Abstract
Over the previous decade, we observed the emergence of the fungal pathogen, Cryptococcus gattii, as a cause of disease in humans and animals in a temperate climate. This outbreak, first documented on Vancouver Island, has since expanded throughout Western North America, with non–travel-associated cases now in British Columbia, Washington, Oregon, and California. Additionally, a secondary outbreak, originating in and still restricted to Oregon, has also occurred. During the past several years, several studies detailing molecular typing, virulence, antifungal susceptibilities, epidemiology, and clinical issues have been published. These studies begin to address the complex dynamics of this novel emergence of a rare and fatal fungus, outline clinical characteristics of human cases, and also opened several new areas that should be explored in the upcoming years.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the threats to human health, infectious diseases remain a significant cause of global morbidity and mortality [1–3]. Advances in healthcare, the onset of the global HIV/AIDS pandemic, the emergence of novel pathogens, and the re-emergence of diseases (ie, increasing incidence and geographic expansion) all contribute to a large worldwide percentage of those affected by bacterial, viral, and fungal diseases [2, 4]. A focal point of disease emergence in the fungal kingdom has been the unprecedented outbreak of Cryptococcus gattii in western North America. Although infections remain relatively rare, the incidences observed in this outbreak are higher than almost anywhere in the world. In fact, only two reports with higher overall incidences have ever been reported, and both were from tropical regions (Aboriginals in the Northern Territory of Australia, and a population in the central province of Papua New Guinea) [5••, 6, 7]. This is the first and only large outbreak documented to arise in a temperate climate to date.
C. gattii is a basidiomycetous yeast species and is estimated to have diverged from its sibling species Cryptococcus neoformans about 37.5 million years ago [8, 9]. Outside of the pathogenic Cryptococcus species complex (C. neoformans and C. gattii), most of the related species are nonpathogenic insect-associated saprophytes [10–12]. However, infections caused by these two species are significant globally, with the predominance of cases estimated to be attributable to C. neoformans. In fact, recent studies report that one million individuals are infected annually in Africa, with more than 620,000 attributable mortalities, accounting for about one third of all HIV/AIDS-associated deaths, surpassing tuberculosis mortality [13].
Historically, C. neoformans serotypes A, B, C, and D were considered one species; more recently, C. gattii was recognized as a unique species, which can be further subdivided into two serotypes (B and C) [14] and four molecular types (VGI-VGIV) [15].
The true prevalence of C. gattii as a cause of disease is not clear, because the organism is not routinely distinguished from C. neoformans in clinical microbiology laboratories. It can be identified by growth on CGB agar, in which C. gattii is canavanine resistant, and able to use glycine as a sole carbon source, triggering a bromothymol blue color reaction [16].
More detailed typing of C. gattii has demonstrated that outbreaks are often comprised of a predominance of one particular molecular type (eg, VGI in Australia and VGII in the North American Pacific Northwest) [17, 18]. Furthermore, unique clonal genotypes are frequently associated with outbreaks (eg, VGIIa, VGIIb, VGIIc) [19••, 20, 21]. Therefore, from clinical and surveillance perspectives, identification of species, molecular type, and genotype are important. In understanding the dynamics of cases in Western North America, such identification becomes especially relevant. As stated above, molecular type VGII accounts for about 95% of all cases in the Pacific Northwest, as far south as northern California. However, recent studies identified that more than 90% of cases from a cohort of patients in Southern California were molecular type VGIII, a type that has also been reported in Mexico [22–25]. The diversity observed in Southern California also indicates that VGIII may have been endemic and unrecognized in the region for longer than appreciated [24], highlighting the complexity of the North American outbreak. Because clinical presentation, optimal therapeutic approach, and outcomes may differ compared to C. neoformans, a thorough understanding of the epidemiology is needed. Recognition of this outbreak, and clinical aspects, are reviewed here.
