Abstract
Purpose of Review
Focusing on studies published within the last decade, we review the literature on the seminal microbiome and male factor infertility. We highlight potential mechanisms by which microbes may impact fertility and underscore key limitations and clinical implications of these studies.
Recent Findings
The seminal microbiome encompasses a metabolically and phylogenetically diverse group of microorganisms. Lactobacillus was consistently associated with normal semen analysis parameters and fertility; Anaerococcus was negatively associated with semen quality. These microbes may participate in a complex cross-talk with the host immune system, thereby modulating local and perhaps systemic inflammatory responses, impacting semen quality.
Summary
Research investigating the intersection between the seminal microbiome and male fertility is still in its infancy. Recent investigations have been exclusively cross-sectional, correlational studies, limiting the clinical applicability of published research. Prospective studies with more sophisticated methodologies are necessary.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human body is home to a complex and diverse ecosystem of microorganisms, the result of an extensive coevolution between humans and the microbial communities that live alongside us. Recent studies on the relationship between the microbiome and human health have revealed fascinating insights into the microbiome’s potential role in the pathogenesis of obesity [1], prostate cancer [2], female infertility [3], and even the success of assisted reproductive technology [3], although investigations exploring the seminal microbiome within the context of male factor infertility have been limited.
Early studies on the seminal microbiome were driven primarily by microscopy, culture-dependent methods, and targeted amplification of microbial DNA of known species. For this reason, the impact of known pathogenic, sexually transmitted microorganisms on the male reproductive tract and semen parameters have been well described [4]. Next-generation sequencing (NGS), which allows for discovery of novel microbes without prior knowledge of sequencing information, has certainly expanded our armamentarium; however, we still do not fully understand the role of the overwhelming majority of non-pathologic microorganisms that reside in the male reproductive tract. Given that the etiology of abnormal semen parameters is not identified in up to 45% of cases, a comprehensive exploration of the seminal microbiome may provide further clarity to our understanding of male factor infertility. [5]
Focusing on studies published within the last decade, we review the literature on the seminal microbiome and male factor infertility. We highlight potential mechanisms by which microbes may impact fertility and underscore key limitations and clinical implications of these studies.
Seminal Microbiome and Infertility
During ejaculation, spermatozoa mix with secretions produced by the seminal vesicles, prostate, and bulbourethral glands. The resulting high-osmolarity, heterogenous fluid contains sugars, protein, minerals—and a diverse group of microorganisms [6]. The precise etiology of these microbes remains unclear, but past studies (healthy human and wild-type mouse models) have suggested they likely originate from a combination of the gut, foreskin, urinary tract, prostate, and seminal vesicles [7]. As studies exploring the seminal microbiome and male factor infertility have been primarily cross-sectional in design, the true impact of these microbes on fertility and infertility remains unclear. Table 1 outlines key studies on the seminal microbiome and male factor infertility using NGS methodologies.
Hou et al. [8••] conducted one of the earliest investigations to leverage NGS to identify differences in the semen microbiome of healthy Asian men, compared with those with male factor infertility. Despite the successful identification of six discrete microbial community clusters, none of these clusters was predictive of a participant’s fertility status. Further statistical testing using a proportional odds model revealed a negative association between the gram-positive, anaerobe Anaerococcus with sperm quality. This finding is consistent with a previous study, which demonstrated a similar relationship between a group of gram-positive cocci, including Anaerococcus, in semen samples of men undergoing infertility workup [9]. Remarkably, there was significant overlap between the bacteria found in semen, and those previously identified in vaginal communities, especially associated with bacterial vaginosis [10].
A year later in 2014, Weng et al. [11] published a similar study investigating Asian men undergoing infertility workup due to a diverse range of etiologies (male factor, female factor, combination, or unknown). Semen samples were stratified by normal or abnormal semen parameters. Perhaps attributed to a more robust sample size, a different patient population and an alternative clustering algorithm, Weng et al. identified just three discrete microbial community clusters: Lactobacillus-, Pseudomonas-, and Prevotella-enriched. Men with abnormal semen parameters were statistically significantly more likely to be enriched in the Pseudomonas or Prevotella clusters, compared to the Lactobacillus cluster. Additionally, increased abundance of Prevotella and decreased abundance of Lactobacillus and Gardnerella were seen in men with two or more abnormal semen parameters.
