Abstract
Secondary cell walls account for the majority of total plant biomass and, as mostly composed of polysaccharides, constitute a promising source of fermentable sugars for the production of biofuels and biomaterials. However, the presence of the aromatic polymer lignin largely precludes the release of monosaccharides during enzymatic hydrolysis of cell wall polysaccharides in the biorefinery. Therefore, it is essential to unraveling the molecular mechanisms underlying lignin metabolism in order to better exploit the potential of lignocellulosic biomass. In the context of the bioeconomy, grasses emerge as a prominent lignocellulosic feedstock due to their high yield potential for biomass production. Still, many aspects of lignin metabolism in grasses, including transcriptional regulation, biosynthesis and polymerization, remain poorly understood, in contrast to eudicots species. Moreover, grasses differ considerably from eudicots in vascular patterning and cell wall composition, suggesting the presence of many grass-specific molecular and biochemical mechanisms that are not found in eudicots and whose knowledge cannot be extrapolated from data obtained with eudicot model plants. Here, we summarize the most recent advances on structural features of grass lignin and on functional characterization of genes directly involved in diverse aspects of lignin metabolism in grasses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grasses are one of the most agronomically and economically important groups of plants, providing the majority of calories that are directly or indirectly consumed by humans. Grass cell walls are not only a major source of nutrients and fibers but also have been proposed as a promising source of fermentable sugars for the production of second generation liquid fuels (Vogel 2008). In this regard, C4 grasses, especially those belonging to the Panicoideae subfamily such as sugarcane, Miscanthus and switchgrass, emerge as potential bioenergy crops due to their high yield potential for biomass production (Weijde et al. 2013). During plant growth, while the cell elongating process is still active, plant cells are surrounded by a primary cell wall, whose components are mainly polysaccharides. Although the overall architecture of eudicot and grass primary cell walls is similar (i.e., cellulose microfibers embedded in a matrix of non-cellulosic polysaccharides), they belong to different types of cell walls, according to the nature and abundance of cell wall components. Type I walls of eudicots and non-commelinid monocots are formed by a xyloglucan-cellulose network encased in a pectin-rich matrix with abundant structural proteins. Type II walls of commelinid monocots, which include grasses, have a glucuronoarabinoxylan-cellulose network embedded in a matrix with relatively low amounts of pectin and structural proteins, but with considerable amounts of cross-linked ferulate esters and other hydroxycinnamates (Carpita and McCann 2008). Once cell elongation is complete, a secondary wall is deposited internally to the primary wall of specialized cell types such as xylary vessels and schlerenchymatic fibers. In the stem of C4 grasses, secondary walls are also deposited in the epidermis, hypodermis and most of the parenchyma cells (Cesarino et al. 2012b). Secondary cell walls (SCW) are mostly composed of polysaccharides (i.e., cellulose and various hemicelluloses) impregnated and coated by the aromatic polymer lignin, which provides strength and hydrophobicity (Marriott et al. 2016). Noteworthy, the proportion and chemical composition of the major components of SCW also varies between eudicots and grasses. Because SCWs account for the majority of total plant biomass, and because grasses are one of the main sources of plant biomass for downstream applications in biorefineries, understanding the molecular mechanisms that control SCW deposition and composition in grasses is an important goal.
One of the major bottlenecks that hinders the efficient conversion of cell wall polysaccharides into fermentable sugars is the presence of the aromatic polymer lignin, which has been considered the most recalcitrant factor in plant biomass (Chen and Dixon 2007; Van Acker et al. 2013; Wilkerson et al. 2014). Lignin is a heterogeneous aromatic polymer mostly found in the walls of terminally differentiated cells of supportive and water-conducting tissues, providing mechanical reinforcement for the plant to stand upright and allowing long-distance water transport (Bonawitz and Chapple 2010). Lignin is made of monomers derived from the phenylpropanoid pathway, whose primary precursor is the aromatic amino acid phenylalanine. Following the deamination of phenylalanine by the entry point enzyme phenylalanine ammonia lyase (PAL), the resulting product cinnamic acid undergoes a series of aromatic ring and propane tail modifications to yield three hydroxycinnamyl alcohols, p-coumaryl, coniferyl, and sinapyl alcohols, differing in their degree of methoxylation (Liu 2012). Once incorporated into the polymer, these monolignols produce p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) units, respectively. In addition to the canonical monolignols, other less abundant units might be incorporated at varying levels, such as hydroxycinnamyl aldehydes, hydroxycinnamyl p-coumarates, and hydroxycinnamate esters (Ralph et al. 2004; Vanholme et al. 2012). These monomers are first synthesized in the cytoplasm and further transported to the apoplast, where they are oxidized by peroxidases and/or laccases for subsequent polymerization through combinatorial radical-radical coupling (Wang et al. 2013). Because lignin polymerization is combinatorial and not controlled by proteins, coupling reactions are thought to be solely under chemical control, being affected by typical physical parameters, among others pH, ionic strength and co-factor supply (Vanholme et al. 2008; Wang et al. 2013; Dima et al. 2015). Finally, the whole lignification program is not only developmentally regulated but also responds to specific environmental conditions, such as biotic and abiotic stresses (Moura et al. 2010; Barros et al. 2015).