Epidemiology
To fully appreciate the epidemiology of C. gattii in Western North America, one must examine not only clinical cases, but also veterinary cases and even isolates collected directly from the environment. Humans become infected through the inhalation of infectious propagules (spores or desiccated yeast) from the environment. Therefore, monitoring the natural habitat is critical [26]. Furthermore, wide ranges of animals have been infected by isolates identical to those infecting humans [27, 28]. Veterinary cases serve as additional resources for data collection; because many animals are often nonmigratory, these cases are frequently sentinels for geographic expansion of outbreaks [29–31].
With the increasing discriminatory power of direct DNA sequence analysis and the continued reduction in costs, sequence analysis for molecular epidemiology has become increasingly applied in the analysis of C. gattii strains [19••, 20, 32–34]. The most widely used sequence-based typing method for the analysis of C. gattii is multilocus sequence typing (MLST) [35]. This method is robust and portable between laboratories. Most often, analysis of several genomic loci is conducted for each isolate [20, 32, 34].
One important application of molecular epidemiology was applied with the emergence of C. gattii in the North American Pacific Northwest. Amplified fragment length polymorphism (AFLP) analysis, then MLST, confirmed that two genotypes were largely responsible for the outbreak, VGIIa/major and VGIIb/minor [17, 21]. Subsequent studies on the outbreak were then employed to confirm these genotypes in the region and also to document that the outbreak had expanded into the United States [20, 27, 36]. Additionally, sequence-based analysis revealed that a third outbreak genotype, unique to Oregon (VGIIc/novel), was also contributing to this ongoing outbreak [19••, 28, 37]. The novel VGIIc genotype is thus far restricted to Oregon, with the exception of one travel-associated case (to Oregon) from Idaho that is slightly divergent from the other VGIIc isolates examined thus far [19••, 37].
Importantly, studies have shown that both the VGIIa and VGIIc genotypes are hypervirulent when compared to other closely related VGII genotypes that are not directly associated with the Pacific NW outbreak, both in macrophages and whole-animal murine models of infection [19••, 20, 38•]. To date, cases of all three of the outbreak genotypes have been reported in humans and other animals. However, there have been no cases of VGIIc in Canada, and VGIIc has yet to be isolated from the environment. Therefore, continued surveillance of human and veterinary cases, and further environmental sampling in the region, is warranted.
In addition to the expansion of the outbreak in the Pacific Northwest, studies now suggest that C. gattii molecular type VGIII is endemic in Southern California. A recent re-examination of Cryptococcus infections in patients with HIV/AIDS from Los Angeles County (collected prior to 2005) [25] demonstrated about 12% C. gattii; more than 90% of these isolates were VGIII. The diversity of genotypes observed was much greater than the levels seen in the Pacific Northwest outbreak region for VGII (> 25 VGIII genotypes). These findings suggest that C. gattii VGIII may have been endemic in Southern California for a longer period of time in comparison to the more clonal VGII population found in the Pacific Northwest, or that VGIII is more actively recombining and/or mutating [24]. More recent analysis of VGIII isolates collected from patients who do not have HIV/AIDS from the same region further indicates that C. gattii is endemic in the area of Southern California, and infecting humans (Byrnes, Marr, Filler, Heitman, unpublished data).
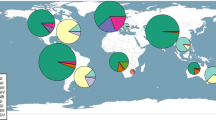
Another recent case of C. gattii infection that developed in a healthy person who lacked travel exposure was documented on Hawaii; the organism implicated in this infection was found to be a unique VGII strain, using MLST (Byrnes and Marr, data unpublished). Hence, these infections have been increasingly documented in Western North America, both in the United States and Canada. A summary of the documented emergence of different C. gattii types is presented in Fig. 1. It is important to recognize that many infections are established using antigen assays, thereby negating distinction of species. Even when the organism is isolated in culture, it has not been identified to species level in clinical microbiology laboratories, with most laboratories; hence, a significant burden of C. gattii infection is likely to be unrecognized globally, with potential prognostic and therapeutic implications.
