Abstract
Long non-coding RNAs (LncRNAs) are one of the many layers of transcription in higher plants. LncRNAs are responsive to biotic and abiotic stresses and regulate genes. In our study, we have identified 905 novel lncRNAs from 8471 drought-responsive, novel transcripts of RNA-Seq reads from two banana cultivars, a drought-tolerant cv. ‘Saba’ (ABB) and -susceptible cv. ‘Grand Naine’ (AAA). Of these 905 lncRNAs, 75 (8.3 %) transcripts were natural antisense RNAs (NATs) and 2 transcripts identified as precursors of microRNA-miR156 and miR166. Among the 905 identified lncRNAs, 216, 150 and 279, 164 lncRNAs were induced and reduced to drought stress, respectively, in tolerant and susceptible in comparison to their equivalent controls. The remaining 22 lncRNA of tolerant cultivars was not regulated by drought stress. Of the 882 drought-responsive lncRNAs, 44 new lncRNAs were identified as induced. Musa lncRNAs were unevenly distributed in 11 chromosomes of Musa acuminata and no lncRNAs were found in chromosome-9 of drought-tolerant cultivar. The average lengths of lncRNAs were 683 nucleotides (nt). Drought-responsive differential expression of lncRNAs was found between +8.11585- and −4.04311-fold. Around 7.9 % of the identified lncRNAs were decoys of 85 conserved microRNAs. These findings will lay a basic platform for effective strategic planning of developing drought-resilient crop varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bananas are considered as an important food security crop, providing staple food for millions of people, especially third world countries. Banana crop productivity was severely affected by drought stress. Drought-induced yield losses were documented to be around 20–65 % in bananas, especially bananas with AAA genome of east Africa (Van Asten et al. 2010). Vast research efforts were made earlier to identify the QTL/network of genes responsible for drought stress tolerance. Till now, many layers of regulatory elements such as protein kinases, stress-induced expression of several protein-coding genes, transcription factors, small regulatory RNAs such as microRNAs, snoRNAs, siRNAs and several unknown factors have been identified (Ponting et al. 2009; Liu et al. 2012; Kornienko et al. 2013). However, still many factors contributing toward stress response are largely not known. Functional long non-coding RNAs in drought stress response have been reported in model plants (Liu et al. 2012; Zhang et al. 2014; Shuai et al. 2014). These lncRNAs are crucial gene transcription regulators in plant response to biotic and abiotic stress (Xin et al. 2011; Zhang et al. 2013). LncRNAs also play a major role in plant developmental processes (Amor et al. 2009).
LncRNAs are not translated into proteins and can regulate gene expression (Kornienko et al. 2013). The characteristic features of lncRNAs were non-homolog to known proteins, >200 nt length, absence of discernible ORFs and no coding potential (Kong et al. 2007; Zhu and Wang 2012). LncRNA biogenesis is fairly understood both in animals and plants. They can be transcribed from intergenic, intragenic, intronic, exonic, UTRs and promoter regions and from both sense and antisense strands (reviewed by Ponting et al. 2009). LncRNAs are mostly localized in nuclear or cytosolic fractions and like every eukaryotic mRNA transcripts lncRNAs also undergo post-transcriptional modifications such as 5′capping, splicing and polyadenylation. LncRNAs comprises natural antisense RNA (NATs) of protein-coding genes, small RNA precursors (microRNA, small nucleolar RNA/SnoRNA and other small regulatory RNA) and structural RNAs (tRNA and ribosomal RNA). LncRNAs were once considered as dark matters of genome and started emerging as crucial gene regulators in the post-genomic era. Genome-wide transcriptome analysis (Microarrays, RNA-Seq) coupled with computational methods were widely used to unearth novel lncRNAs of animal and plant species (Xin et al. 2011; Sun et al. 2012; Zhang et al. 2014). Deep sequencing methods (RNA-Seq) led to unbiased identification of several thousands of non-coding RNAs including lncRNAs in model plants like Arabidopsis and maize. The utilization of these resources for orthologous lncRNA identification in other plant species is likely not possible, since