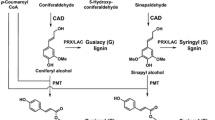
Despite intensively characterized over the past few years, lignin metabolism still yields unexpected discoveries. Here, we summarize the most recent findings on structural aspects of grass lignin and on functional characterization of genes involved in different aspects of lignin metabolism (Fig. 1), including the transcriptional regulation of lignin deposition.
Schematic representation of the phenylpropanoid pathway showing the most recent discoveries on side pathway reactions and gene functions in grasses, which are colored. The arrow representing the conversion of caffeoyl shikimate to caffeic acid is dashed because CSE function was not confirmed in grasses by means of reverse genetics. PAL phenylalanine ammonia-lyase, PTAL bifunctional phenylalanine tyrosine ammonia-lyase, C4H cinnamate 4-hydroxylase, 4CL 4-coumarate:CoA ligase, C3H p-coumarate 3-hydroxylase, HCT p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase, CSE caffeoyl shikimate esterase, CCoAOMT caffeoyl-CoA O-methyltransferase, CCR cinnamoyl-CoA reductase, CAD cinnamyl alcohol dehydrogenase, COMT caffeic acid O-methyltransferase, F5H ferulate 5-hydroxylase, PMT p-coumaroyl-CoA:monolignol transferase, CHS chalcone synthase, CHI chalcone isomerase, F3′H flavonoid 3′-hydroxylase, OMT O-methyl transferase, FPGS folylpolyglutamate synthase, MTHFR methylenetetrahydrofolate reductase, PRX peroxidase, LAC laccase
New findings on structural aspects of grass lignin
In a major recent finding, the flavonoid tricin has been implicated as the first lignin monomer from outside the monolignol biosynthetic pathway, specifically in monocot lignins. Tricin [5,7-dihydroxy-2-(4-hydroxy-3,5-dimethoxyphenyl)-4H-chromen-4-one] is a member of the flavone subclass of flavonoid compounds and is widely distributed in herbaceous plants, where it is found either in free or conjugated forms such as tricin-glycosides and tricin lignans (Li et al. 2016). Incorporation of tricin into lignin was first described by Del Río et al. (2012a, b), who characterized the lignin structure in wheat straw by a combination of analytical pyrolysis, 2D nuclear magnetic resonance spectroscopy (2D-NMR) and derivatization followed by reductive cleavage (DFRC). Subsequently, the presence of tricin in lignin fractions was also verified in a variety of other monocot plants such as maize (Lan et al. 2015, 2016a), sugarcane (Del Río et al. 2015), coconut coir fibers (Rencoret et al. 2013), Arundo donax (You et al. 2013) and bamboo (Wen et al. 2013). Recently, a quantitative method based on thioacidolysis followed by LC–MS characterization was developed and applied to evaluate the occurrence and content of lignin-integrated tricin in various seed-plant species (Lan et al. 2016b). This study revealed relatively high amounts of lignin-integrated tricin in species of the family Poaceae, with the highest amount of integrated tricin in members of the subfamily Pooideae such as oat (33.11 mg/g on lignin basis), wheat (32.72 mg/g on lignin basis) and Brachypodium distachyon (28.01 mg/g on lignin basis). Interestingly, tricin incorporation into lignin is wider than previously suggested, since tricin was also found in the lignin of monocots outside the order Poales (e.g., order Arecales, family Arecaceae), of non-commelinid monocots (e.g., order Asparagales, family Orchidaceae) and even in one species of eudicots (Medicago sativa, family Fabaceae) (Lan et al. 2016b). Additionally, by using a biomimetic system with peroxidase/hydrogen peroxide, Lan et al. (2015) demonstrated that tricin cross couples with monolignols to form tricin-(4´-O-β)-linked dimers. Moreover, analysis of gel permeation chromatography-fractionated acetylated maize lignin using NMR revealed tricin incorporation into high molecular weight fractions, demonstrating that tricin is fully compatible with lignification in vivo (Lan et al. 2015, 2016a). These experiments unambiguously confirmed that tricin is an authentic lignin monomer in monocots. Another important conclusion from this study was that, as tricin apparently incorporates into lignin exclusively via 4´-O-β-coupling, it would necessarily localize at one terminus of the lignin chain and, thus, it functions as a nucleation site for lignification in monocots (Lan et al. 2015). Previously, a role as nucleation site had been suggested for ferulic acid (Ralph et al. 1995), which is esterified to arabinosyl residues of arabinoxylan chains and later cross-linked to G units via ether bonds (Riboulet et al. 2009). These findings finally helped resolving the dilemma that, differently from eudicots with similar syringyl-guaiacyl compositions, monocot lignin chains do not appear to be initiated via monolignol homodehydrodimerization (Lan et al. 2015).
