescu HI, Lin CW, Evans CH. Synovial activation of chondrocytes: evidence for complex cytokine interactions. Agents Actions. 1991;34:285–288.
Blom AB, van Lent PL, Libregts S, Holthuysen AE, van der Kraan PM, van Rooijen N, van den Berg WB. Crucial role of macrophages in matrix metalloproteinase-mediated cartilage destruction during experimental osteoarthritis: involvement of matrix metalloproteinase 3. Arthritis Rheum. 2007;56:147–157.
Bondeson J, Wainwright SD, Lauder S, Amos N, Hughes CE. The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res Ther. 2006;8:R187.
Chen AC, Bae WC, Schinagl RM, Sah RL. Depth- and strain-dependent mechanical and electromechanical properties of full-thickness bovine articular cartilage in confined compression. J Biomech. 2001;34:1–12.
Crawford A, Frazer A, Lippitt JM, Buttle DJ, Smith T. A case of chondromatosis indicates a synovial stem cell aetiology. Rheumatology (Oxford). 2006;45:1529–1533.
Deans RJ, Moseley AB. Mesenchymal stem cells: biology and potential clinical uses. Exp Hematol. 2000;28:875–884.
De Bari C, Dell’Accio F, Vandenabeele F, Vermeesch JR, Raymackers JM, Luyten FP. Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. J Cell Biol. 2003;160:909–918.
Di Cesare PE, Carlson CS, Stollerman ES, Chen FS, Leslie M, Perris R. Expression of cartilage oligomeric matrix protein by human synovium. FEBS Lett. 1997;412:249–252.
Dodge GR, Hawkins D, Boesler E, Sakai L, Jimenez SA. Production of cartilage oligomeric matrix protein (COMP) by cultured human dermal and synovial fibroblasts. Osteoarthritis Cartilage. 1998;6:435–440.
Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177.
Fife RS, Caterson B, Myers SL. Identification of link proteins in canine synovial cell cultures and canine articular cartilage. J Cell Biol. 1985;100:1050–1055.
Fortier LA, Brofman PJ, Nixon AJ, Mohammed HO. Disparate chondrocyte metabolism in three-dimensional fibrin cultures derived from autogenous or commercially manufactured fibrinogen. Am J Vet Res. 1998;59:514–520.
Fowler MR, Nathan CO, Abreo F. Synovial metaplasia, a specialized form of repair. Arch Pathol Lab Med. 2002;126:727–730.
Gelse K, von der Mark K, Aigner T, Park J, Schneider H. Articular cartilage repair by gene therapy using growth factor-producing mesenchymal cells. Arthritis Rheum. 2003;48:430–441.
Gronthos S, Simmons PJ. The growth factor requirements of STRO-1-positive human bone marrow stromal precursors under serum-deprived conditions in vitro. Blood. 1995;85:929–940.
Hamerman D, Smith C, Keiser HD, Craig R. Glycosaminoglycans produced by human synovial cell cultures. Coll Relat Res. 1982;2:313–329.
Homminga GN, Buma P, Koot HW, van der Kraan PM, van den Berg WB. Chondrocyte behavior in fibrin glue in vitro. Acta Orthop Scand. 1993;64:441–445.
Hunziker EB. Growth-factor-induced healing of partial-thickness defects in adult articular cartilage. Osteoarthritis Cartilage. 2001;9:22–32.
Hunziker EB, Rosenberg LC. Repair of partial-thickness defects in articular cartilage: cell recruitment from the synovial membrane. J Bone Joint Surg Am. 1996;78:721–733.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408.
Maurice H, Crone M, Watt I. Synovial chondromatosis. J Bone Joint Surg Br. 1988;70:807–811.
Miyamoto H, Sakashita H, Wilson DF, Goss AN. Synovial chondromatosis of the temporomandibular joint. Br J Oral Maxillofac Surg. 2000;38:205–208.
Mochizuki T, Muneta T, Sakaguchi Y, Nimura A, Yokoyama A, Koga H, Sekiya I. Higher chondrogenic potential of fibrous synovium- and adipose synovium-derived cells compared with subcutaneous fat-derived cells: distinguishing properties of mesenchymal stem cells in humans. Arthritis Rheum. 2006;54:843–853.
Nishimura K, Solchaga LA, Caplan AI, Yoo JU, Goldberg VM, Johnstone B. Chondroprogenitor cells of synovial tissue. Arthritis Rheum. 1999;42:2631–2637.