lncRNAs were poorly conserved among plant species including model plants such as Arabidopsis and Oryza sativa (Xin et al. 2011; Hao et al. 2015). Hence, de novo identification of plant lncRNAs is highly demanded in each plant species. Novel lncRNAs detection strategies were mainly focused on key characteristic features of lncRNAs. Transcripts with more than 200 nt length and shorter ORFs with lack of coding potential are considered as core criteria for selection of lncRNAs from several transcriptional units (TU) of RNA-Seq reads (Boerner and McGinnis 2012; Zhang et al. 2014). Algorithms were developed to assess the coding potential of given transcripts using key features of biologically meaningful sequences and it can effectively discriminate coding and non-coding RNAs (Kong et al. 2007). Xin et al. (2011) identified 125 putative stress-responsive lncRNAs in response to powdery mildew infection and heat stress in wheat leaves. Twenty-two putative salt- and water-deficit stress-responsive lncRNA were identified in Arabidopsis (Xin et al. 2011). Zhang et al. (2014) identified a large number of lncRNAs involved in the sexual reproduction of rice. Till now, several hundreds of plant lncRNAs were predicted from different plant species, especially from model plants such as Arabidopsis thaliana, O. sativa, Populus trichocarpa, and Zea mays. A total of 5571 non-coding RNAs (ncRNAs) were deposited in plant ncRNA database (Yi et al. 2015).
Banana lncRNAs remain poorly characterized, and no genome-wide screening of potential lncRNAs in banana has been reported. Moreover, comparative analysis of expression of stress-responsive lncRNAs under control and drought stress conditions between cultivars expected to help us to identify their contribution in drought-tolerance mechanisms Therefore, the present study is focused on identification of novel drought-responsive lncRNAs from drought-tolerant and susceptible banana cultivars followed by profiling of differentially expressed lncRNAs in both cultivars during drought conditions. In plants, the majority of the lncRNAs were either Natural antisense RNAs (NATs) or small RNA precursors (Zheng et al. 2013; Wang et al. 2005). NATs are well characterized in plants for their role in degradation of sense transcripts via the RNAi pathway (Wang et al. 2005). In banana, several NATs of Musa acuminata were identified after complete genome sequences of DH-Pahang, an M. acuminata cultivar, was made available (D’Hont et al. 2012). All predicted Musa NATs are available to the research community at PlantNATsdb (Chen et al. 2012). NATs can be transcribed either from single locus (cis-acting NATs) or from different loci (trans-acting) to target one or more sense transcripts (Henz et al. 2007). Plant lncRNAs were recently reported as endogenous target mimics of miRNAs (Wu et al. 2013). As target mimics, lncRNAs block microRNA negative regulation of authentic targets (Meng et al. 2012). Therefore, it is paramount to identify lncRNAs and to study the expression level during drought stress to delineate the regulatory pathways involved. Hence, identification of these regulatory RNAs in drought stress will improve our conceptual knowledge of bringing out drought-resistant varieties for crop improvement.
Materials and methods
Drought imposition
Banana (Musa spp.) plants of two cultivars, the drought-tolerant ‘Saba’ (ABB genome) and drought-susceptible ‘Grand Naine’ (AAA genome), were used. These cultivars were selected based on the screening for drought tolerance conducted at field conditions (Surendar et al. 2013). 50 uniform-sized cultivars (≈1.0 kg and 90 days old, National Research Centre for Banana, India) were planted into cement pots (48 cm diameter, 38 cm depth, one plant per pot) that were filled with a fumigated pot mixture (35 kg) in the ratio of 1:1:1 (v/v/v, sand:farmyard manure:red soil), maintained at field conditions (temperature 39 °C/27 °C and humidity 40 %/85 % day/night) and sufficiently watered with tap water for 60 days for plant acclimatization. After 60 days, progressive soil moisture-deficit stress was imposed on one set of 25 plants of both cultivars simultaneously by withholding water for 24 days continuously; the other set of plants was normally watered (irrigated control).