Another remarkable feature of grass lignins is the significant level of monolignol p-coumaroylation, whose levels can reach up to 17% of total isolated lignin by weight in mature maize stem (Ralph et al. 1994). It is reasonably accepted that monolignols are enzymatically acylated inside the cell through esterification of p-coumaric acid to the monolignol γ-position, and further transported to the cell wall, where the p-coumarate ester conjugates undergo radical coupling and cross-coupling reactions (Ralph 2010). It has been suggested that p-coumaroylation of sinapyl alcohol is important to allow the radicalization of these monolignols via peroxidase that otherwise would oxidize those substrates more slowly (Hatfield et al. 2008; Ralph 2010). Whereas monolignol acylation by p-coumarates is characteristic to both C3 and C4 grasses, other naturally acylated lignins are found in nature, such as p-hydroxybenzoylation in palms and Populus species (Meyermans et al. 2000; Lu et al. 2015) and acetylation in palms, kenaf, sisal and abaca (Del Río et al. 2007). Recently, low levels of lignin γ-acylation with acetate groups have also been found in grass species such as bamboo, wheat straw, elephant grass and sugarcane (Del Río et al. 2007, 2012a, b, 2015). Interestingly, while lignin γ-acetylation occurs predominantly on S units in most plants, γ-acetylation was mainly found on G units in the lignin of grasses. These findings suggest that monolignol acetylation in grasses is performed by so far uncharacterized acetyl transferases with a higher affinity towards coniferyl alcohol (Del Río et al. 2015).
Newly characterized genes involved in many aspects of lignin metabolism in grasses
Biosynthetic genes
The phenylpropanoid pathway starts with the deamination of phenylalanine by the entry point enzyme phenylalanine ammonia lyase (PAL), resulting in the production of trans-cinnamic acid, which is subsequently converted into p-coumaric acid via the para-hydroxylation of the aromatic ring by cinnamate-4-hydroxylase (C4H). Monocots harbor a subset of bifunctional PAL proteins with additional tyrosine ammonia lyase (TAL) activity, enabling the direct conversion of tyrosine into p-coumaric acid and, thus, bypassing the reaction catalyzed by C4H (Rosler et al. 1997; Watts et al. 2006). Substrate specificity of such enzymes is determined by a single residue position: PAL proteins having phenylalanine at position 123 (Phe123) specifically utilize phenylalanine as substrate, while PAL proteins harboring a histidine residue at this position (His123) utilize both phenylalanine and tyrosine (Watts et al. 2006). In Brachypodium, PAL isoforms were targeted by RNA interference and large reductions in transcript abundance for two of the eight putative BdPAL genes were identified in stem tissues, one of them, namely BdPAL1, was predicted to have TAL activity based on the presence of His130 (Cass et al. 2015). Accordingly, stem extracts of BdPAL RNAi lines targeting multiple BdPAL genes showed reductions in both PAL and TAL activities compared with the WT, suggesting that at least one of Brachypodium BdPAL (likely BdPAL1) is a bifunctional PTAL protein with the ability to deaminate both phenylalanine and tyrosine (Cass et al. 2015). However, the in planta role of PTAL has not been unambiguously demonstrated in any plant species until recently when Barros et al. (2016) identified a single homotetrameric bifunctional ammonia-lyase (PTAL) among eight BdPAL enzymes in Brachypodium. BdPTAL1 down-regulation and 13C isotope labelling experiments showed that the TAL activity of BdPTAL1 contributes to nearly half of the total lignin deposited in Brachypodium, with a preference for S-lignin and wall-bound coumarate biosynthesis. Furthermore, isotope dilution experiments suggest that lignin biosynthesis via l-phenylalanine is distinct from that via l-tyrosine, supporting the existence of parallel pathways due to the organization of lignin biosynthetic enzymes in different metabolons (Barros et al. 2016).