In 2019, Baud et al. [12] analyzed a European cohort of healthy men with normal and abnormal semen analysis parameters. Consistent with the previously described results, three microbial community clusters emerged: Prevotella-enriched, Lactobacillus-enriched, and polymicrobial. Increased abundance of Lactobacillus and Staphylococcus was associated with normal sperm morphology and overall normal semen analysis parameters, respectively. To identify microbial communities that are more likely to be interacting with one another, a co-occurrence network analysis was applied to the data and three functional clusters emerged. Functional clusters 1 and 2 were composed of typical vaginal flora members with similar oxygen requirements (strict or facultative anaerobes), whereas skin floras were typical members of functional cluster 3. The results of this co-occurrence network analysis support the idea that the male genitourinary tract is composed of different microenvironments, allowing for a metabolically diverse group of microbes to exist.
Monteiro et al. [13•] took a unique methodological approach to explore the role of the seminal microbiome and infertility, by pooling samples based on semen analysis findings (normal semen analysis, oligoasthenoteratozoospermia, asthenoteratozoospermia, and seminal hyperviscosity without teratozoospermia) and then analyzing the pooled samples. Although there was no evidence of infection, samples in the oligoasthenoteratozoospermia and hyperviscosity pools demonstrated increased abundance of classically pathogenic organisms including Pseudomonas, Klebsiella, and Neisseria. These findings suggest that these microorganisms may be negatively impacting semen quality through a different, likely more insidious, mechanism than what would otherwise be associated with a fulminant infection by these microbes.
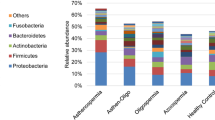
Recent evidence also suggests that the testes may not be a completely sterile environment. Alfano et al. performed a study [14••] exploring testicular tissue bacterial microbiota in normozoospermic men prior to orchiectomy (n = 5) and in men with idiopathic non-obstructive azoospermia (n = 10) undergoing microdissection testicular sperm extraction. Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria were the dominant phyla in normozoospermic men, whereas men with idiopathic non-obstructive azoospermia showed decreased taxa richness (the count of different species in a phylogenetic genus) driven primarily by reductions in Bacteroidetes and Proteobacteria.
These studies highlight just some of the important insights we have learned from applying NGS technologies to analyze the seminal microbiome. Overall, results of clustering analyses were consistent between these studies, with inconsistencies reasonably explained by differences in patient populations and methodologies. A select group of microbes, including Anaerococcus and other, more typically pathogenic microbes may be primarily responsible for contributing to male factor infertility. Regardless, the importance of commensal microbes cannot be overstated, given that one of the most consistent findings in almost all of the aforementioned studies was the association of Lactobacillus with normal semen analysis parameters and fertility.
Potential Mechanisms
Mechanistic studies exploring how the seminal microbiome may impact fertility are lacking, though there is reasonable evidence from closely related areas of study to suggest these microbes may participate in a complex cross-talk with the host immune system, thereby modulating local and systemic inflammatory responses and semen quality.
In a study [15•] of 22 men with human immunodeficiency virus (HIV) and 27 uninfected men, Liu et al. explored the relationship between the seminal microbiome, plasma cytokines, and HIV. Men with HIV demonstrated reductions in semen microbial diversity and richness, which could be restored following just 6 months of antiretroviral therapy. Studies of the gastrointestinal microbiome have definitively demonstrated an association between decreased microbial diversity with immune activation and poor health outcomes, but it is unclear if such a relationship holds true with respect to seminal microbiome diversity [16]. Uninfected men did not show an association between semen microbes and plasma cytokine levels, whereas HIV-infected men showed an association between semen bacterial load and plasma pro-inflammatory cytokines including TNF-alpha, IL-6, IL-10, IL-17, and IL-1b, though only IL-1b correlated with both semen microbes and HIV RNA viral load. These findings suggest an intimate relationship between the immune system and seminal microbes. This immune system cross-talk may contribute to infertility, as a previous study has demonstrated an association between idiopathic infertility with increased levels of IL-10 and decreased levels of IL-1b. [17]
Alternatively, microbes may directly contribute to an inflammatory genitourinary and seminal microenvironment. A diverse array of microbes can directly produce reactive oxygen species, especially in response to attacks from other, competing microbes or even host defenses [18]. An overabundance of reactive oxygen species may shift the balance of pro- and anti-oxidants found in semen towards inflammation, resulting in destruction of protective sperm lipid and protein plasma membranes [19].