The leaf samples were collected from three independent plants of irrigated controls and drought-stressed plants of two cultivars. At the end of the 24th day of drought condition, the volumetric soil water content (measured at 5 cm depth from the top of the soil level with ML2X probe/HH2 Moisture Meter, Delta-T Devices, Cambridge, Great Britain) was approximately 4.8 % (50 mm) and 9.1 % (200 mm) for the stressed plants, compared with approximately 30.6 % (50 mm) and 33.5 % (200 mm) for the control plants. Leaf samples of each treatment were frozen with liquid nitrogen and stored at −70 °C until use for cDNA library constructions.
Construction of cDNA library and Illumina deep sequencing
The cDNA library was constructed using an mRNA-Seq assay for paired-end transcriptome sequencing. The library construction and sequencing were performed by the Genotypic Technology, Bangalore, India, according to the Illumina TruSeq RNA library protocol outlined in “TruSeq RNA Sample Preparation Guide” (Part # 15008136; Rev. A; Nov 2010). Briefly, the total RNA was isolated from control and drought-stressed leaf samples of tolerant and susceptible cultivars. RNA was fragmented for 4 min at elevated temperature (94 °C) in the presence of divalent cations and reverse transcribed with Superscript II Reverse transcriptase by priming with random hexamers. Second-strand cDNA was synthesized in the presence of DNA polymerase I and RnaseH, and 200–700 nt fragments were incubated with RNA Fragmentation Reagent. The cDNA was cleaned up using Agencourt Ampure XP SPRI beads (Beckman Coulter). Illumina adapters were ligated to the cDNA molecules after end repair and addition of “A”-base. SPRI cleanup was performed after ligation. The library was amplified using 11 cycles of PCR for enrichment of adapter-ligated fragments. The prepared library was quantified using Nanodrop and validated for quality by running an aliquot on High Sensitivity Bioanalyzer Chip (Agilent). The library was loaded onto the channels of an Illumina HiSeq™ GAII Analyzer instrument for 4 gigabase in-depth sequencing, which was used to obtain more detailed information about gene expression. Each paired-end library had an insert size of 200–700 bp. The average read length of 90 bp was generated as raw data.
De novo assembly and sequence clustering
The clean reads were obtained from raw data by filtering out adaptor-only reads from reads containing more than 5 % unknown nucleotides (nt), and low-quality reads (reads containing more than 50 % bases with Q value ≤20). Then, de novo assembly of the clean reads was performed to generate non-redundant unigenes. We used de novo assembled sequence using Cufflinks v2.0.1 method. Sequence directions of the resulting unigenes were determined by performing BLASTX searches against protein databases, with the priority order of NR (non-redundant protein sequences in NCBI), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes database (KEGG), and COG (E value ≤1e–5) if conflicting results were obtained. The expression levels of unigenes were measured as the number of clean reads mapped to its sequence. The number of clean reads mapped to each annotated unigene was calculated and then normalized to RPKM and adjusted by a normalized factor.
Differential gene expression (DGE)
Banana RNA-Seq reads were mapped to the reference genome with TopHat2 (Trapnell et al. 2009), transcripts were assembled and their relative abundances were calculated using Cufflinks. The summation of FPKM (fragments per kilobase of transcript per million mapped reads) values for every transcript associated with a particular gene gives the expression measurement in FPKM. The differential gene expression is calculated by the Cuffdiff program using the ratio of control vs. treated FPKM values for every gene (Reference). We have done the reference-based DGE analysis with the Banana transcriptome gene transfer information downloaded from ensemble (ftp://ftp.ensemblgenomes.org/pub/plants/release-18/gtf/musa_acuminata). The following cutoff was assigned for up-regulated and down-regulated genes: FC <−1 = down-regulated, −1 ≤ FC ≤ 1 = neutral and FC >1 = up-regulated. (FC is log fold change to the base 2).
