Abstract
Computer-aided liver diagnosis helps doctors accurately identify liver abnormalities and reduce the risk of liver surgery. Early diagnosis and detection of liver lesions depend mainly on medical imaging techniques such as magnetic resonance imaging (MRI) and computed tomography (CT). Segmentation and identification of hepatic lesions in these images are very challenging because these images often come with low resolution and severe noise. Many new machine learning and image analysis techniques have been gradually used on this topic, and their performance is still limited. An automatic and accurate model that incorporates tracking, detection, and diagnosis of hepatic lesions in the 3D volumes of CT and MRI is still lacking. This paper aims to review different models for the automatic detection and diagnosis of the hepatic lesion with CT and MRI and discusses the medical background of liver tumors and the standard elements of the CAD liver diagnosis system. In addition, the concept of federated learning has been introduced, and the fused information from multi-modality (CT and MRI) and the robust and complex features that represent liver lesions accurately have been discussed. More specifically, this paper presents a comprehensive study of the latest work on liver tumor detection and diagnosis, which identifies the contributions of these different approaches and the recommendation model suggested for practical use. Furthermore, this paper was intended to encourage researchers from the medical community, image processing, and machine learning community to pay much attention to the use of deep and federated learning, spiking neuron model, bio-inspired optimization algorithms, fuzzy logic, and neutrosophic logic to address the problems of segmentation and prediction/classification for real-time diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The leading cause of disease and death worldwide is liver disease. Liver diseases, including viral hepatitis (HBV, HCV), non-alcoholic fatty liver disease (NAFLD), and alcoholic liver disease, afflict about 300 million people (ALD). Additionally, the number of people with alcoholic and non-alcoholic fatty liver disease is rising alarmingly. The second most common and deadly type of cancer is liver cancer [1,2,3,4,5,6,7]. China is home to more than 383,000 liver cancer fatalities yearly or 51% of all liver cancer fatalities globally [8,9,10].
Medical imaging techniques like CT and MRI are the gold standard for finding, assessing, and diagnosing liver tumors [11]. Typically, a doctor’s job is to quantify the liver lesions visible in these photographs and describe their size. This measurement could indicate an object’s size, mean density, or relative placement. The most commonly used software that hospitals employ can take such actions. These tools, however, frequently lack user friendliness or demand a substantial amount of human input. Even if the procedure is ineffective, it is highly subjective, and the same doctor frequently marks the same thing differently, producing different findings. In order to analyze medical images and support physicians in diagnosis and treatment, it is crucial to create an automated computer-assisted diagnosis (CAD) model. Contrarily, CAD technologies have aided tools for radiologists rather than replacing them entirely [12].
1.1 Clinical Background
The liver is the largest organ in the human body and serves several functions. According to [13], most liver cancers originate in other organs. This results in a large blood volume flowing into the liver, rendering it susceptible to secondary tumor occurrence. Tumors must be localized and diagnosed in order for treatment to be effective.
Cancer is the general name for cells developing out of control in one body part. Cancer cells grow and proliferate in order to produce more cells than the needs of the human body. These additional cells are called tumors. Tumors may be benign or malignant [13]. Types of malignant tumors are (Hepatocellular-carcinoma (HCC), Cholangiocarcinoma (CC), and Metastases), and types of benign tumors are (Hemangioma, Focal-nodular-hyperplasia (FNH), Adenoma, and Cyst). Figure 1a shows a sample of hepatic focal lesions with different sizes and shapes for different patients.
Tumors occur in a variety range of shapes, sizes, and contrasts. They can be highly diffused and regularly obscured by normal tissue. Depending on the morphological features of the tumor, clinicians attempt to estimate the probability of malignancy. Their shape categorizes tumors into round, oval, lobular, and irregular shapes [14]. Figure 1b shows a sample of such tumor shapes.
1.2 Liver Tumor Diagnosis and Its Challenges
The hepatic diagnosis via CAD model usually follows two stages; computer-assisted detection and computer-assisted diagnosis, as shown in Fig. 2.
Pre-processing, liver segmentation, and lesion detection compensate for the detection stage. Due to several reasons, including low contrast of soft tissue, noise level, uneven liver form and size, the similar intensity with other organs, fuzzy boundary, and variation in lesion shape, defining the lesion boundary during the detection stage is particularly difficult. Along the z-axis, the voxel spacing ranges from 0.45 to 6.0 mm [15,16,17]. These problems make automatic and accurate liver and lesion segmentation very difficult.
Traditional imaging segmentation methods, such as edge detection, regional transformation, watershed transformation, fuzzy logic, and others, are widely used. However, these methods have certain disadvantages, which means that they cannot be used for the exact results. Deep learning (DL)-based segmentation has recently gained popularity with many different types of architecture, including (SurvivalNet, ResNet, GAN, CNN, etc.). DL can predict key regional characteristics and segment organs with accuracy. However, DL requires a lot of time, has between 30 and 150 hidden layers, and requires that the parameters be changed [18,19,20,21].
The diagnostic stage, on the other hand, includes feature extraction, feature selection, and classification/prediction. In numerous medical imaging applications, extracting texture/shape information is crucial. Each ROI that has been segmented in the earlier stage has features taken from it. Differential characteristics of each ROI sent to the classifier distinguish between liver lesions (benign and malignant). The selection of features for the best and hidden knowledge rather than employing all actual facts is a serious problem with massive medical data. For feature selection, numerous alternative search techniques are employed (forward selection search, selection random search, relief, fisher, mutual information, etc.). All of these methods are still hindered by the recession and sticking at local minima as well as the high computational cost [22]. Additionally, there are restrictions on stochastic training, time consumption, becoming stuck in local minima, lack of strength, and tweaking parameters with machine learning prediction/classification algorithms (such as; radius, learning rate, decay rates, size of the network, etc). Additionally, they cannot be learned progressively, making it unable to dynamically remedy recognition errors.
The automatic segmentation and detection of liver cancers remains difficult for the CAD system due to issues like:
-
Liver and tumor irregularity and similarity to other organs.
-
Two common characteristics in CT/MRI images are severe noise and low resolution.
-
The algorithms used for liver diagnosis may consume time and error regions.
-
The detection and diagnosis process should not take more than a few minutes.
-
CT images are gray instead of color. Even an intensity change may not clearly display lesions.
-
The diagnostic process in hospitals should not require unrealistic resources, such as supercomputers or grid computing. The entire process should be run on individual computers.
1.3 Key Scientific Problems
The segmentation process is used to segment the liver, localize and delineate lesions. However, the classical segmentation methods have no exact solutions for all cases, high time computation, and stuck in local minima. The feature extraction process analyzes ROIs that depict lesions at only a single scale, thereby losing many dominant characteristics. Feature selection methods have high time computation in the feature selection process, are stuck in local minima, and do not agree to get the optimal solution. The employed classifiers have stochastic training, time, strength, and tuning parameter limitations.
The following vital problems can be concluded from the above issues as follow:
-
Difficult to segment liver and localize lesions in abdominal CT/MRI imaging techniques. The liver has irregularity and variation in both shape and scale for each patient and has similar intensity with neighbor organs. In addition, liver lesion segmentation is considered a more challenging task due to appearances, have no clear boundary, and visible edges.
-
The diagnosis approach needs to keep the model continues learning, adapt parameters automatically by federated learning, needs spikes neuron signals to carry more information than classical coding features, needs optimized and in-depth features for real-time diagnosis model.
This review aims to provide a comprehensive survey of the current state of the art and discuss open problems and challenges for future work for live tumor diagnosis via the CAD system. Moreover, this paper suggested a recommendation model for liver tumor diagnosis based on federated deep neural network, spiking neuron model, bio-inspired optimization algorithms, and fuzzy and neutrosophic sets. We expect that this survey will attract researchers working on different diagnostic fields of CT/MRI further to investigate effective and efficient approaches to new challenges. This paper is also intended to encourage researchers from the medical community, image processing, and machine learning community to pay much attention to deep federated learning, spiking neuron models, bio-inspired evolutionary algorithms, fuzzy and neutrosophic logic the problems of segmentation and classification.
The rest of this paper is as follows. Section 2 reviews different related work for liver tumor detection and diagnosis; various cases of deep learning, swarm optimization, and neutrosophic logic are introduced for analyzing liver tumors from CT/MRI scans. Section 3 suggests a recommendation model for liver tumor diagnosis. Finally, the conclusion is given in Sect. 4.