The biochemical production pathway leading to the canonical monolignols was thought to be fully defined over a decade ago. Surprisingly, caffeoyl shikimate esterase (CSE), an enzyme central to the lignin biosynthetic pathway, was recently identified and shown to catalyze the conversion of caffeoyl shikimate into caffeate in Arabidopsis (Vanholme et al. 2013; Vargas et al. 2016). The activity of CSE combined with that of p-coumaroyl/caffeoyl-CoA ligase (4CL) produces caffeoyl-CoA, bypassing the second reaction of HCT. CSE orthologues have been widely found in other plant species, suggesting that this enzymatic step is conserved within the plant lineage. Nevertheless, grass genomes seem to largely lack a functional orthologue of Arabidopsis CSE: while putative orthologues were found in the genome of switchgrass (Panicum virgatum) and rice, they were absent in important bioenergy crops such as maize, sorghum and sugarcane (Vanholme et al. 2013; Vicentini et al. 2015). Although the expression of putative switchgrass CSE genes was down-regulated during lignification of suspension cell cultures (Shen et al. 2013), evidence for the involvement of CSE in lignin biosynthesis in switchgrass was recently reported. While studying early lignin pathway enzymes in switchgrass, Escamilla-Treviño et al. (2013) found that recombinant PvHCTs were largely inefficient in converting caffeoyl shikimate into caffeoyl-CoA in the presence of CoA. In addition, stem protein extracts were not able to carry out the same reaction but, instead, hydrolyzed the shikimate ester to generate caffeic acid. CSE activity in crude protein extracts of switchgrass stem tissues was further confirmed in a subsequent study of the same group, while only weak esterase activity was observed for protein extracts of Brachypodium and maize (Ha et al. 2016). Altogether, these results suggest the involvement of CSE in the shikimate shunt during lignin biosynthesis in switchgrass, while the classical dual role of HCT is unlikely to occur. Nevertheless, the step catalyzed by CSE may not be essential for lignification in all grasses.
Tricin is a flavonoid compound recently shown to be an authentic lignin monomer in monocots. Although this molecule has been extensively studied due to its benefits to human health, the biochemical pathway leading to tricin biosynthesis in monocots remained largely unknown until recently. The flux towards flavonoids starts with the activity of chalcone synthase (CHS), which catalyzes the condensation of three molecules of malonyl-CoA with p-coumaroyl-CoA to produce naringenin chalcone, which is then isomerized by chalcone isomerase (CHI) to naringenin, the common precursor for all flavonoids (Tohge et al. 2013). From naringenin, the 3′,5′-dimethoxyflavone nucleus is produced before O-linked conjugations. Previously, the proposed pathway towards tricin would involve the conversion of apigenin into tricetin by the activity of a flavonoid 3′,5′-hydroxylase (F3′5′H), followed by the sequential O-methylation of tricetin, which has been demonstrated only in vitro (Zhou et al. 2006). However, the fact that endogenous F3′5′H genes essential for tricin biosynthesis remained unidentified in grasses and that the occurrence of tricetin in plants is scarce suggested the existence of a different pathway. Recently, the biosynthetic route towards tricin was finally resolved in rice in two consecutive papers from the same research group. First, a bona fide flavone synthase II (FNSII) that catalyzes the direct conversion of flavanones to flavones was identified (Lam et al. 2014). This enzyme, cytochrome P450 93G1, desaturates naringenin into apigenin, which is further 3′-hydroxylated to generate luteolin via flavonoid 3′-hydroxylase (F3′H/CYP75B4) activity. This hydroxyl group is then methylated by an O-methyl transferase (OMT) to form chrysoeriol. In a subsequent paper, a chrysoeriol 5′-hydroxylase was identified and shown to generate selgin, the immediate precursor of tricin, through 5′-hydroxylation of chrysoeriol (Lam et al. 2015). Finally, selgin is methylated at the 5′-position to form tricin. This study has shown that chrysoeriol, and not tricetin, is an intermediate for the biosynthesis of tricin in vivo, and suggests that the recruitment of chrysoeriol 5′-hydroxylase activity by the ancestor of grasses was a key evolutionary step leading to the prevalence of tricin and its derivatives in this group of plants (Lam et al. 2015).