Distribution of Cryptococcus gattii infections in Canada and the United States. From 1999–2011, C. gattii infections have been documented in British Columbia (BC), Alberta (AB), Washington (WA), Oregon (OR), California (CA), Hawaii (HI), and North Carolina (NC). Molecular types (VGI-VGIII) and outbreak genotypes (VGIIa, VGIIb, and VGIIc) are listed. All molecular types/genotypes were reported in human cases, with the exception of VGIIb-WA, VGIIb-CA, and VGI-HI. These types have only been found in animal cases to date but are presumed to be endemic in the respective regions and consequently could infect humans. All cases have occurred in western North America with the exception of well-documented travel-associated cases in Alberta and North Carolina
Clinical Issues
Risks
One recent and comprehensive retrospective study from the British Columbia Center for Disease Control (CDC) analyzed demographic and clinical features of reported cases, hospitalizations, and deaths attributed to C. gattii during 1999–2007 [5••]. A total of 218 cases were reported with an average annual incidence of 5.8 per million persons. Most patients presented with a pulmonary finding (76.6%) or isolated lung cryptococcoma (75.4%). Overall, there were 19 deaths in the cohort, contributing to a mortality rate of 8.7%. About 40% of the patients had some underlying disease; 62% of those with C. gattii infection had no prior underlying health concerns [5••].
A study group in the United States initially described the first recognized cohort of 20 patients in Washington and Oregon, and noted that about half of the patients presented with only pulmonary disease, without central nervous system (CNS) findings, with about half of the patients described as “previously healthy”—having no prior medical diagnoses or immunocompromising states [39]. The US Centers for Disease Control and Prevention (CDC) has now summarized a larger cohort within the Pacific Northwest, reporting similarities, but also marked differences compared to what was observed in Canada [40••]. From 2004 to 2010, there were 52 documented cases, with more than 70 cases in the United States to date [40••]. In the United States, only 19% of patients were previously healthy hosts; underlying diseases in patients who presented with C. gattii infection include a diverse constellation of mildly to overtly immunocompromised states (eg, chronic obstructive pulmonary disease, hematologic malignancies, myelodysplastic syndromes, autoimmune disorders, and prior receipt of organ transplant). There have been few patients with HIV/AIDS who have developed documented C. gattii infection in this region, although these numbers are likely severely underestimated because of lack of routine isolate identification. In the US cohort, 20% of patients were thought to have died from C. gattii infection and 13% died with C. gattii infection [40••].
Most recently, a study from the British Columbia CDC documented that oral steroids, pneumonia, lung conditions, age ≥50 years, smoking, HIV infection, and history of invasive cancer were all significant factors for infection [41•]. This shows that although many without risk factors contract illness, several factors are associated with increased risk of infection. This study should be used as a foundation for expanding upon the examination of risk factors throughout the region, specifically as it relates to infections in the United States.
These studies are relatively small, but they present some important clinical observations that should be further defined. Although this organism historically was noted to cause disease in primarily healthy hosts residing in endemic regions, in North America, it appears that 20% to 50% of patients who have developed this infection actually have an underlying immunosuppressive condition, or underlying pulmonary disease. The actual prevalence of C. gattii infection in patients with HIV/AIDS remains unclear, owing largely to lack of routine microbial diagnosis. It is likely that the diversity in hosts infected with C. gattii will at least partially mimic the population residing in the new endemic region; one can conclude that C. gattii causes invasive infection in both healthy hosts and patients with a multitude of immunocompromising conditions.
Presentation and Management
Most often, C. neoformans is recognized after infection disseminates from the lungs to involve the CNS, with typical findings of meningoencephalitis. A minority of patients are diagnosed with disease isolated to the lung. In contrast, C. gattii infection more frequently presents with isolated pulmonary disease, with about half of cases involving the lungs alone. In fact, several of the cases of isolated pulmonary disease were uncovered with biopsy and/or resection of focal lesions thought likely to be malignancy (Marr, unpublished observation).
Findings of meningoencephalitis, including fever, headache, mental status changes, and meningeal irritation develop in a proportion of patients who present with disease involving the CNS. Infection can be very inflammatory, with very high cerebrospinal fluid (CSF) opening pressures and cellular profiles denoting active infection. Radiography frequently demonstrates multiple small or large cryptococcomas, with biopsy typically revealing yeasts and active or granulomatous inflammation.