2 Current Status and Trends
The segmentation of liver lesions is regarded as a more complex process due to the wide range of lesion tissue appearances. In addition, because of the low contrast of liver lesions, this segmentation is often performed on a noisy background. CT/MRI abdominal imaging presents an additional problem from the diagnostic point of view: the uneven enhancement caused by anatomical variations between lesions Further, certain lesions lack clearly visible boundaries, making the use of edge-based segmentation approaches as the sole segmentation method ineffective. Unfortunately, hepatic lesion segmentation has received low interest from researchers compared to many proposed methods of liver segmentation. This can be related to the challenges involved with the segmentation of liver lesions and an exciting lack of public datasets, limiting the development of lesion segmentation and diagnostic techniques.
The methods for segmenting images have been the subject of various studies, and depending on how automated they are, they can be divided into full, semi-automated, and interactive. Histogram-based [23,24,25], region growing [12, 26, 27], active contour models or level sets [28, 29], graph cuts [30,31,32], watershed [31,32,33], fuzzy logic [34, 35], and most recently deep learning algorithms [31] are the different types of computerized liver and lesion segmentation approaches that are currently available. Regarding applicability, appropriateness, performance, and computational expense, each of these strategies has benefits and disadvantages. However, extracting liver lesions from CT/MRI images, particularly those with low contrast, remains difficult. In histogram-based segmentation methods, Soler suggested the first method of liver lesion segmentation [32]. Several authors then used a threshold technique to detect liver lesions. A thresholding technique is simple but powerful. The key drawback is that the threshold is quite noisy and does not take spatial features into account [33].
The advantages and disadvantages of various segmentation methods are shown in Table 1. However, extracting liver tumors from CT/MRI images, particularly those with low contrast, remains challenging.
The objective of the detection stage is to delineate and segment certain ROIs that may contain abnormalities from the original abdominal CT/MRI scans and increase the efficiency of diagnostic procedures.
In the histogram-based segmentation approach, Soler suggested the first method of liver lesion segmentation [32]. Several authors then used a threshold technique to detect liver lesions. A thresholding technique is simple but powerful. The key drawback is that the threshold is quite noisy and does not take into consideration spatial properties [33].
Choudhary et al. proposed the semi-automatic segmentation approach. The watershed algorithm for liver segmentation and adaptive multi-thresholding based on minimum cross-entropy was used for tumor segmentation with high accuracy and less computation [36]. Patil et al. proposed a tumor segment approach using adaptive threshold, global threshold, and morphological operators for liver segmentation. The second phase tumor is segmented using an adaptive hybrid threshold with morphological operators, fuzzy c-mean, and region-growing techniques. This approach improves the accuracy of segmentation for various CT image quality [25]. Campadelli et al. proposed significant edges to segment liver volume from coronal, sagittal, and axial phases. A prominent histogram peak identifies the region of the liver [37]. Campadelli et al. presented another study of abdominal organ segmentation based on morphology operators, threshold, neural network, level set, model fitting, and data-driven methods [38].
Dixit and Pruthi proposed a simple method for the global threshold and morphological operator segmentation process. However, the global threshold cannot take advantage of the image characteristics [24]. Kumar and Moni proposed adaptive threshold operations and morphology. Each ROIs depict tumor region was automatically extracted using a fuzzy c-mean method [39]. Mahjoub proposed an approach based on the histogram threshold, the algorithm for connected labeling components, morphological operators, and detection of contours. This approach depends on prior knowledge, such as liver position and shape [23]. Akram et al. the adaptive threshold was used, and morphological operations were applied. This approach has achieved a precision of 96% for 100 patients [40].
Region growing (RG)-based segmentation approach, the foundation of RG is the presumption that adjacent pixels within an area have the same intensity value. One of the most essential algorithms for regional segmentation is the RG algorithm. By examining adjacent pixels of a group of pixels known as seed points, it performs image segmentation by assessing if the pixels can be grouped into the seed point cluster. The advantages of this method are that it is the simplest and can accurately separate areas with similar features. The main drawbacks of the RG are that it is sensitive to seed point location and threshold, fails in the presence of weak boundaries, resulting in leakage, and takes much time to scan each pixel prosperity. As a result, the segmentation must go through a number of pre- and post-processing processes. These issues make the image segmentation more susceptible to fragmentation. RG problems can be solved by integrating RG with different algorithms such as watershed, fuzzy c-means, connected component algorithm [25, 41]. Finally, liver segmentation cannot be handled only using a regional global approach; it is generally coupled with other approaches.
To automatically find the seed point, Massieh et al. presented the automatic RG approach with the FCM method [42]. In order to achieve the maximum contrast stretching filter performance with 84% precision, Mostafa et al. introduced RG to isolate the liver region from scans with morphological operators [43]. A hybrid strategy to segment the liver utilizing the RG algorithm and post-processing techniques was proposed by Rusko et al. (pre-defined intensity range to image threshold and smooth histogram to reach the full significance). The tiny portions were then eliminated using image erosion. The liver’s intensity was not significantly different from that of other organs. However, the segmentation of the lesions was a problem for this method [44]. The proposed method by Deokar and Hambarde relies on the median filter to enhance the quality of the image; RG and morphological operators are employed to produce a good liver segmentation region [26].
The watershed-based segmentation approach (WS) is utilized to separate tumors from liver images, one of the most challenging processing tasks. This type of issue is frequently handled through watershed transformation. Finding the “watershed lines” in an image that divide the various regions is the main objective of the WS approach. Three images segmentation dimensions—“minimum,” “catchment basin,” and “watershed line”—are used by WS to express topography. Although the WS approach has the advantage of continuous ROI borders and may be used in conjunction with other algorithms to improve segmentation accuracy, its shortcomings and critiques are due to over-segmentation and time consumption.
Lim et al. [45] used prior knowledge in the morphological filter algorithm to automatically segment the liver and detect initial liver contour utilizing a morphological label and clustering operator. The main problem of this approach requires an amount of manual parameter setting at each step. Yim and Foran proposed WS and active contour methods for liver segmentation to obtain hepatic liver. Comparison with different methods showed that active contour was able to detect lesions compared to the WS algorithm [46]. WS algorithm causes segmentation. Over segmentation, the problem was solved by reducing the number of false regions resulting by [47]. Abdalla et al. applied a hybrid approach by combining the level set with the WS for liver segmentation from abdominal CT scans. They achieved accuracy reached 92.1% [48]. Sudhamani and Raju extracted the liver using the WS method and the RG algorithm. However, their technique relies on the initial seed point for human expert assistance in identifying the suspected region [33]. For tumor segmentation, Jean et al. introduced the WS method with random Markov fields. This algorithm is fast and gives decent segmentation results [49].
A contour-based segmentation approach is used to segment liver parenchyma and lesions from CT/MRI images. An active contour, also referred to as “snacks or deformable models,” is based on contour. Active contour and level sets models are popular liver lesion segmentation methods. However, they have drawbacks due to complicated initialization and usually utilize a speed function that assumes good contrast images.
Pan and Dawant were the first to employ level sets for liver segmentation [50]. This method is highly regarded and extensively used for segmentation and a final optimization step. The main disadvantage is the high computational time for the segmentation process [51]. Massoptier and Casciaro integrated a gradient vector flow active contour to get the liver from the CT scan. This method proves that high textures and low contrast content in images are insufficient [52]. Zidan et al. proposed a level set method to segment abdominal CT into a set of interesting ROIs. The challenge with this algorithm is to define a suitable speed function and its parameters [48]. Li et al. suggested a level set that combined edge energy and minimized likelihood energy. The target density distribution is ascertained using energy reduction, and the suggested edge detector maintains the ramp for weak borders. The dataset consists of 18 scans, and the findings indicated that the Jaccard distance error was 14.4 ± 5.3% [28]. The three-step method developed by Li et al. begins with an image enhancement phase that involves filtering the anisotropic diffusion of the curvature and enhancing the image using a statistical model based on primary component analysis (PCA). The mean shape model was shifted to a coarse point in a test image using the threshold and Euclidean distance in the second phase. Finally, using a deformable graph cut, the liver was found. 50 images from the two common databases Sliver07 and 3Dircadb are used in the suggested method. This method successfully detects hepatic parenchyma in abdominal CT scans, according to trials [53].
The cluster-based Segmentation approach is a fully automatic segmentation that does not require human intervention and considers data uncertainty and ambiguity. Because of the diversity of the liver and tumors from patient to patient, several clustering approaches, such as k-means clustering, fuzzy c-means, self-organizing map (SOM), and others, were used to segment CT/MRI images extract ROIs that represent tumors. The drawbacks of these methods are that the results contain many false positive regions that need post-processing steps to focus on liver tumors and cannot handle the Indeterminacy and effects by initial clusters. Anter et al. improved liver segmentation using hybrid fuzzy c-means and neutrosophic sets (NS). The CT image is converted to NS domain represented by True (T), Indeterminacy (I), and False (F). Then the threshold adapted using the FCM algorithm [54]. Massoptier used k-means to classify the lesions against the parenchyma inside a liver envelope [55].