Identification of drought-responsive banana long non-coding RNAs
A total of 8471 transcript sequences annotated as uncharacterized or with no sequence homology to annotated transcripts and existing protein databases were initially extracted from differential gene expression calculated from RNA-seq data of drought-tolerant and -susceptible cultivars. To identify potential long non-coding RNAs 6009 transcripts with >200 nt were selected (Zhang et al. 2014) for further analysis. A total of 4037 non-redundant potential lncRNA transcript units were loaded at Coding Potential Calculator (CPC) to discriminate coding and non-coding RNAs (Kong et al. 2007; Wang et al. 2014). Transcript units with CPC score <0 were considered as long non-coding RNAs. To predict and visualize spliced lncRNAs, tools like ASFinder (Min 2013) and ASTALAVISTA (Foissac and Sammeth 2007) were used.
Prediction of potential microRNA target mimics from lncRNA sequences
The target mimics the mechanism of lncRNA–microRNA and their potential roles in gene expression were recently reported in plants (Wu et al. 2013). To explore the possibility of predicted lncRNA as microRNA targets, all lncRNA sequences were submitted at psRobot (Wu et al. 2012) and mapped to microRNAs of model plant Arabidops thaliana. Penalty score threshold was 3.0 and no permitted gaps were used as strict parameters for target prediction.
Results and discussion
Identification of novel drought-responsive long non-coding RNAs (lncRNAs) in banana cultivars
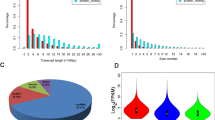
To obtain drought stress-responsive gene expression, leaf cDNA library for transcriptome analysis was prepared from RNA of control and drought-stressed samples of drought-tolerant (‘Saba’) and -susceptible (‘Grand Naine’) cultivar. A four-gigabase depth/deep sequencing was done using Illumina Hi-seq method. The reliable sequence reads were mapped to the reference genome (M. acuminata) with TopHat2 resulting in 119183 transcript assemblies or transcriptional units (Unpublished data). Annotation of these TU to M. acuminata and Viridiplantae plant species showed that 8471 TUs exhibiting no sequence similarity to annotated transcripts or known proteins were classified as hypothetical proteins. These TUs were shown to be equally responsive (p < 0.05) to drought stress as other functionally known drought-responsive mRNA transcripts. In this study, to identify putative lncRNAs, 6041 out of 8471 TUs consisting of ≥210 nts were selected and duplicate sequences, known protein homologs of NCBInr, were further removed. A total of 2491 and 1546 transcript sequences each from drought-tolerant and -susceptible cultivars were loaded to Coding Potential Calculator (CPC) to assess their coding potential. A total of 905 transcripts, respectively, from 556 of 2491 TUs of tolerant and 349 of 1546 TUs of susceptible cultivars with CPC score <0 were selected as long non-coding RNAs (lncRNAs) (Supplementary data. 1). The remaining 3132 TUs with CPC score >0 were considered as either protein-coding sequences or weak protein-coding sequences which were discarded from further analysis. Of 905 transcripts, 455 transcripts are strong lncRNAs, while the remaining sequences were considered as weak non-coding sequences as their CPC score lies between −1 and 0 as suggested by Kong et al. (2007). The length of the lncRNAs were ranged from 200 to 5196 nt, the majority of which (92 %) were approximately 300–600 nt in length (Fig. 1). The average size of all lncRNAs is 683 nt, which is higher than the average size of the lncRNAs of Arabidopsis (Liu et al. 2012). All the predicted lncRNAs were further queried to plant lncRNAs (5,571 records) of model plants, such as A. thaliana, O. sativa, P. trichocarpa, and Z. mays, to check banana lncRNAs conservation among these plant species. Banana lncRNAs have no sequence homology to any of the lncRNAs of these model plants and hence considered as novel lncRNAs of banana cultivars. To classify these novel lncRNAs, transcripts were further aligned to all classes of RNA families recorded at Rfam (Griffiths-Jones 2003) and it was found that one lncRNA (MUSA-S-NC321) corresponded to microRNA-166 precursors. Although tRNAs and snoRNAs were considered as two of the largest classes of non-protein-coding RNAs (Schattner et al. 2005), no significant sequence similarity was found between banana lncRNAs and tRNAs and snoRNAs of other species. To make stringent classification, lncRNAs were once again mapped to all species of plant small RNA families available at the PNRD database (Yi et al. 2015) separately, such as miRNA stem loop precursors, mature miRNA, snoRNAs, and trans-acting siRNA (tasiRNA). One lncRNA, MUSA-T-NC506, was found to have significant homology to microRNA-156 considered as microRNA-156 precursors.