Although many of the studies exploring the relationship between the semen microbiome and infertility have shown a positive association between Lactobacilli with semen quality, there have not been any mechanistic studies related to potentially beneficial microbes and fertility. One study demonstrated that a strain of Lactobacillus gasseri successfully protects primary mouse embryonic fibroblast cells from oxidative stress by inducing expression of oxidative stress-related genes [20]. Lactobacillus gasseri may contribute to a clinically meaningful antioxidant effect, as nematodes fed this probiotic demonstrated prolonged longevity [21]. Another potential mechanism for the role of Lactobacillus in fertility may be through the production of lactic acid, contributing to the stability of seminal pH (typically 7.2–8). Alterations in seminal pH have been reported to have negative consequences on sperm motility and capacitation [22]. Although the major contributors to the buffering capacity of semen are proteins and bicarbonate, other low-molecular weight components, such as lactic acid, are believed to contribute to about 50% of the remaining buffering capacity [23].
Limitations and Clinical Implications
NGS techniques have certainly advanced our understanding of the relationship between the seminal microbiome and male fertility; however, a focus on exclusively cross-sectional, correlational studies focused on investigations of semen analysis parameters has limited the impact and clinical applicability of published research on this topic. The studies we highlight in this review reveal insights into the relationship between members of the seminal microbiome and semen analysis parameters. It remains unclear if seminal microbes are driving these deleterious changes, or if they are just a result of an altered genitourinary microenvironment that has already been primed for subfertility. Studies that investigate not just “who” is present (classic phylogenetic analysis using NGS), but “what” these microbes are doing (transcriptomics, metabolomics) may shed light on this relationship and help guide future, mechanistic, investigations. As the field continues to grow, it will be interesting to see the concordance between standard semen analysis parameters such as semen white blood cell count and, for example, pro-inflammatory cytokines such as IL-10 and IL-1b within the context of the semen microbiome. Furthermore, future studies may also benefit from incorporating more sophisticated measures of sperm quality, including sperm DNA fragmentation index, especially given the ability of microbes to modulate the generation of reactive oxygen species in their micro (and macro) environments.
Given the consistency in the results of the studies we described here, there is minimum concern for contamination at the time of semen collection, though this may emerge as a larger issue as more research is performed in this space. At this time, there is no role for seminal NGS in the clinical workup for infertility. However, as our understanding of the relationship between seminal microbes and male fertility evolves, the next generation of clinical testing may feature an assessment of semen microbiota health. These tests will likely generate additional questions. What is the role of antibiotic therapy in a man without the presence of classically pathogenic microorganisms but with the presence of a group of microorganisms that is associated with subfertility, such as Anaerococcus? Similarly, what is the role of probiotic therapy for the subfertile man and his partner?
Conclusions
Research investigating the intersection between the seminal microbiome and male fertility is still in its infancy. Studies leveraging NGS technology have made it possible to explore this relationship with a degree of granularity that has previously been impossible, both financially and methodologically. A commensal genitourinary microbiome, one enriched in Lactobacillus, likely plays a protective role in fertility, perhaps actively through the generation of microbiota-derived metabolites that protect sperm or passively by occupying a niche that would otherwise be occupied by a deleterious group of microbes. Mechanistic preclinical and prospective clinical studies will be necessary to better characterize the relationship between the seminal microbiome and male fertility.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Osadchiy V, Martin CR, Mayer EA. The gut-brain axis and the microbiome: mechanisms and clinical implications. Clin Gastroenterol Hepatol. 2019;17(2):322–32.
Golombos DM, Ayangbesan A, O'Malley P, Lewicki P, Barlow L, Barbieri CE, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, Pilot Study. Urology. 2018;111:122–8.
Sirota I, Zarek SM, Segars JH. Potential influence of the microbiome on infertility and assisted reproductive technology. Semin Reprod Med. 2014;32(1):35–42.
Gimenes F, Souza RP, Bento JC, Teixeira JJ, Maria-Engler SS, Bonini MG, et al. Male infertility: a public health issue caused by sexually transmitted pathogens. Nat Rev Urol. 2014;11(12):672–87.
Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, Dohle G, et al. European Association of Urology guidelines on male infertility: the 2012 update. Eur Urol. 2012;62(2):324–32.
Owen DH, Katz DF. A review of the physical and chemical properties of human semen and the formulation of a semen simulant. J Androl. 2005;26(4):459–69.
Mandar R. Microbiota of male genital tract: impact on the health of man and his partner. Pharmacol Res. 2013;69(1):32–41.