Using hybrid FCM and level set, Li and Luo proposed a fully automatic method for segmenting the liver. The images were first pre-processed. The liver region was then extracted and refined using FCM with the level set [29]. An automated CT scanning method for liver and tumour segmentation was suggested by Kumar and Moni. The method uses Alternative Fuzzy c-means (AFCM), a modified form of FCM, to segment tumours [39]. In order to narrow the search area for segmentation, Ben-Cohen et al. used a matching template to locate the lesion location in the CT scan using an important segmentation mask. Lesions have been divided using the mean shift algorithm and the adaptive RG. There are 127 CT scans from the Sheba Medical Center in the database. The outcome was 83%, with an error rate of 0.08 [56]. For the segmentation of the liver and associated tumour, Alahmer and Ahmed proposed two-phase methods. The liver’s binary mask is used to first segment the liver. Following that, rapid FCM was used to divide the image into three classes. In order to eliminate small items beyond the liver region, the clustering procedure was improved using morphological operators. Then, tumours from the segmented liver were divided using region growth [57].
Probabilistic atlas-based segmentation approach. Typical method for including prior knowledge in the segmentation process. To segment liver tumours, the authors [58, 59] used a probabilistic atlas (PA) and a statistical shape model (SSM). Both PA and SSM rely on previously collected liver data. SSM performs better than PA when working with enhanced CT/MRI pictures, whereas PA can only handle non-contrast images. The potential hepatic deformations that an SSM can depict are also restricted, and its initialization is difficult yet crucial. Low-level procedures, an affine probabilistic atlas, and a multi-atlas-based liver segmentation were used by Platero and Tobar. Due to registrations, atlas selections based on ROIs, and coarse segmentation, this method offers extremely accurate segmentation [60]. With the use of an active geodesic contour, patient-specific contrast-enhancement characteristics given to an adaptive convolution, and correction for shape and placement issues, Linguraru et al. used probabilistic atlases to segment the liver. The accuracy achieved from this approach variant from 92.7 to 96.2% [61].
Neutrosophic Sets (NS)-based Segmentation Approach. While a fuzzy set was simply used to address uncertainty, NS previously dealt with both indeterminacy and uncertainty. The fuzzy set uses a real number µA(X) \(\in\) [0, 1] to represent the membership of set A defined on universe X. In real-time applications that depending on Indeterminacy, neutrosophic sets should be used. Fuzzy logic cannot be considered for these applications because it is limited to the truth and false percentage. So, it is hard for the fuzzy set to solve problems like this [62].
Neutrosophic logic (NL) shows a percentage of “indeterminacy” due to unexpected parameters hidden in some propositions. NL is an extension of fuzzy logic, includes a new component termed “indeterminacy” which carries more information than fuzzy logic. Applications that use neutrosophic sets outperform those that use fuzzy logic. NL is so new that it is worth exploring in many fields [62].
Anter et al. proposed a hybrid NS and adaptive FCM approach for CT liver segmentation. This approach achieved accuracy reached 94% [63]. Cheng et al. proposed neutrosophic logic and a modified fuzzy clustering approach for CT segmentation [62]. Zhang proposed WS based on NS to Image Segmentation. The CT images were interpreted into different components, objects referred to T and background referred to F. This approach is suitable for handling blurred edges objects [64]. Anter et al. applied a new hybrid approach based on NS and particle swarm optimization (PSO) to the liver tumor segmentation. The CT image was enhanced, and the noise was removed using neutrosophic sets components (T,I, and F).
PSOFCM technique used to segment liver parenchyma and lesions from abdominal CT scan. The results showed that this approach was robust and precise with quick convergence and time-consuming [12]. Siri and Latte proposed NS and Fast Marching Method (FMM) for liver segmentation. First, noise has been reduced by a median filter, and CT images were expressed in a neutrosophic domain in order to extract the liver. Morphological operations were then performed on the subset of Indeterminacy (I). The fast-marching method automatically identifies the starting points in the liver section to detect the exact liver boundary. This method achieved a high accuracy reached to 91.61% [65]. In addition, Siri and Latte improved their work by combining NS efforts to extract liver tumors. Without user intervention, the Chan-Vese model was used to detect the initial contour in the liver. This approach achieved high results of 95.59%. Experiments have shown that this approach is robust and effective [66].
Swarm Optimization-based Segmentation Approach. Particle-swarm-optimization (PSO), Genetic-algorithm (GA), Firefly algorithm (FFA), Grey-wolf optimization (GWO), Crow-search algorithm (CSA), Social-spider-optimization algorithm (SSO), and other bio-inspired meta-heuristic optimization algorithms have been used to tackle a variety of problems. When standard techniques are unsuccessful or take too long to discover a good solution, meta-heuristic and optimization techniques have been applied. The bio-inspired optimization techniques have a high convergence speed and avoid becoming stuck in local minima, as shown by Mostafa et al. [27]. The disadvantages of these methods are; they work in a random behavior and give inconsistent results.
Bio-inspired and meta-heuristic algorithms can be integrated with classical methods such as fuzzy c-means, watershed, region growing, etc., to solve its main drawbacks. For example, in FCM, clusters need to be specified before the clustering process, very slow convergence, and the initial selection of the centroids effect on the results performance. The main problem is the seed point in the region growing, and time-consuming to check each pixel intensity. All these problems can be solved by integrating with computational intelligence optimization algorithms. The integration results show that the hybridized approach can improve image segmentation efficiency [67].
To segment and detect abnormalities in the image using PSO, Venkatesan and Parthiban suggested two basic methods: FCM and maximum entropy (FCM-PSO). By comparing the segmentation findings, cluster distances, and intra- and inter-cluster distances, the analysis is carried out. The FCM-PSO shortens the calculation time and produces precise results [68]. In order to categorize liver parenchyma pixels, tumours, and other organs in the abdominal CT scans in various homogenous regions, Anter et al. developed a new segmentation approach based on NS, FCM, and PSO. The CT picture has been improved and the noise removed before being transferred to the NS domain. The improved image was then sent to PSOFCM for segmentation of the liver tumour from the abdominal CT scan. The outcomes demonstrate that NS- PSOFFCM was accurate and reliable, with quick convergence to the best solution and shorter computation times [12].
In addition, bio-inspired and meta-heuristic algorithms can be integrated with conventional and classical methods to solve its problems, affecting the results performance such as threshold method, k-means, watershed, region growing, etc. Mostafa et al. proposed an algorithm for clustering problems using artificial bee colony (ABC) to segment liver from CT images. ABC calculates the cluster centroids in the image. The extracted regions are integrated to give an initial estimate of the liver area. RG was used to refine the liver segmented. The results employed on 38 images achieved excellent segmentation accuracy, reached 93.73% [69]. Mostafa et al. enhanced their work using the grey wolf optimization algorithm (GWO). This approach combines GWO, liver statistic, RG, and medium shift clustering. GWO has been used to calculate the pre-defined number of clusters centroids. RG has been used to boost the segmented liver. In order to segment lesions, the mean shift clustering technique was finally used. The results showed that the total precision achieved was 94.08% [27].
Singh and Gupta proposed a segmentation approach for the liver using k-means and Ant Colony Optimization (ACO). ACO was used for the optimization of poorly selected k-clusters. Quantitative analyzes were carried out based on precision and specificity. The experimental result showed that high accuracy and specificity, 98.74, 99.20, respectively. This method is fully automatic compared to the standard k- means and achieves better segmentation even if the boundary is not clear [70]. El-Masry et al. proposed an automated CT liver image clustering approach based on the invasive weed optimization (IWO) evolutionary algorithm. Without prior information on the number of naturally occurring groups in the images, IWO was used [71]. By integrating the PCM method with PSO, Zhang et al. greatly increased segmentation effectiveness. Research revealed that segmentation greatly outperformed the FCM algorithm in terms of effect and efficiency [72]. To solve the challenge of segmenting liver tumours, Ali et al. suggested combining the PSO and FCM algorithms. This method’s benefits include quick convergence to the best solution, increased efficiency, and more accurate segmentation [54].
Multi-discriminative markers are used in Eapen et al. for segmenting the liver (prior domain information, intensity model, and regional characteristics of the liver in the graph-cut image segment). An edge-adaptive weight function using swarm intelligence as inspiration governs the energy reduction of the conventional graph-cut model. Public data sets are used to qualitatively and statistically validate the model (MICCAI 2007, 3D-IRCAD). Quantitative evaluation of segmentation results was performed with an accuracy of 80.8%, 82.5% on MICCAI, IRCAD, respectively [73]. Sayed and Hassanien proposed the segmentation of the CT liver based on PSO and an adaptive algorithm. The accuracy obtained reached 94 % [74].