To identify lncRNAs as drought-responsive natural antisense RNA (NATs) in banana, the unigene ID of each lncRNAs was mapped to M. acuminata NATs present in plant natural antisense RNA database (PlantNATsdb) (Chen et al. 2012). The results show that 75 out of 905 (8.3 %) lncRNAs were able to form NAT pair with Musa sense/protein-coding transcripts. Collectively, 395 NAT pairs were formed through 54 cis and 339 trans interaction with different locus of sense/protein-coding transcripts (Supplementary data. 3). Both cis and trans NATs can be classified according to their relative orientation and degree of NATs. In our study, 72 % of cis-NATs group was presented by nearby tail-end–tail (3′ close to 3′) pair according to Osato et al. (2007). Similarly, 75 % the trans-NAT pairs had continuous complementary region more than 100 nt and classified as 100 nt pair followed by the HC pair. Chen et al. (2012) showed that 90 % of the non-coding RNAs were NATs in Arabidopsis. NAT pairs either prevent translation of sense RNA into protein or process double-stranded RNA into small interfering RNAs (siRNAs) as reported by Zhang et al. (2012) in Arabidopsis. When Musa NATs were mapped to siRNA precursors, no correspondence was observed between NATs class lncRNAs and siRNAs, suggesting that these lncRNAs may not produce siRNAs. To identify splice variants from overlapping lncRNA transcripts that are mapped to the same genomic locus and have internal variable exon/intron boundaries, initially all predicted lncRNAs were mapped to 11 individual chromosome sequences of M. acuminata using ASFinder algorithms (Min, 2013). No splice clusters were formed from lncRNA sequences of both cultivars.
Distribution of novel lncRNAs in Musa chromosomes
Drought-responsive banana lncRNAs are not conserved among plant species, as no sequence homology between banana lncRNAs and total plant lncRNAs of other species including model plants, Arabidopsis and O. sativa was observed. However, a total of 7674 Musa protein-coding genes were conserved in Arabidopsis and O. sativa (Hont et al. 2013). Our study corroborates the findings of Hao et al. (2015) who stated that lncRNAs were less conserved in comparison to mRNA transcripts. Earlier, Xin et al. (2011) reported that wheat lncRNAs are not conversed among plant species. However, genomic distribution of these lncRNAs is crucial for genetic manipulation to reduce crop loss due to drought stress. By mapping these sequences with reference genomic sequences, we found a maximum of 19 % predicted lncRNAs of tolerant cultivar distributed in chromosome4 (chr4), whereas in drought-susceptible cultivar, ‘Grand Naine’, a maximum of 25 % lncRNAs were found on chromosome-8 (Fig. 2). No lncRNAs were found on chromosome 9 in tolerant cultivar, suggesting chr9 may not be responsible for biogenesis of these lncRNAs, whereas chr9 of susceptible cultivar contributed 21 % of lncRNAs. The uneven distribution pattern of lncRNAs was earlier noticed by Hao et al. (2015) in lncRNAs of cucumber chromosomes. To our knowledge, this is the first important study on identification of Musa lncRNAs and their genomic distribution in banana chromosomes.