The current Infectious Diseases Society of North America (IDSA) treatment recommendations for C. gattii infection largely approximate the typical approach taken for C. neoformans, with induction courses of amphotericin B formulations, combined with 5-flucytosine, and maintenance fluconazole therapy being the primary treatment of infection involving the CNS; some patients with isolated pulmonary disease can be managed with azole therapy alone [42]. However, subtle differences are now just becoming apparent. Management of all CNS cryptococcal diseases requires aggressive management of intracranial hypertension, and this is especially true for C. gattii infection. It has been reported that intracranial hypertension occurs in more than 50% of immunocompromised patients, and in even more immunocompetent hosts [43]. During the course of therapy, it is not uncommon for these patients to evolve focal neurologic deficits secondary to inflammatory meningitis and ventricular obstruction, even causing a paradoxical deterioration in the setting of objective improvement of infection (eg, clearance of CSF cultures). In this setting, sterile arachnoiditis causing hydrocephalus can develop [44]. Pressure management, including placement of intraventricular shunts, is critical. Administration of corticosteroids has been reported to be associated with good outcomes, especially in patients with sterile inflammation causing neurologic decline late in the course of infection [44, 45].
Recently, it has been reported that some C. gattii isolates have disparate susceptibilities to certain azole antifungals; specifically, it appears that some VGIIa and VGIIc isolates recovered from the Pacific Northwest outbreak have demonstrated relatively high minimum inhibitory concentrations to fluconazole [37, 46]. The clinical significance of this observation has yet to be documented, although we are personally aware of numerous cases in which fluconazole therapy, especially low-dose therapy, failed to elicit good clinical outcomes (Marr, personal observation). The optimal approach to treatment of C. gattii infection has not yet been defined.
Conclusions
In response to the North American C. gattii outbreak, a multidisciplinary C. gattii working group was established to address the epidemiology, clinical features, and basic science questions surrounding this emergence [40••, 47•, 48]. Very little is currently known about how or why humans develop disease. It likely involves unique host factors, including possible genetic predisposition(s). In addition, the origins of the outbreak genotypes remain elusive. Substantial progress has been achieved in addressing the molecular epidemiology and expansion of the outbreak, and the phenotypic characteristics that make these genotypes unique. However, many critical questions remain to be addressed in order to understand the dynamics of this unprecedented C. gattii emergence. Among these, expanded environmental sampling, further phenotypic characterization of associations with host animals and plants, and genome sequencing of C. gattii mitochondrial and nuclear genomes, in addition to those available for the VGI isolate WM276 and the VGIIa/major outbreak isolate R265 [49], should be conducted.
In addition, a pressing need exists for prospective clinical studies that capture more information than case description, clinical course, therapies, detailed underlying host risks, and the like. This would most likely be facilitated by the establishment of a network for prospective surveillance and reporting of clinical outcomes of C. gattii infections, to allow for follow-up during and after treatment, and the collection of fungal isolates and patient blood samples. A centralized clinical database and sample repository will allow analyses of epidemiology, therapeutic efficacy, clinical risks, and patient outcomes. Furthermore, a linked repository could serve as a critical resource for basic studies of fungal isolates and host immunity/genetic predisposition(s). Understanding aspects of this novel fungal emergence, via examinations of both host and pathogen, will foster translational research aimed at enhancing diagnosis, prognosis, and patient outcomes and serve as a paradigm for examinations of emerging pathogens. These studies, combined with basic research and environmental analyses, will allow for an enhanced understanding of this outbreak and generally how and why C. gattii infects hosts to cause life-threatening disease.
Why C. gattii emerged in a temperate climate for the first time remains unclear, and it is not known how far the outbreak will expand and if new risk areas may occur in other temperate regions. Furthermore, if the outbreak will inexorably expand down the coast or alternatively discontinuously jump to a new region remains unclear. This makes the Pacific Northwest outbreak a focal point for studies and a paradigm for novel emergent fungal diseases. To this end, it is critical that clinical laboratories and veterinary diagnostic laboratories begin to assign species identity to clinical isolates, thereby distinguishing C. neoformans and C. gattii.