Machine Learning (ML)-based Segmentation Approach. ML was proposed to achieve accurate tumor segmentation. ML methods are stable and can account for the diversity of liver tumor topologies and characteristics, but they typically need many training samples or a lengthy training period [31]. Many ML techniques are used for determining the ROIs of the liver and tumor. Danciu et al. proposed a support vector machine (SVM) method to liver segmentation with wavelet characteristics [75]. SVM is an interesting classifier that does not require much training. Kumar et al. proposed a neural network to classify FCM results, identifying which regions belong to the tissue of the liver [76]. Aldeek et al. used the Bayesian model to extract the liver from the CT image for a robust and reproducible semi-automatic technique. The proposed approach generates 87% accuracy for 44 cases [77]. Susomboon et al. applied approach used four steps: In the first step, the intensity-based threshold was applied to obtain the regions. The second step, region-based, was used for texture-based classification to classify all regions based on textures. Third step, an initial liver region was determined for each patient. In the fourth step, a confidence interval of 95% determined from the region’s intensities to detect the liver ROI, the proposed approach achieved 86% accuracy [78].
Deep Learning (DL)-based Segmentation Approach. DL is a very well algorithm that is employed to complete numerous crucial tasks, particularly in medical imaging. DL can predict significant regional features and accurately segment organs. Although DL is simple to train, it requires more data and meticulous results interpretation. Since it has more layers than traditional machine learning, which would fail if it had more than three layers, DL is still in its early stages. The majority of systems today have between 30 and 150 layers. Outside of medicine, DL has significantly increased the performance of computer algorithms, and in the near future, it is anticipated that the performance of medical applications will also significantly increase [18]. DL algorithms may be applied to tumour identification and segmentation in medical images.
A fully automated method for segmenting liver tumours was created by Chlebus et al. using a 2D convolutional deep neural network with shape-based post-processing. The detection and segmentation assessment was carried out on the LiTS data set. The proposed method can detect 77% of lesions accurately [79]. Bellver et al. proposed the liver segmentation method and its CT lesions using Convolutional Neural Networks (CNNs). The network used a cascaded architecture to focus on the liver region to segment the lesions [80]. Christ et al. proposed a two-step HCC malignancy prediction model, and the first step is automatically segmenting HCC tumor lesions using a fully convolutional neural cascade network (CFCN). A 3D neural network (SurvivalNet) was then used to predict the malignancy of the HCC lesions. This model assessed 31 patients with HCC and achieved a 65% accuracy [81]. Han has developed a deep convolutional neural network (DCNN) to produce the center slice segmentation map. The model has 32 layers and uses both the U-Net and the residual net (ResNet). The model was trained using the 130 slices and reached a 67% Dice score [82]. Amitai and Greenspan proposed fully convolutional (FCN) and conditional generative adversarial (GAN) networks to segment the CT liver. Dataset consists of 25 CT scans. The results showed a high detection performance of 92.3% using True Positive Rate (TPR) and 0.25 using False Positive Rate (FPR) per case [83]. Using 338 3D CT scans for training and testing, Kalinovsky, Liauchuk, and Tarasau suggested a deep learning strategy to identify CT cancer. Slice-wise lesion identification in 2D images using semantic segmentation, slice-wise lesion detection in 2D images using the sliding window approach, and lesion detection in full 3D CT scans using semantic segmentation are all examples of deep CNN-based algorithms that have been developed and put into use. Compared to previous methods built on traditional image analysis, our technique performs better [84]. Todorok et al. used 3D multi-phase contrast-enhanced liver CT data from 75 cases to develop a method for recognizing CT liver cancers with a deep CNN. The proposed DCNN approach outperformed the Bayesian model in detection accuracy [31]. Yasaka et al. presented a deep learning technique for hepatic mass differentiation using CNN. The training was supervised using 55536 image sets, and in test set 100 liver mass image was tested. The median precision obtained was 84%. Experimental results showed that deep learning with CNN achieved high diagnostic performance in diagnosing the CT liver process [85].
According to an approach developed by Christ et al., fully convolutional neural networks and dense 3D conditional random fields (CRFs) can autonomously segment the liver and lesions (CFCNs). The results showed that the liver and lesions can be segmented using CFCN to achieve Dice scores for the liver of over 94% with computation times in the low 100s. Additionally, the Cascaded Unet technique that Christ et al. proposed for automatic liver and lesion segmentation had a precision of 93.1%. [81]. Dou et al. proposed 3D deeply supervised CT liver segmentation network (3D-DSN). The 3D-DSN proposed benefits from an utterly revolutionary architecture. The results have been assessed using the public data set MICCAI- SLiver07. The experiments showed that this approach achieved competitive results with higher speed and performance [86]. In the same year, In order to correctly segment livers with large shape fluctuations or with fuzzy borders with surrounding organs or lesions, Hu et al. trained a 3D-CNN model [87]. Additionally, a two-stage approach for the pancreatic segmentation job was suggested by Roth et al. During the detection phase, the holistic CNN creates a 3D bounding box of CT volumes with axial, sagittal, and coronal perspectives. In order to produce a more precise final segmentation of the pancreatic, the second stage entails merging the mid-level information of CNNs trained to segment the interior and border of the pancreas into the bounding box suggested in the first stage [88]. An automatic CNN-based method for segmenting lesions in CT images was proposed by Li, Jia, and Hu. AdaBoost, Random Forests (RF), and SVM, three well-known machine learning algorithms, were contrasted with the CNN model. On 30 CT scans, accuracy was achieved at 82.67 and 84.34 percent, respectively. The outcomes showed that CNNs performed better than other methods and are promising for segmenting liver tumours [89]. Vivanti et al. developed a fully automated approach for segmenting liver tumors using deformable baseline scan registration and tumor delineation and training CNN as a voxel classifier on all baselines. The findings revealed an average overlap error of 16.26 % in 67 tumors from 21 patients using ground-truth Segmentation and a standard deviation (Std = 10.33) [90].
Table 2 summarizes the various proposals for liver and tumor segmentation. In addition, several other aspects of the works, such as segmentation methods and segmentation accuracy, are shown.
3 Proposed Model for Liver Tumor Diagnosis
A dynamic CAD model is suggested for liver tumor diagnosis using multi-modality CT and MRI in this review. A unique opportunity for developing advanced new methods may be provided for a real-time diagnosis. In addition, the above limitations have been addressed to design a CAD model that can automatically diagnose liver lesions based on bio-inspired optimization algorithms, chaos theory, deep convolutional neural network, and spiking neuron model for liver tumor segmentation and diagnosis as well as neutrosophic logic can be applied to decrease and handle the Indeterminacy and uncertainty. This kind of methodology has never been proposed and applied for liver tumor localization and diagnoses presenting high characteristics to the best of the author’s knowledge. This paper is potentially helpful in developing a CAD model for liver diagnosis and treatment to help physicians provide critical information for surgical planning and early tumor detection and analysis. In addition, explore the effects of treatment methods such as (radiotherapy and chemotherapy) on the hepatic tumor, which can efficiently help determine the effective doses and fractions of the therapy. In addition, this model may be possible to guide surgical doctors to directly locate live tumors and destroy tumors with minimally invasive techniques, such as ablation and embolization. Furthermore, this model could assist physicians in classifying high-risk patients and make a novel diagnosis. Moreover, this model will open up new opportunities for medical imaging applications by proposing highly accurate models with minimal human intervention, extending their use and ultimately leading to improved patient care.
The suggested model composes of mainly five phases; (1) DICOM data pre-processing, (2) Liver parenchyma segmentation and lesion detection and delineation from the multi-modality CT/MRI abdominal images, (3) Features extraction and subset feature selection from each lesion (ROIs) based on evolutionary spiking deep convolutional neural network, (4) Diagnosis and decision making based on federated deep spiking neural network, (5) Assessing the response of hepatic lesion to treatment based on deep spiking neural network, and (6) Evaluation criteria.
3.1 Pre-processing Phase
Multi-modality CT and MRI series images have been collected from different modalities. Moreover, other available multi-center benchmark datasets will be used to ensure the proposed model’s performance and stability [86, 91, 92]. First, these scans will be pre-processed; the initial step is DICOM series registration to align all CT/MRI images to a common space. Then, grayscale inhomogeneity results from the bias field of the CT/MRI scanner will correct. The subsequent combination of multiple images from multiple imaging modalities to improve features involves identifying and selecting features focusing on the relevance of features for diagnosis and evaluation purposes of a liver lesion. In addition, different filters will use to enhance contrast, smooth, remove noise and high frequencies and improve the overall visibility of features of a specific sign of malignancy.