Drought stress regulation of novel lncRNAs in drought-tolerant and -susceptible cultivars
Except 22 lncRNAs, all other lncRNAs (882) reported in our study were differentially expressed to drought stress. The expression profiles of these 905 lncRNAs obtained between control and drought stress samples of both drought-tolerant and -susceptible cultivars showed induced expression of 216, 150 lncRNAs, respectively, in drought-tolerant and -susceptible cultivars in comparison to their respective equivalent controls (Supplementary data. 2). Similarly, 279 and 164 showed reduced expression during drought stress (Table 1). Further expression pattern of lncRNAs explored from FPKM ratios (Control vs. Drought stressed) of RNA-Seq showed expression of 44 lncRNAs were found only under drought conditions. Of 44 new lncRNAs, 13 lncRNAs were induced in drought-tolerant cultivar and 31 lncRNAs were induced in drought-susceptible cultivars. No sequence similarity was found between any of the new lncRNAs of both cultivars. The magnitude of fold change in the expression of lncRNA under drought condition was observed between +8.11585 to −4.04311 and +4.27558 to −3.89842 in tolerant and susceptible cultivars, respectively. The Musa lncRNA expression pattern was almost similar to drought-responsive mRNAs expression in this study.
To investigate all other drought-regulated common lncRNAs between two cultivars, the sequence homology of lncRNAs was analyzed between these cultivars. None of the lncRNA sequences showed more than 95 % identity with at least 70 % query coverage and therefore considered as different lncRNAs. The dissimilarity between lncRNA sequences may be because lncRNAs from drought stress-responsive lncRNAs of ‘ABB’ genome containing tolerant cv. ‘Saba’ are different from lncRNAs of ‘AAA’ genome containing drought-susceptible cv. ‘Grand Naine’. Hence, the expression of drought-responsive banana lncRNAs is dependent of genomic constitution of cultivars.
Banana lncRNAs as microRNA targets
LncRNAs can either be targeted to nonsense-mediated mRNA decay pathway (Kalyna et al. 2012) or to play direct functional role as transcription regulator, although the exact function is yet to be known. Recent experimental evidences revealed that lncRNAs can act as microRNA targets or target mimics in plants (Wu et al. 2013). To explore the possibility of banana lncRNAs as target mimics of microRNA, all the lncRNAs were analyzed against 1581 miRNA sequences of model plant, A. thaliana (PNRD), according to Wu et al. (2012). Interestingly, 72 out of 905 drought-responsive Musa lncRNAs were predicted to be ‘decoys’ of 85 conserved miRNAs (Arabidopsis) (Supplementary data. 4). Seven of the lncRNAs predicted to be miRNA decoys were found in both cultivars under drought conditions. Expression of these decoys between drought-tolerant and -susceptible cultivars was different (Fig. 3). The corresponding microRNAs, miR854a, miRf10376-npr, miR156a, miRf10448-npr, miRf10448-npr, miRf10746-npr, and miR5658 have 433 authentic targets altogether. Some of the authentic targets are well characterized for their role in drought stress known as dehydration-responsive protein (ath-miR854a), arogenate dehydrogenase2 (ath-miR5658), squamosa promoter binding protein-like2 (ath-miR156a), and glutathione peroxidase (ath-miR5658).