•• Hou D, Zhou X, Zhong X, Settles ML, Herring J, Wang L, et al. Microbiota of the seminal fluid from healthy and infertile men. Fertil Steril. 2013;100(5):1261–9 This study is among the earliest to apply next-generation sequencing methodology to analyze the semen microbiome, demonstrating an association between Anaerococcus and negative sperm quality.
Kiessling AA, Desmarais BM, Yin HZ, Loverde J, Eyre RC. Detection and identification of bacterial DNA in semen. Fertil Steril. 2008;90(5):1744–56.
Swidsinski A, Dorffel Y, Loening-Baucke V, Mendling W, Verstraelen H, Dieterle S, et al. Desquamated epithelial cells covered with a polymicrobial biofilm typical for bacterial vaginosis are present in randomly selected cryopreserved donor semen. FEMS Immunol Med Microbiol. 2010;59(3):399–404.
Weng SL, Chiu CM, Lin FM, Huang WC, Liang C, Yang T, et al. Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PLoS One. 2014;9(10):e110152.
Baud D, Pattaroni C, Vulliemoz N, Castella V, Marsland BJ, Stojanov M. Sperm microbiota and its impact on semen parameters. Front Microbiol. 2019;10:234.
• Monteiro C, Marques PI, Cavadas B, Damiao I, Almeida V, Barros N, et al. Characterization of microbiota in male infertility cases uncovers differences in seminal hyperviscosity and oligoasthenoteratozoospermia possibly correlated with increased prevalence of infectious bacteria. Am J Reprod Immunol. 2018;79(6):e12838 This study pooled a large number of semen samples based on semen analysis findings and then analyzing the pooled samples, demonstrating a more cost-effective but equally efficacious method to perform next-generation sequencing-based analysis of the semen microbiome.
•• Alfano M, Ferrarese R, Locatelli I, Ventimiglia E, Ippolito S, Gallina P, et al. Testicular microbiome in azoospermic men-first evidence of the impact of an altered microenvironment. Hum Reprod. 2018;33(7):1212–7 This study demonstrates that the testicular microbiome is alterted in men with azoospermia.
• Liu CM, Osborne BJ, Hungate BA, Shahabi K, Huibner S, Lester R, et al. The semen microbiome and its relationship with local immunology and viral load in HIV infection. PLoS Pathog. 2014;10(7):e1004262 This study explores the relationship between the seminal microbiome and the immune system within the context of an HIV-infected patient population.
Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–6.
Havrylyuk A, Chopyak V, Boyko Y, Kril I, Kurpisz M. Cytokines in the blood and semen of infertile patients. Cent Eur J Immunol. 2015;40(3):337–44.
Dong TG, Dong S, Catalano C, Moore R, Liang X, Mekalanos JJ. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc Natl Acad Sci U S A. 2015;112(7):2181–6.
Sanocka D, Kurpisz M. Reactive oxygen species and sperm cells. Reprod Biol Endocrinol. 2004;2:12.
Kobatake E, Nakagawa H, Seki T, Miyazaki T. Protective effects and functional mechanisms of Lactobacillus gasseri SBT2055 against oxidative stress. PLoS One. 2017;12(5):e0177106.
Nakagawa H, Shiozaki T, Kobatake E, Hosoya T, Moriya T, Sakai F, et al. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell. 2016;15(2):227–36.
Zhou J, Chen L, Li J, Li H, Hong Z, Xie M, et al. The semen pH affects sperm motility and capacitation. PLoS One. 2015;10(7):e0132974.
Wolters-Everhardt E, Dony JM, Peters WH, De Pont JJ. Buffering substances of human semen. Fertil Steril. 1987;48(1):159–61.
Funding
SVE is supported by a Research Scholar Award from the Urology Care Foundation and the American Urological Association. EAM is supported by grants from the National Institutes of Health including DK048351 and DK082370. These organizations played no role in the collection, analysis, and interpretation of data, in the writing of the manuscript, and in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
SVE is a consultant for Metuchen Pharmaceuticals. JNM is a consultant for Antares Pharma, Boston Scientific, and Endo Pharmaceuticals. VO and EAM declare no competing interests.
Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Male Sexual Dysfunction and Disorder
Rights and permissions
About this article
Cite this article
Osadchiy, V., Mills, J.N., Mayer, E.A. et al. The Seminal Microbiome and Male Factor Infertility. Curr Sex Health Rep 12, 202–207 (2020). https://doi.org/10.1007/s11930-020-00273-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11930-020-00273-5