References
Papers of particular interest, published recently, have been highlighted as: •Of importance •• Of major importance
Cohen ML. Changing patterns of infectious disease. Nature. 2000;406:762–7.
Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–9.
Morens DM, Folkers GK, Fauci AS. Emerging infections: a perpetual challenge. Lancet Infect Dis. 2008;8:710–9.
King DA, Peckham C, Waage JK, et al. Epidemiology. Infectious diseases: preparing for the future. Science. 2006;313:1392–3.
•• Galanis E, MacDougall L. Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999–2007. Emerg Infect Dis. 2010;16:251–7. This important study documents the epidemiological details of the C. gattii outbreak within British Columbia from the onset in 1999 through 2007. Interestingly, the authors found that although VGIIa predominates (>95% of cases), it does not seem to cause greater illness or death when compared to other strains. More and larger studies will be necessary to determine if clinical outcomes are different based on isolate types.
Fisher D, Burrow J, Lo D, Currie B. Cryptococcus neoformans in tropical northern Australia: predominantly variant gattii with good outcomes. Aust N Z J Med. 1993;23:678–82.
Seaton RA. The management of cryptococcal meningitis in Papua New Guinea. P N G Med J. 1996;39:67–73.
Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–33.
Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–6.
Casadevall A, Perfect J. Cryptococcus neoformans. Washington: ASM; 1998.
Findley K, Rodriguez-Carres M, Metin B, et al. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot Cell. 2009;8:353–61.
Heitman J, Kozel T, Kwon-Chung J, et al. Cryptococcus: from human pathogen to model yeast. Washington: ASM; 2011.
Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–30.
Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–87.
Boekhout T, Theelen B, Diaz M, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907.
Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J Clin Microbiol. 1982;15:535–7.
Kidd SE, Guo H, Bartlett KH, et al. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot Cell. 2005;4:1629–38.
Sorrell TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–68.
•• Byrnes 3rd EJ, Li W, Lewit Y, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. This study uses high-resolution molecular typing to illustrate a clear expansion of the C. gattii outbreak throughout Washington and Oregon. Additionally, virulence of the novel VGIIc genotype is characterized, showing high virulence (similar to VGIIa) in both macrophage and murine models of infection.
Fraser JA, Giles SS, Wenink EC, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–4.
Kidd SE, Hagen F, Tscharke RL, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101:17258–63.
Castanon-Olivares LR, Lopez-Martinez R, Barriga-Angulo G, Rios-Rosas C. Crytococcus neoformans var. gattii in an AIDS patient: first observation in Mexico. J Med Vet Mycol. 1997;35:57–9.
Olivares LR, Martinez KM, Cruz RM, et al.: Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Med Mycol: (2009) 1-9.
Byrnes EJ, 3 rd, Li W, Ren P, et al.: A diverse population of Cryptococcus gattii molecular type VGIII in Southern Californian HIV/AIDS patients. PLoS Pathog Under Review (2011)
Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V. Cryptococcus gattii in AIDS patients, southern California. Emerg Infect Dis. 2005;11:1686–92.
Kidd SE, Chow Y, Mak S, et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol. 2007;73:1433–43.
Byrnes 3rd EJ, Bildfell RJ, Dearing PL, et al. Cryptococcus gattii with bimorphic colony types in a dog in western Oregon: additional evidence for expansion of the Vancouver Island outbreak. J Vet Diagn Invest. 2009;21:133–6.
Byrnes 3rd EJ, Bildfell RJ, Frank SA, et al. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199:1081–6.
Duncan C, Bartlett KH, Lester S, et al.: Surveillance for Cryptococcus gattii in horses of Vancouver Island, British Columbia, Canada. Med Mycol In Press (2011)
Duncan C, Schwantje H, Stephen C, et al. Cryptococcus gattii in wildlife of Vancouver Island, British Columbia, Canada. J Wildl Dis. 2006;42:175–8.
Stephen C, Lester S, Black W, et al. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 2002;43:792–4.
Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–38.