Monolignol acylation genes
Until recently, the enzyme responsible for the biosynthesis of the p-coumaroylated monolignols in grasses remained unknown. In 2012, Withers and colleagues reported the identification of a grass-specific enzyme from rice that acylates monolignols with p-coumarate in vitro (Withers et al. 2012). The enzyme, named p-coumaroyl-CoA:monolignol transferase (OsPMT), is part of the BAHD acyltransferase family and was identified through a co-expression analysis using genes involved in monolignol biosynthesis. The recombinant enzyme was shown to catalyze the transesterification between monolignols, especially sinapyl and p-coumaroyl alcohols, and p-coumaroyl-CoA, producing monolignol p-coumarates (Withers et al. 2012). Two years later, two independent groups provided genetic evidence that confirmed the role of PMT in grass lignification. Petrik et al. (Petrik et al. 2014) reported the identification of an orthologue of OsPMT in Brachypodium and its functional characterization in planta through the generation of RNAi-BdPMT and overexpression plants. Brachypodium plants with reduced BdPMT expression had levels of p-coumarate on lignin as low as 10% of that of the wild type, while the amount of p-coumarate esterified to arabinosyl units on arabinoxylans remained unchanged. Conversely, BdPMT-overexpressing plants showed increased levels of lignin p-coumaroylation, but again the levels of p-coumarate on arabinosyl units remained unchanged. In a complementary work, a hydroxycinnamyl transferase that couples p-coumarate to monolignols (pCAT) was identified through a proteomics approach and subsequently down-regulated in maize (Marita et al. 2014). By analyzing isolated cell walls of mature stems and leaves, it was shown that lignin content was not altered in pCAT-RNAi plants but the levels of p-coumarate were decreased and lignin composition was altered. Maize plants with the lowest levels of p-coumarate also showed decreased incorporation of syringyl units. Altogether, these results suggest that BAHD acyltransferases specifically acylate monolignols with p-coumarate but do not work on hemicelluloses, and that modification of PMT expression might affect lignin structure and biomass properties.
Brown midrib genes
The brown midrib constitutes a group of cell wall mutants isolated in maize (bm mutants) and sorghum (bmr mutants, to differentiate from bloomless mutants) by spontaneous and chemical mutagenesis. This nomenclature was given due to the characteristic reddish-brown coloration of the vascular tissue in the leaves and stems associated with altered lignification (Sattler et al. 2010). Until recently, two allelic classes of maize bm mutants and three classes of sorghum bmr mutants have been identified and mapped: bm1 and bm3, encoding cinnamyl alcohol dehydrogenase (CAD) and a caffeic O-methyl transferase (COMT), respectively, and bmr2, bmr6 and bmr12, encoding 4-coumarate: coenzyme A ligase (4CL), CAD and COMT, respectively (Vignols et al. 1995; Halpin et al. 1998; Bout and Vermerris 2003; Saballos et al. 2009, 2012). This group of mutants has been extensively studied and has largely contributed to the understanding of lignin metabolism and the molecular basis of lignocellulose recalcitrance in grasses. Apart from helping in the identification of lignin biosynthetic genes, brown midrib mutants hold promise of identifying new sets of genes that might be either involved in different aspects of lignin metabolism (e.g., transcriptional regulation, polymerization) or might be indirectly involved in lignin biosynthesis. Accordingly, recent mapping and molecular characterization of bm2 and bm4 mutants in maize are good examples of the later and highlights the great potential of brown midrib mutants as tools for gene discovery and pathway characterization. The bm2 gene was mapped to a region of chromosome 1 containing a putative methylenetetrahydrofolate reductase (MTHFR) gene, whose expression levels were significantly low in bm2 plants (Tang et al. 2014). This gene was shown to encode a functional MTHFR based on the complementation of a yeast strain that is unable to convert 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, a co-substrate for homocysteine re-methylation to methionine. Thus, it was expected that MTHFR plays a role in the production of the methyl donor S-adenosyl-l-methionine (SAM) and that affecting its