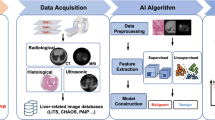
Figure 3a shows the data collection process with different modalities CT and MRI from multi-center datasets to check the stability and reliability of the proposed model. Figure 3b shows the sequence of pre-processing steps for imaging data.
3.2 Liver Segmentation, Lesion Detection, and Localization Phase
3.2.1 Pre-segmentation
In this phase, the pre-processed images will transform to the neutrosophic domain to increase segmentation efficiency and decrease noise-level and non-uniform neutrosophy is a philosophy field that is a generalization of intuitionistic fuzzy sets para-consistent sets, and tautological sets. The neutrosophic set (NS) and its characteristics are briefly described in [34, 65]. These issues that fuzzy logic cannot handle may be well solved by neutrosophic logic. NS investigates the neutrosophic logical values of propositions represented by (Percentage of true (T), Indeterminacy (I), and False (F)).
To use neutrosophy theory, an image must be transformed to a neutrosophic-domain \(P_{NS}\), each pixel in the image neutrosophic-domain can be shown as T, I, and F, which means t% true, i% indeterminate, and f% false, where t varies in T, i varies in I, and f varies in F.
The neutrosophic components are \(T(i,j),I(i,j),F(i,j)\), and each pixel P(i,j) in the image domain is converted to the NS-domain \(P_{NS} (i,j)\) and can be computed as follows:
where \(\overline{g(i,j)}\) indicates the local mean value of the window size, and \(Ho(i,j)\) indicates the homogeneity value.
3.2.2 Post-segmentation
After the image is transformed to the NS domain, the neutrosophic truth set (T) is selected for the post-processing, representing the objects of the image. The indeterminacy set (I) that represents the edges of the image is removed, and the false set (F) representing the background is also removed.
In this phase, the liver will be segmented, and lesions will be localized using the improved fuzzy c-means (FCM), as shown in Fig. 4. Due to its overall performance, FCM is an unsupervised learning approach and a prevalent technique for statistical data analysis utilized in many domains. In FCM, a data point can belong to any class with varying degrees of membership. FCM adopts fuzzy partitions of given data between 0 and 1. A meta-heuristic swarm intelligence algorithm will be used to guide the fast fuzzy c-means (FFCM) method for the best clusters. Bio-inspired swarm algorithms have been employed in situations where traditional approaches either fail to find a suitable solution or take a too long to find it.
The limitations and shortcomings of the FCM, such as (a) the number of clusters must be predetermined, (b) the results depend on the initial centroids selection, (c) the FCM lacks sufficient convergence speed, especially in the emergency situations, and (d) the FCM assigns membership ratings to each pixel. As a result, every voxel belongs to every cluster, and (e) updating the cluster centres and membership grades repeatedly requires a lot of time. The swarm intelligence algorithms can work to handle these limitations of the FCM.
3.3 Meta-heuristic Optimization Algorithms
A heuristic is a method of solving a problem in computing that does not require the exhaustive application of a technology. The use of a meta-heuristic approach is advantageous for solving search and optimization problems. Thus, the meta-heuristic technique often (i) lacks formal verification of convergence to the optimal solution (ii) is computationally quicker than exhaustive search, and (iii) typically seeks a near-optimal solution rather than the actual ideal solution [93].
Stochastic procedures are widely used by iterative meta-heuristic techniques to change one or more initial candidate solutions. Due to the inherent practicality of many real-world optimization problems, classical algorithms may not always be applicable or effective in solving such concerns. The main factor driving meta-heuristic techniques’ present appeal is their capacity to handle a range of complexity levels in relation to practical problems and provide a largely workable solution [67].
Most meta-heuristic techniques are inspired by natural, physical, or biological principles and attempt to simulate them at a basic level using various operators. A meta-heuristic in mathematical optimization is a higher-level technique to discover an excellent solution to an optimization problem, mainly when partial or faulty information or limited computation capacity.
Meta-heuristics are frequently referred to using the following words bio-inspired and nature-inspired. They can, however, be further subdivided into evolutionary algorithms, swarm-intelligence, and physical phenomenon. Meta-heuristics follow a similar series of operations and may thus be described within standard generic stages, as seen in Algorithm 1.
The phases of the proposed model for liver and tumor segmentation utilizing the fuzzy swarm intelligence algorithm are depicted in Fig. 4. The primary issue with the SI is the random behavior of agents in the search space, which produces inconsistent results and takes a long time to converge to the optimal solution. As a result, chaos theory will be used to solve this problem. The advantages of chaos theory include the capacity to avoid local minima, quick convergence, non-invertible maps, and the ability to do comprehensive search faster than stochastic search, which is based on probabilities, non-repetition, and ergodicity of chaotic behavior [94]. These benefits can significantly enhance the overall performance of meta-heuristic algorithms in terms of local minima avoidance and fast convergence.
Algorithm 1: Generic steps of meta-heuristic |
|---|
1. Init. a possible solutions. |
2. Init. all parameters (No. agents, No. iterations). |
3. Init. fitness value to all agents in search space. |
4. Eval. the candidate solution. |
5. Repeat |
a. Generate new candidate solutions. |
b. Eval. the new candidate solutions. |
6. Until satisfy the criteria. |
3.4 Diagnosis-Based Deep Spiking Neural Network
In this phase deep spiking neural network based on the spiking neuron model, deep convolutional neural network (DCNN), and bio-inspired SI algorithm will be used to characterize and diagnose liver lesions. The hybrid spiking neuron model and deep convolutional neural network (Spiking-DCNN) will extract features from the segmented liver tumors in multi-modality CT/MRI. Then, the features are fused to increase diagnosis performance, and the bio-inspired SI optimization algorithm will use to select the optimal and significant features from the whole extracted features. Finally, the spiking neuron model will predict liver tumors and obtain diagnosis decisions, as shown in Fig. 5.
Deep learning (DL) has emerged as a primary method for completing many complex tasks, demonstrating exceptional performance in a wide range of computer vision and medical applications. This achievement, however, comes at the expense of gathering and processing vast amounts of data, which are sometimes inaccessible due to privacy concerns. As shown in Fig. 6, federated learning has recently been introduced to train parameters on local hospital datasets and store them in a repository before sharing only the trained parameters with a centralized model, which is then responsible for updating other local real-time parameters models.
The Spiking-DCNN is an excellent generalization model because it can continuously learn from data and pursue the growing data pattern by recursively modifying the system structure and meta-parameters.
3.4.1 Spiking Neuron Model
The spiking neural network (SNN), in contrast to traditional neural artificial networks (ANNs), uses neurons biological realistic models to carry out computation in an effort to close the gap between neuroscience and machine learning. An ANN’s neurons exhibit a single, static, continuous-valued activation as their defining feature. Spikes, which are discrete events that occur at certain times in time rather than continuous values, are used by SNN to operate. The timing of the spikes is how neurons in an SNN communicate with one another.
SNN requires many fewer operations and is the best candidate for spatio-temporal data processing. SNN has become the focus of several recent applications as a single layer in many pattern recognition areas [95, 96].
The spiking neuron model potentially used and recommended is the Leaky Integrate-and-Fire (LIF) model. The previous study found that coding with spike-timing contains meaningful information and has high computing power in biological systems [97]. Deep SNN training, on the other hand, remains challenging. Spiking neurons’ transfer functions are often non-differentiable, prohibiting back-propagation. SNN also does not scale well in terms of computing complexity to actual high-dimensional pictures of size. Furthermore, spike-timing-dependent plasticity (STDP) does not enable learning hierarchical models that reflect several layers in a visual environment, such as edges or parts of objects, essential to a deep learning model.
3.4.2 Deep Convolutional Neural Network (DCNN)
Using DCNN as a framework for image classification issues has proven to be very successful. A feed-forward classification comes after a sequence of convolution and pooling layers in a DCNN. This network type has shown remarkable performance in object identification, segmentation, object recognition, and bioinformatics. It is made up of three main parts: a fully connected layer, a spatial sampling/pooling layer, and a convolutional layer [98].
The primary visual cortex, the initial cortical region in the visual brain system, shares similarities with the early layers of the DCNN in terms of representational properties. Convolution kernels are used as a CNN’s weights. A convolution layer convolves a portion of the preceding layer with a group of weight kernels to produce an array of output maps. The output maps are expressed by the model below.
where \(f\) indicate the neurons activation function, \(x^{k}\) is activation value of the neurons in the output maps, \(x^{i}\) is the activation of the neurons in the previous layers, and \(W^{k}\) represent the set of weight kernels. As explained in [95], Back-propagation training is used to train the convolutional weight kernels and fully connected weights for the final output layer of the DCNN.