Expression of target mimics/decoys commonly found in control and drought-stressed leaf samples of drought-tolerant banana cv. ‘Saba’ and susceptible cv. ‘Grand Naine’ from RNA-Seq data. Expression of almost all the microRNA decoys identified from tolerant cultivar was induced during drought stress except miRf10376-npr, while all miRNA decoy expression was reduced by drought stress in the susceptible cultivar
Expression of miRNA decoys of tolerant cultivar were induced during drought stress in comparison to its equivalent control, except decoy of miRf10376-npr, while all predicted miRNA decoys were down-regulated in drought-susceptible cultivars under drought stress in the present study. Induced expression of glutathione peroxidase (antioxidant enzyme), squamosa promoter binding protein-like (transcription factor), and dehydration-responsive protein during stress was shown to enhance plants stress tolerance (Zivkovic et al. 2010; Shinozaki and Yamaguchi-shinozaki 2007). Drought-induced expression of miR156 in tolerant cv. Saba (Muthusamy et al. 2014) was expected to disrupt the functional activity of authentic target squamosa promoter binding protein-like genes through its post-transcriptional regulation mechanism. However, squamosa promoter binding protein-like2 was observed to be induced in drought-tolerant plants, but not in susceptible plants (unpublished). In the present study, miR156a decoy lncRNA was up-regulated in tolerant plants and down-regulated in susceptible plants under drought conditions. Hence, we hypothesize that Musa lncRNA acts as a decoy for miR156a in the present study to regulate the positive expression of the Squamosa promoter binding-like 2 genes, despite the fact that the expression is also dependent on many other factors. Zhang et al. (2014) reported that two rice lncRNAs were involved in sequestering of rice miRNAs (OsmiR160 and OsmiR164). They also reported that the functional pair of lncRNA–miRNA is an important regulator of floral and seed development in rice. Similarly, the authentic targets such as dehydration-responsive protein, arogenate dehydrogenase2 and glutathione peroxidase were found to be induced in drought-tolerant cultivar. However, the functional relationships between the respective lncRNA–miRNA and positive regulation of the targets genes could not be established, since the expression pattern of miR854a, miR5658, and ath-miR5658 in banana leaves for drought stress is not available.
To conclude, 97.5 % of the Musa lncRNAs identified from drought-responsive transcripts of drought-tolerant and -susceptible banana cultivars showed significant differential expression to drought stress, of which 8.3, 7.9 % were identified as NATs and microRNA decoys. Most of the stress-responsive lncRNAs exhibit distinct expression pattern between cultivars depending upon genomic constitution of cultivars. This report will lay a basic platform in ruling out the drought-resistant regulatory pathways that will bring out successful crop improvement strategies.
References
Amor BB, Wirth S, Merchan F et al (2009) Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res 19:57–69. doi:10.1101/gr.080275.108.1
Boerner S, McGinnis KM (2012) Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS One 7:e43047. doi:10.1371/journal.pone.0043047
Chen D, Yuan C, Zhang J et al (2012) PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acid Res 40:D1187–D1193. doi:10.1093/nar/gkr823
D’Hont A, Denoeud F, Aury J et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488(7410):213–217. doi:10.1038/nature11241
Foissac S, Sammeth M (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acid Res 35:W297–W299. doi:10.1093/nar/gkm311
Griffiths-Jones S (2003) Rfam: an RNA family database. Nucleic Acid Res 31:439–441. doi:10.1093/nar/gkg006
Hao Z, Fan C, Cheng T et al (2015) Genome-wide identification, characterization and evolutionary analysis of long intergenic noncoding RNAs in cucumber. PLoS One 10:e0121800. doi:10.1371/journal.pone.0121800
Henz SR, Cumbie JS, Kasschau KD et al (2007) Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol 144:1247–1255. doi:10.1104/pp.107.100396
Kalyna M, Simpson CG, Syed NH et al (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acid Res 40:2454–2469. doi:10.1093/nar/gkr932
Kong L, Zhang Y, Ye ZQ et al (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acid Res 35:W345–W349. doi:10.1093/nar/gkm391
Kornienko AE, Guenzl PM, Barlow DP, Pauler FM (2013) Gene regulation by the act of long non-coding RNA transcription. BMC Biol 11:59. doi:10.1186/1741-7007-11-59