Pacifici M, Koyama E, Iwamoto M, Gentili C. Development of articular cartilage: what do we know about it and how may it occur? Connect Tissue Res. 2000;41:175–184.
Pei M, He F, Vunjak-Novakovic G. Synovium-derived stem cell-based chondrogenesis. Differentiation. 2008; Doi: 10.1111/j.1432-0436.2008.00299.x.
Pei M, Seidel J, Vunjak-Novakovic G, Freed LE. Growth factors for sequential cellular de- and re-differentiation in tissue engineering. Biochem Biophys Res Commun. 2002;294:149–154.
Pei M, Solchaga LA, Seidel J, Zeng L, Vunjak-Novakovic G, Caplan AI, Freed LE. Bioreactors mediate the effectiveness of tissue engineering scaffolds. FASEB J. 2002;16:1691–1694.
Recklies AD, Baillargeon L, White C. Regulation of cartilage oligomeric matrix protein synthesis in human synovial cells and articular chondrocytes. Arthritis Rheum. 1998;41:997–1006.
Sakaguchi Y, Sekiya I, Yagishita K, Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52:2521–2529.
Sakimura K, Matsumoto T, Miyamoto C, Osaki M, Shindo H. Effects of insulin-like growth factor I on transforming growth factor beta1 induced chondrogenesis of synovium-derived mesenchymal stem cells cultured in a polyglycolic acid scaffold. Cells Tissues Organs. 2006;183:55–61.
Shintani N, Hunziker EB. Chondrogenic differentiation of bovine synovium: bone morphogenetic proteins 2 and 7 and transforming growth factor beta1 induce the formation of different types of cartilaginous tissue. Arthritis Rheum. 2007;56:1869–1879.
Shirasawa S, Sekiya I, Sakaguchi Y, Yagishita K, Ichinose S, Muneta T. In vitro chondrogenesis of human synovium-derived mesenchymal stem cells: optimal condition and comparison with bone marrow-derived cells. J Cell Biochem. 2006;97:84–97.
Sim FH, Dahlin DC, Ivins JC. Extra-articular synovial chondromatosis. J Bone Joint Surg Am. 1977;59:492–495.
Theoret CL, Barber SM, Moyana T, Townsend HG, Archer JF. Repair and function of synovium after arthroscopic synovectomy of the dorsal compartment of the equine antebrachiocarpal joint. Vet Surg. 1996;25:142–153.
Wexler SA, Donaldson C, Denning-Kendall P, Rice C, Bradley B, Hows JM. Adult bone marrow is a rich source of human mesenchymal ‘stem’ cells but umbilical cord and mobilized adult blood are not. Br J Haematol. 2003;121:368–374.
Xue C, Takahashi M, Hasunuma T, Aono H, Yamamoto K, Yoshino S, Sumida T, Nishioka K. Characterisation of fibroblast-like cells in pannus lesions of patients with rheumatoid arthritis sharing properties of fibroblasts and chondrocytes. Ann Rheum Dis. 1997;56:262–267.
Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153:35–46.
Yokoyama A, Sekiya I, Miyazaki K, Ichinose S, Hata Y, Muneta T. In vitro cartilage formation of composites of synovium-derived mesenchymal stem cells with collagen gel. Cell Tissue Res. 2005;322:289–298.
Yoshimura H, Muneta T, Nimura A, Yokoyama A, Koga H, Sekiya I. Comparison of rat mesenchymal stem cells derived from bone marrow, synovium, periosteum, adipose tissue, and muscle. Cell Tissue Res. 2007;327:449–462.
Acknowledgments
We thank Suzanne Smith for editing the manuscript and A.B. Billings for performing a post-hoc power analysis for the samples.
Author information
Authors and Affiliations
Corresponding author
Additional information
One or more of the authors (MP) has received funding from the Musculoskeletal Transplant Foundation.
Each author certifies that his or her institution has approved the animal protocol for this investigation and that all investigations were conducted in conformity with ethical principles of research.
About this article
Cite this article
Pei, M., He, F., Kish, V.L. et al. Engineering of Functional Cartilage Tissue Using Stem Cells from Synovial Lining: A Preliminary Study. Clin Orthop Relat Res 466, 1880–1889 (2008). https://doi.org/10.1007/s11999-008-0316-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11999-008-0316-2