3.4.3 A New Approach Based on Deep Spiking Neural Networks (Deep-SNNs)
The success of deep neural networks (DNNs), which are modelled after the cortical visual processing systems, has lately improved as a result of the accessibility of more potent computational hardware (GPU accelerators) and large training data sets. Despite their efficiency, the significant computational cost of building and testing such massive networks has limited their use to servers and the cloud. There is a need for specific hardware and novel computing approaches to create gadgets with cognitive capabilities.
Brain-like asynchronous event-based calculations power SNN. SNN focuses its computational effort on presently active network components, efficiently saving power on the remaining components and attaining lower power consumption orders than their ANN counterparts [95, 96]. As a result, there is a need to design practical learning algorithms that can use the particular features of SNN (event-driven, low-ANN power, high-learning) while maintaining the qualities of the DNNs model (general-purpose, scalable to more significant issues with higher accuracy).
A new generation of neural networks has developed, combining the multilayer structure of DNN and the type of information transfer used in SNN. Deep-SNN is an excellent choice for studying neural computing and diverse coding methods in the brain. DNN is a non-spiking network that communicates through continuous-valued activations. Although DNN’s energy efficiency is expected to be improved, SNN presents a unique potential in this respect because of the sparsity of spike occurrences in time. Spiking networks offer the added benefit of being naturally sensitive to the temporal patterns of information transmission in biological brain systems. Deep-SNN architectures are suitable for developing an efficient, brain-like representation [99].
In this review, a learning model based on spiking information and a deep convolutional neural network (Spike-DCNN) is suggested for liver tumor diagnosis, as shown in Fig. 7. A temporal-coding layer will be followed by a cascade of subsequent convolutional (feature extraction and descriptors) and pooling layers. The first layer transforms the input image into an asynchronous spikes train, with visual information encoded in the spikes’ temporal order. The second layer is regarded as extracting main visual features using the Difference-of-Gaussian (DoG) filter and shape morphology. The neurons in convolutional layers then integrate the incoming pulses and spike immediately after reaching their threshold. STDP is used in these layers to learn visual features.
The first layer applies a DoG filter with size w1D × w2D on the input lesion image and encodes the image to spike trains based on pixel intensity. Three convolutional and pooling layers, a feature selection (FS) layer-based swarm intelligence, and an SNN classifier pick the neuron with the highest activity (spike frequency) as the image’s class
Learning takes place sequentially only in the convolutional layers. When a new image of a liver lesion is shown, neurons in the convolutional layer compete with others, and those that fire first trigger STDP and learn the input pattern. A simplified STDP such as the one below may be used:
where i and j refer to the index of post- and presynaptic neurons, \(t_{i }\) and \(t_{j}\) represent the spike times, \(\Delta w_{ij}\) indicates the synaptic weight modification, and \(b^{ + }\) and \(b^{ - }\) specify the learning rate.
Visual information compression and translation invariance are made possible by pooling layers. Visual features get more broad and intricate as they go across the network. To reduce computation time and increase the effectiveness of the classifier, it may be suggested to use swarm intelligence-based feature selection in the last layer to select the best and optimal features from the complete set of complicated features. Additionally, the swarm intelligence will advise integrating with a spiking neural network to enable a particular type of learning (weight modification) that depends on the relative timing of spikes between pairs of directly connected neurons when the information required for weight modification is locally available. Finally, an SNN classifier may be utilized to differentiate between liver lesions based on neuronal activity with global receptive fields.
The very foundations of complex system identification may be affected by this model. As a result, the scope of its applications will cover straightforward clustering-based methods for pattern recognition, image segmentation, and other tasks, as well as more advanced modelling, classification, and prediction issues in a range of medical applications.
3.5 Assessing the Response of Hepatic Lesion to Treatment
According to the World Health Organization (WHO), quantitative imaging plays a critical role in monitoring the response of malignant hepatic lesions to treatment, which may be measured by mass size. In clinical practice, objective tumor size measurement is widely employed to detect tumor response to therapy. In clinical studies, objective response based on tumor size change may be converted to an early clinical endpoint in place of overall survival. The CT/MRI imaging criteria are ideal for determining tumor response to treatment. This model may help explore the effectiveness of different treatment modalities, including (surgical resection, ablation, radiotherapy, and chemotherapy) in the treatment of liver tumors. This can be applied by assessing the liver mass before and after the treatment and for post-treatment follow-up using fuzzy swarm intelligence, Spiking-DCNN, and morphological characteristics of the hepatic lesion for multi-time-point images for the same subject.
4 Conclusion
This paper discussed the medical background to the liver, tumor types, and hepatic imaging techniques. A general overview has been presented of the CAD system. The paper summarizes the various approaches used to create a CAD system for the segmentation of liver tumors and diagnosis and the limitation of the abdominal CT/MRI images. Several conclusions can be drawn from these previous works. A comprehensive comparison of different approaches to a wide range of data sets with many problems to test their advantages and disadvantages can help develop new effective approaches to various problems.
The segmentation phase is used to segment and delineate liver and tumors. The classical segmentation methods do not have exact solutions for all cases, high time computation, and stuck in local minima. The feature extraction phase used to analyze ROIs that depict tumors at only a single scale loses many dominant characteristics. Feature selection methods have high time computation in the feature selection phase, are stuck in local minima, and do not agree to get the optimal solution. The classifiers used have limitations on stochastic training, time consumption, lack of strength, and tuning parameters.
The limitations of the CAD system are addressed to diagnose benign and malignant tumors of the liver automatically. In addition, this paper suggests a new model-based bio-inspired optimization algorithms, federated learning, spiking neuron model, and deep convolutional neural network for liver tumor segmentation and diagnosis and neutrosophic theory was introduced to decrease and handle the Indeterminacy and uncertainty of data. The interaction of individuals in the population may be used to identify optimum features of the complex search space using meta-heuristic and bio-inspired optimization methods. Furthermore, the segmentations would automatically simplify and speed up physicians who are unable to obtain the segmentation of liver tumors without a long and tedious manual process guided by bio-inspired optimization algorithms to obtain the optimal solution and accelerate the convergence rate.
Data Availability
Data is available from the authors upon reasonable request.