MacDougall L, Kidd SE, Galanis E, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 2007;13:42–50.
Meyer W, Aanensen DM, Boekhout T, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47:561–70.
Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5.
Upton A, Fraser JA, Kidd SE, et al. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J Clin Microbiol. 2007;45:3086–8.
Iqbal N, Debess EE, Wohrle R, et al. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii from the Pacific Northwest of the United States. J Clin Microbiol. 2009;48:539–44.
• Ma H, Hagen F, Stekel DJ, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A. 2009;106:12980–5. In this study, Ma et al. show that the VGIIa genotype has a high proliferation rate inside macrophages. They also show that this phenotype correlated to both in vivo virulence and a tubular mitochondrial phenotype. Via microarray analysis, their findings suggest that the VGIIa mitochondrial genome might be implicated in virulence.
West SK, Byrnes EJ, 3 rd , Mostad S, et al.: Emergence of Cryptococcus gattii in the Pacific Northwest United States. Presented at the 48th Annual ICAAC/IDSA 46th Annual Meeting, Washington, DC, October 25–28, 2008 Oral presentation, Session 179 Abstract M-1849 Page 658 of the Abstract Book. (2008)
•• CDC: Emergence of Cryptococcus gattii—Pacific Northwest, 2004–2010. MMWR Morb Mortal Wkly Rep (2011) 59: 865–868. This report is the first comprehensive public health study of the C. gattii outbreak within the United States. In total, more than 50 human cases are documented. It is also shown that although the mortality rate in Canada is less than 9%, the mortality rate in the United States ranges from 20% to 33%.
• Macdougall L, Fyfe M, Romney M, et al. Risk Factors for Cryptococcus gattii Infection, British Columbia, Canada. Emerg Infect Dis. 2011;17:193–9. This study documents risk factors for C. gattii infection in people diagnosed in British Columbia, Canada. The authors found that receipt of oral corticosteroids, pneumonia, lung conditions, ≥50 years of age, smoking, HIV infection, and history of invasive cancer were all significant factors associated with infection. Although this infection has classically been reported to cause infection in otherwise healthy people, patients with infection in North America have had a multitude of underlying diagnoses.
Perfect JR, Dismukes WE, Dromer F, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis. 2010;50:291–322.
Graybill JR, Sobel J, Saag M, et al. Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. The NIAID Mycoses Study Group and AIDS Cooperative Treatment Groups. Clin Infect Dis. 2000;30:47–54.
Lane M, McBride J, Archer J. Steroid responsive late deterioration in Cryptococcus neoformans variety gattii meningitis. Neurology. 2004;63:713–4.
Phillips P, Chapman K, Sharp M, et al. Dexamethasone in Cryptococcus gattii central nervous system infection. Clin Infect Dis. 2009;49:591–5.
Hagen F, Illnait-Zaragozi MT, Bartlett KH, et al.: In vitro antifungal susceptibilities and AFLP genotyping of a worldwide collection of 350 clinical, veterinary and environmental Cryptococcus gattii isolates. Antimicrob Agents Chemother (2010).
• Datta K, Bartlett KH, Baer R, et al. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15:1185–91. This paper highlighted and reviewed the emergence of the outbreak, overviewed previous findings, and recommended future goals. Importantly, this paper was a product of a multidisciplinary and international C. gattii working group of the Pacific Northwest.
Datta K, Bartlett KH, Marr KA: Cryptococcus gattii: Emergence in Western North America: Exploitation of a Novel Ecological Niche. Interdiscip Perspect Infect Dis 2009: 176532.
D’Souza CA, Kronstad JW, Taylor G, et al.: Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio (2011) 2.
Disclosure
Conflicts of interest: E.J. Byrnes III—none; K.A. Marr—consultancies, Astellas, Basilea, Merck, Pfizer, grants, Astellas and Merck.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Byrnes, E.J., Marr, K.A. The Outbreak of Cryptococcus gattii in Western North America: Epidemiology and Clinical Issues. Curr Infect Dis Rep 13, 256–261 (2011). https://doi.org/10.1007/s11908-011-0181-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11908-011-0181-0