Liu J, Jung C, Xu J et al (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24:4333–4345. doi:10.1105/tpc.112.102855
Meng Y, Shao C, Wang H, Jin Y (2012) Target mimics: an embedded layer of microRNA-involved gene regulatory networks in plants. BMC Genomic 13:197. doi:10.1186/1471-2164-13-197
Min XJ (2013) ASFinder: a tool for genome-wide identification of alternatively splicing transcripts from EST-derived sequences. Int J Bioinform Res Appl 9:221–226. doi:10.1504/IJBRA.2013.053603
Muthusamy M, Uma S, Backiyarani S, Saraswathi MS et al (2014) Computational prediction, identification, and expression profiling of microRNAs in banana. J Hortic Sci Biotech 89:208–214
Osato N, Suzuki Y, Ikeo K, Gojobori T (2007) Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics 176:1299–1306. doi:10.1534/genetics.106.069484
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641. doi:10.1016/j.cell.2009.02.006
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acid Res 33:W686–W689. doi:10.1093/nar/gki366
Shinozaki K, Yamaguchi-shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227. doi:10.1093/jxb/erl164
Shuai P, Liang D, Tang S et al (2014) Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Exp Bot 65:4975–4983. doi:10.1093/jxb/eru256
Sun L, Zhang Z, Bailey TL et al (2012) Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinform 13:331. doi:10.1186/1471-2105-13-331
Surendar KK, Devi DD, Ravi I et al (2013) Water Stress in Banana—a Review. Bull Env Pharmacol Life Sci 2:1–18
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi:10.1093/bioinformatics/btp120
Van Asten PJA, Fermont AM, Taulya G (2010) Drought is a major yield loss factor for rainfed East African highland banana. Agric Water Manag 98:541–552. doi:10.1016/j.agwat.2010.10.005
Wang XJ, Gaasterland T, Chua NH (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6:R30. doi:10.1186/gb-2005-6-4-r30
Wang H, Chung PJ, Liu J et al (2014) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24:444–453. doi:10.1101/gr.165555.113.2006
Wu HJ, Ma YK, Chen T et al (2012) PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acid Res 40:W22–W28. doi:10.1093/nar/gks554
Wu HJ, Wang ZM, Wang M, Wang XJ (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161:1875–1884. doi:10.1104/pp.113.215962
Xin M, Wang Y, Yao Y et al (2011) Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol 11:61. doi:10.1186/1471-2229-11-61
Yi X, Zhang Z, Ling Y et al (2015) PNRD: a plant non-coding RNA database. Nucleic Acid Res 43:D982–D989. doi:10.1093/nar/gku1162
Zhang X, Xia J, Lii YE et al (2012) Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol 13:R20. doi:10.1186/gb-2012-13-3-r20
Zhang J, Mujahid H, Hou Y et al (2013) Plant long ncRNAs: a new frontier for gene regulatory control. Am J Plant Sci 04:1038–1045. doi:10.4236/ajps.2013.45128
Zhang YC, Liao JY, Li ZY et al (2014) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15:512. doi:10.1186/s13059-014-0512-1
Zheng H, Qiyan J, Zhiyong N, Hui Z (2013) Prediction and identification of natural antisense transcripts and their small RNAs in soybean (Glycine max). BMC Genomic 14:280. doi:10.1186/1471-2164-14-280
Zhu QH, Wang MB (2012) Molecular functions of long non-coding RNAs in plants. Genes (Basel) 3:176–190. doi:10.3390/genes3010176
Zivkovic S, Popovic M, Dragisic-Maksimovic J et al (2010) Dehydration-related changes of peroxidase and polyphenol oxidase activity in fronds of the resurrection fern Asplenium ceterach L. Arch Biol Sci 62:1071–1081. doi:10.2298/ABS1004071Z
Acknowledgments
This work was supported, in part, by the Global Crop Diversity Trust of the Food and Agriculture Organization (FAO), Rome, Italy, and the NPTC-Functional Genomics component of the Indian Council for Agricultural Research (ICAR), India. The authors thank the Director of the National Research Centre for Banana for providing technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11816_2015_363_MOESM2_ESM.xlsx
Supplementary data. 2 Musa drought-responsive, novel lncRNA expression and coding potential from RNA-Seq data (XLSX 136 kb)
Rights and permissions
About this article
Cite this article
Muthusamy, M., Uma, S., Backiyarani, S. et al. Genome-wide screening for novel, drought stress-responsive long non-coding RNAs in drought-stressed leaf transcriptome of drought-tolerant and -susceptible banana (Musa spp) cultivars using Illumina high-throughput sequencing. Plant Biotechnol Rep 9, 279–286 (2015). https://doi.org/10.1007/s11816-015-0363-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-015-0363-6