References
Almotairi KH, Hussein AM, Abualigah L, Abujayyab SK, Mahmoud EH, Ghanem BO, Gandomi AH (2023) Impact of artificial intelligence on COVID-19 pandemic: a survey of image processing, tracking of disease, prediction of outcomes, and computational medicine. Big Data Cogn Comput 7(1):11
Azizi S, Soleimani R, Ahmadi M, Malekan A, Abualigah L, Dashtiahangar F (2022) Performance enhancement of an uncertain nonlinear medical robot with optimal nonlinear robust controller. Comput Biol Med 146:105567
Singh M, Pujar GV, Kumar SA, Bhagyalalitha M, Akshatha HS, Abuhaija B, Gandomi AH (2022) Evolution of machine learning in tuberculosis diagnosis: a review of deep learning-based medical applications. Electronics 11(17):2634
Nadimi-Shahraki MH, Taghian S, Mirjalili S, Abualigah L (2022) Binary aquila optimizer for selecting effective features from medical data: a covid-19 case study. Mathematics 10(11):1929
Shehab M, Abualigah L, Shambour Q, Abu-Hashem MA, Shambour MKY, Alsalibi AI, Gandomi AH (2022) Machine learning in medical applications: a review of state-of-the-art methods. Comput Biol Med 145:105458
Al-Shourbaji I, Kachare PH, Abualigah L, Abdelhag ME, Elnaim B, Anter AM, Gandomi AH (2023) A deep batch normalized convolution approach for improving COVID-19 detection from chest X-ray images. Pathogens 12(1):17
Abualigah L, Diabat A, Sumari P, Gandomi AH (2021) A novel evolutionary arithmetic optimization algorithm for multilevel thresholding segmentation of covid-19 ct images. Processes 9(7):1155
Zhou J, Sun HC, Wang Z, Cong WM, Wang JH, Zeng MS, Han GH (2018) Guidelines for diagnosis and treatment of primary liver cancer in China (2017 Edition). Liver Cancer 7(3):235–260
Wang FS, Fan JG, Zhang Z, Gao B, Wang HY (2014) The global burden of liver disease: the major impact of China. Hepatology 60(6):2099–2108
Lam S, Doran S, Yuksel HH, Altay O, Turkez H, Nielsen J, Mardinoglu A (2020) Addressing the heterogeneity in liver diseases using biological networks. Brief Bioinform. https://doi.org/10.1093/bib/bbaa002
Alzu’bi D, Abdullah M, Hmeidi I, AlAzab R, Gharaibeh M, El-Heis M, Abualigah L (2022) Kidney tumor detection and classification based on deep learning approaches: a new dataset in CT scans. J Healthc Eng
Anter AM, Hassenian AE (2018) Computational intelligence optimization approach based on particle swarm optimizer and neutrosophic set for abdominal CT liver tumor segmentation. J Comput Sci 25:376–387
Pesapane F, Codari M, Sardanelli F (2018) Artificial intelligence in medical imaging: threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp 2(1):35
Anter AM, Hassenian AE (2018) Normalized multiple features fusion based on PCA and multiple classifiers voting in CT liver tumor recognition. In: Advances in soft computing and machine learning in image processing. Springer, Cham, pp 113–129
Li X, Chen H, Qi X, Dou Q, Fu C, Heng P (2018) H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging 37(12):2663–2674
Gotra A, Sivakumaran L, Chartrand G, Vu KN, Vandenbroucke-Menu F, Kauffmann C, Tang A (2017) Liver segmentation: indications, techniques and future directions. Insights Imaging 8(4):377–392
Moghbel M, Mashohor S, Mahmud R, Saripan MIB (2018) Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev 50(4):497–537
Erickson BJ, Korfiatis P, Kline TL, Akkus Z, Philbrick K, Weston AD (2018) Deep learning in radiology: does one size fit all? J Am Coll Radiol 15(3):521–526
Liu Z, Song YQ, Sheng VS, Wang L, Jiang R, Zhang X, Yuan D (2019) Liver CT sequence segmentation based with improved U-Net and graph cut. Expert Syst Appl 126:54–63
Vorontsov E, Cerny M, Régnier P, Di Jorio L, Pal CJ, Lapointe R, Tang A (2019) Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiology 1(2):180014
Yasaka K, Abe O (2018) Deep learning and artificial intelligence in radiology: current applications and future directions. PLoS Med 15(11):e1002707
ElSoud MA, Anter AM (2016) Computational intelligence optimization algorithm based on meta-heuristic social-spider: case study on CT liver tumor diagnosis. Comput Intell 7(4):466–475
Mahjoub MA (2011) Automatic liver segmentation method in CT images. Can J Image Process Comput Vis 2(8):92–95
Dixit V, Pruthi J (2014) Review of image processing techniques for automatic detection of tumor in human liver. Int J Comput Sci Mob Comput 3(3):371–378
Patil S, Udupi VR, Patole D (2013) A robust system for segmentation of primary liver tumor in CT images. Int J Comput Appl 75(13):6–10
Deokar SM, Hambarde SM (2014) Detection of liver cancer in CT scan images. Int J Innov Technol Adapt Manag 1(6):1–4
Mostafa A, Hassanien AE, Hefny HA (2017) Grey wolf optimization-based segmentation approach for abdomen CT liver images. In: Handbook of research on machine learning innovations and trends. IGI Global, pp 562–581
Li C, Wang X, Eberl S, Fulham M, Yin Y, Chen J, Feng DD (2013) A likelihood and local constraint level set model for liver tumor segmentation from ct volumes. IEEE Trans Biomed Eng 60(10):2967–2977
Li X, Luo S, Li J (2013) Liver segmentation from CT image using fuzzy clustering and level set. Int J Signal Inf Process 4(03):36–42
Beichel R, Bornik A, Bauer C, Sorantin E (2012) Liver segmentation in contrast enhanced CT data using graph cuts and interactive 3D segmentation refinement methods. Med Phys 39(3):1361–1373
Todoroki Y, Han XH, Iwamoto Y, Lin L, Hu H, Chen YW (2017) Detection of liver tumor candidates from CT images using deep convolutional neural networks. In: International conference on innovation in medicine and healthcare. Springer, Cham, pp 140–145
Soler L, Delingette H, Malandain G, Montagnat J, Ayache N, Koehl C, Marescaux J (2001) Fully automatic anatomical, pathological, and functional segmentation from CT scans for hepatic surgery. Comput Aided Surg 6(3):131–142
Sudhamani MV, Raju GT (2014) Segmentation and classification of tumour in computed tomography liver images for detection, analysis and preoperative planning. Int J Adv Comput Res 4(1):166–171
Gaber T, Ismail G, Anter A, Soliman M, Ali M, Semary N, Snasel V (2015) Thermogram breast cancer prediction approach based on Neutrosophic sets and fuzzy c-means algorithm. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp 4254–4257. IEEE
Anter AM, Hassenian AE (2019) CT liver tumor segmentation hybrid approach using neutrosophic sets, fast fuzzy c-means and adaptive watershed algorithm. Artif Intell Med 97:105–117
Choudhary A, Moretto N, Ferrarese FP, Zamboni GA (2008) An entropy based multi-thresholding method for semi-automatic segmentation of liver tumors. In: MICCAI workshop, vol 41, no 1, pp 43–94
Campadelli P, Casiraghi E, Lombardi G (2007) Automatic liver segmentation from abdominal CT scans. In: 14th international conference of the EEE on image analysis and processing (ICIAP 2007), pp 731–736
Campadelli P, Casiraghi E, Pratissoli S, Lombardi G (2009) Automatic abdominal organ segmentation from CT images. Electron Lett Comput Vis Image Anal 8(1):1–14
Kumar SS, Moni RS (2010) Diagnosis of liver tumor from CT images using curvelet transform. Int J Comput Sci Eng 2(4):1173–1178
Akram MU, Khanum A, Iqbal K (2010) An automated system for liver CT enhancement and segmentation. ICGST-GVIP J 10(4):17–22
Anter AM, El Souod MA, Azar AT, Hassanien AE (2014) A hybrid approach to diagnosis of hepatic tumors in computed tomography images. Int J Rough Sets Data Anal 1(2):31–48
Abdel-massieh NH, Hadhoud MM, Amin KM (2010) A novel fully automatic technique for liver tumor segmentation from CT scans with knowledge-based constraints. In: 10th international conference of the IEEE on intelligent systems design and applications (ISDA2010), pp 1253–1258
Mostafa A, Hefny H, Ghali N, Hassanien AE, Schaefer G (2012) Evaluating the effects of image filters in CT Liver CAD system. In: International conference of the IEEE on biomedical and health informatics (BHI 2012), IEEE-EMBS, pp 448–451
Rusko L, Bekes, G, Nemeth G, Fidrich M (2007) Fully automatic liver segmentation for contrast-enhanced CT images. MICCAI Wshp. 3D Segment Clin 2(7):143–150
Lim SJ, Jeong YY, Ho YS (2005) Segmentation of the liver using the deformable contour method on CT images. In: Advances in multimedia information processing-PCM 2005. Springer, Berlin, pp 570–581
Yim PJ, Foran DJ (2003) Volumetry of hepatic metastases in computed tomography using the watershed and active contour algorithms. In: 16th international conference of the IEEE symposium on computer-based medical systems, pp 329–335
Liu J, Wang Z, Zhang R (2009) Liver cancer CT image segmentation methods based on watershed algorithm. In: International conference of the IEEE on computational intelligence and software engineering (CiSE2009), pp 1–4
Zidan A, Ghali N, Hassamen AE, Hefny H (2012) Level set-based CT liver image segmentation with watershed and artificial neural networks. In: 12th international conference of the IEEE on hybrid intelligent systems (HIS2012), pp 96–102
Stawiaski J, Decenciere E, Bidault F (2008) Interactive liver tumor segmentation using graph-cuts and watershed. In: 11th international conference on medical image computing and computer assisted intervention. In Workshop on 3D segmentation in the clinic: a grand challenge II. Liver Tumor Segmentation Challenge. MICCAI, New York, USA, pp 1–12
Pan S, Dawant BM (2001) Automatic 3D segmentation of the liver from abdominal CT images: a level-set approach. In: International Society for Optics and Photonics on Medical Imaging, pp 128–138
Lim SJ, Jeong YY, Ho YS (2006) Automatic liver segmentation for volume measurement in CT Images. J Vis Commun Image Represent 17(4):860–875
Massoptier L, Casciaro S (2007) Fully automatic liver segmentation through graph-cut technique. In: 29th annual international conference of the IEEE on engineering in medicine and biology society (EMBS 2007), pp 5243–5246
Li G, Chen X, Shi F, Zhu W, Tian J, Xiang D (2015) Automatic liver segmentation based on shape constraints and deformable graph cut in CT images. IEEE Trans Image Process 24(12):5315–5329
Ali AR, Couceiro MS, Anter AM, Hassanian AE (2014) Evaluating an evolutionary particle swarm optimization for fast fuzzy c-means clustering on liver ct images. IGI, Computer Vision and Image Processing in Intelligent Systems and Multimedia Technologies, pp 1–21
Massoptier L, Casciaro S (2008) A new fully automatic and robust algorithm for fast segmentation of liver tissue and tumors from CT scans. Eur Radiol 18(8):1658–1665
Ben-Cohen A, Klang E, Diamant I, Rozendorn N, Amitai MM, Greenspan H (2015) Automated method for detection and segmentation of liver metastatic lesions in follow-up CT examinations. J Med Imaging 2(3):034502
Alahmer H, Ahmed A (2015) Computer-aided classification of liver lesions using contrasting features difference. In: ICMISC 2015: 17th international conference on medical image and signal computing, pp 27–28 November 2015, London
Lu YC, Kemper AR, Gayzik S, Untaroiu CD, Beillas P (2013) Statistical modeling of human liver incorporating the variations in shape, size, and material properties. Stapp Car Crash J 57:285–311
Lu YC (2014) Probabilistic analysis of the material and shape properties for human liver (Doctoral dissertation, Virginia Tech)
Platero C, Tobar MC (2014) A multiatlas segmentation using graph cuts with applications to liver segmentation in CT scans. Comput Math Methods Med
Linguraru MG, Sandberg JK, Li Z, Shah F, Summers RM (2010) Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Med Phys 37(2):771–783
Cheng HD, Guo Y, Zhang Y (2011) A novel image segmentation approach based on neutrosophic set and improved fuzzy c-means algorithm. New Math Nat Comput 7(01):155–171
Anter AM, Hassanien AE, ElSoud MAA, Tolba MF (2014) Neutrosophic sets and fuzzy C-means clustering for improving CT liver image segmentation. In: Proceedings of the fifth international conference on innovations in bio-inspired computing and applications (IBICA2014), Springer, New York, pp 193–203
Zhang M (2010) Novel approaches to image segmentation based on neutrosophic logic, PhD, Utah State University. http://digitalcommons.usu.edu/etd/795/, July, 2014
Siri SK, Latte MV (2017) A novel approach to extract exact liver image boundary from abdominal CT scan using neutrosophic set and fast marching method. J Intell Syst. https://doi.org/10.1515/jisys-2017-0144
Siri SK, Latte MV (2017) Combined endeavor of Neutrosophic Set and Chan-Vese model to extract accurate liver image from CT scan. Comput Methods Programs Biomed 151:101–109
Anter AM, Bhattacharyya S, Zhang Z (2020) Multi-stage fuzzy swarm intelligence for automatic hepatic lesion segmentation from CT scans. Appl Soft Comput 96:106677
Venkatesan A, Parthiban L (2013) Hybridized algorithms for medical image segmentation. Int J Eng Adv Technol 2(3):305–307
Mostafa A, Fouad A, Elfattah MA, Hassanien AE, Hefny H, Zhu SY, Schaefer G (2015) CT liver segmentation using artificial bee colony optimisation. Procedia Comput Sci 60:1622–1630
Singh I, Gupta N (2015) Optimized liver segmentation using ant colony optimization. Int J Adv Res Electron Commun Eng 4(9):2434–2439
El-Masry WH, Emary E, Hassanien AE (2014) Automatic liver CT image clustering based on invasive weed optimization algorithm. In: 2014 international conference on engineering and technology (ICET), pp 1–5. IEEE
Zhang L, Zhang L, Mou X, Zhang D (2011) FSIM: a feature similarity index for image quality assessment. IEEE Trans Image Process 20(8):2378–2386
Eapen M, Korah R, Geetha G (2015) Swarm intelligence integrated graph-cut for liver segmentation from 3D-CT volumes. Sci World J
Sayed GI, Hassanien AE (2016) Abdominal CT liver parenchyma segmentation based on particle swarm optimization. In: The 1st international conference on advanced intelligent system and informatics (AISI2015), November 28–30, 2015, BeniSuef, Egypt. Springer, Cham, pp 219–228
Danciu M, Gordan M, Florea C, Vlaicu A (2012) 3D DCT supervised segmentation applied on liver volumes. In: 35th international conference of the IEEE in telecommunications and signal processing (TSP), pp 779–783
Kumar SS, Moni RS, Rajeesh J (2012) Liver tumor diagnosis by gray level and contourlet coefficients texture analysis. In: International conference on IEEE in computing, electronics and electrical technologies (ICCEET2012), pp 557–562
Aldeek N, Alomari R, Al-Zoubi M, Hiary H (2014) Liver segmentation from abdomen Ct images with Bayesian model. J Theoret Appl Inf Technol 60(3):483–490
Susomboon R, Raicu DS, Furst J (2007) A hybrid approach for liver segmentation. In: Proceedings of MICCAI workshop on 3D segmentation in the clinic: a grand challenge, pp 151–160
Chlebus G, Schenk A, Moltz JH, van Ginneken B, Hahn HK, Meine H (2018) Deep learning based automatic liver tumor segmentation in CT with shape-based post-processing. In: 1st conference on medical imaging with deep learning (MIDL 2018), Amsterdam, The Netherlands, pp 1–9
Bellver M, Maninis KK, Pont Tuset J, Giró Nieto X, Torres Vinals J, Van Gool L (2017) Detection-aided liver lesion segmentation using deep learning. In: Advances in neural information processing systems 30 (NIPS 2017): NIPS proceedings, pp 1–5
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Sommer WH (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 415–423
Han X (2017) Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv preprint arXiv:1704.07239
Amitai MM, Greenspan H (2017) Virtual PET images from CT data using deep convolutional networks: initial results. In: Simulation and synthesis in medical imaging: second international workshop, SASHIMI 2017, held in conjunction with MICCAI 2017, Québec City, QC, Canada, September 10, 2017, Proceedings, vol 10557. Springer, New York, p 49
Kalinovsky A, Liauchuk V, Tarasau A (2017) Lesion detection in ct images using deep learning semantic segmentation technique. Int Arch Photogramm Remote Sens Spatial Inf Sci 42:13
Yasaka K, Akai H, Abe O, Kiryu S (2017) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286(3):887–896
Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng PA (2016) 3D deeply supervised network for automatic liver segmentation from CT volumes. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 149–157
Hu P, Wu F, Peng J, Liang P, Kong D (2016) Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol 61(24):8676
Roth HR, Lu L, Farag A, Sohn A, Summers RM (2016) Spatial aggregation of holistically-nested networks for automated pancreas segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 451–459
Li W, Jia F, Hu Q (2015) Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J Comput Commun 3(11):146
Vivanti R, Ephrat A, Joskowicz L, Karaaslan OA, Lev-Cohain N, Sosna J (2015) Automatic liver tumor segmentation in follow up CT studies using convolutional neural networks. In: Proc. patch-based methods in medical image processing workshop, vol 2
Anter AM, Hassanien AE, ElSoud MA, Azar AT (2015) Automatic liver parenchyma segmentation system from abdominal CT scans using hybrid techniques. Int J Biomed Eng Technol 17(2):148–167
Ge F, Wang S, Liu T (2007) New benchmark for image segmentation evaluation. J Electron Imaging 16(3):033011
Hussain K, Salleh MNM, Cheng S, Shi Y (2019) Metaheuristic research: a comprehensive survey. Artif Intell Rev 52(4):2191–2233
Anter AM, Ali M (2020) Feature selection strategy based on hybrid crow search optimization algorithm integrated with chaos theory and fuzzy c-means algorithm for medical diagnosis problems. Soft Comput 24(3):1565–1584
Tavanaei A, Ghodrati M, Kheradpisheh SR, Masquelier T, Maida A (2019) Deep learning in spiking neural networks. Neural Netw 111:47–63
Kheradpisheh SR, Ganjtabesh M, Thorpe SJ, Masquelier T (2018) STDP-based spiking deep convolutional neural networks for object recognition. Neural Netw 99:56–67
Teeter C, Iyer R, Menon V, Gouwens N, Feng D, Berg J, Szafer A et al (2018) Generalized leaky integrate-and-fire models classify multiple neuron types. Nat Commun 9(1):709
Rawat W, Wang Z (2017) Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput 29(9):2352–2449
Pfeiffer M, Pfeil T (2018) Deep learning with spiking neurons: opportunities and challenges. Front Neurosci 12:774–792
Funding
Not Applicable.
Author information
Authors and Affiliations
Contributions
AMA: Supervision, Conceptualization, Methodology, Investigation, Validation, Writing- Original draft preparation. LA: Supervision, Conceptualization, Methodology,, Investigation, Writing- Original draft preparation. All authors read and approved the fnal manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Research Involving Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Anter, A.M., Abualigah, L. Deep Federated Machine Learning-Based Optimization Methods for Liver Tumor Diagnosis: A Review. Arch Computat Methods Eng 30, 3359–3378 (2023). https://doi.org/10.1007/s11831-023-09901-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11831-023-09901-4