function in planta might influence the availability of this substrate for the methyltransferases involved in lignin biosynthesis (i.e., CCoAOMT and COMT). This enzymatic function is consistent with bm2 lignin phenotype, with significant reduced levels and altered composition compared to the wild type (Tang et al. 2014). Interestingly, the bm4 mutant is affected in a different step of the same biochemical route as the bm2 (MTHFR) gene, and the same research group had reported its functional characterization. The bm4 gene encodes a putative folylpolyglutamate synthase (FPGS), which functions in C1 metabolism to polyglutamylate substrates of folate-dependent enzymes and is ultimately involved in the production of SAM (Li et al. 2015). Mutation in bm4 leads to a modest decrease in lignin content and an overall increase in the S/G ratio when compared to the wild type, which illustrates the importance of FPGS in lignin biosynthesis (Li et al. 2015). The recent advances in DNA sequencing tools and molecular biology will allow further exploitation of brown midrib mutants produced through chemical mutagenesis, providing new resources for basic research and for breeding.
Genes involved in lignin polymerization
Monomer oxidation (i.e., the formation of radicals) is an essential step for the deposition of lignin in plant cell walls. Two types of enzymes have been classically implicated in lignin polymerization, class III peroxidases and laccases, based on their localization in lignification sites and on their catalytic capacity to oxidize lignin precursors in vitro (Lewis and Yamamoto 1990; Cesarino et al. 2012a). However, since both types of enzymes belong to large multigene families with functionally redundant members and broad in vitro substrate specificity, unambiguous determination of the involvement of an individual peroxidase or laccase in lignification is still a major challenge. In grasses, there were no reports, to our knowledge, on natural mutants or transgenic plants affected in a peroxidase or a laccase gene until very recently. By combining co-expression analysis, tissue-specific expression analysis and a genetic complementation approach, Cesarino et al. (2013) correlated a laccase gene, SofLAC, to the lignification process in sugarcane. SofLAC, which is the putative orthologue of the Arabidopsis AtLAC17 gene, was identified as a candidate gene because it was coordinately expressed with several sugarcane homologues of phenylpropanoid genes in a co-expression network constructed with sugarcane cDNA libraries. In situ hybridization analysis demonstrated that SofLAC mRNA was localized in the highly lignified cell types of sugarcane stems, mainly in inner and outer portions of schlerenchymatic bundle sheaths. Finally, the expression of SofLAC under the control of the Arabidopsis AtLAC17 promoter restored the lignin content but not the lignin composition of the Arabidopsis lac17 mutant, suggesting that SofLAC is likely to play a role in lignin polymerization in sugarcane (Cesarino et al. 2013). Similarly, a recent report provided evidence for the participation of laccases in the lignification of B. distachyon (Wang et al. 2015b). Experiments of in situ hybridization and immunolocalization showed that two Brachypodium laccases, LACCASE5 and LACCASE6, which are the putative orthologues of Arabidopsis AtLAC17 and AtLAC4, respectively, are expressed in lignifying tissues. A mutant for LACCASE5, identified in a target-induced local lesion in genome mutant collection, showed 10% less lignin, a slight increase in the levels of S units and a significant increase in ferulic acid ester linked to the mutant cell walls. Interestingly, no effects on lignin metabolism were observed in a mutant for LACCASE6. These results confirm that at least LACCASE5 is involved in monolignol oxidation and lignin polymerization in the model grass B. distachyon (Wang et al. 2015b). The identification and functional characterization of plant laccases is important because the manipulation of such genes has been long considered a potential strategy to reduce plant biomass recalcitrance through genetic engineering (Wang et al. 2015a).
Transcriptional regulation of lignin deposition in grasses
The regulatory mechanisms controlling lignin deposition are embedded within the complex transcriptional network controlling the whole program of SCW deposition, which employs a hierarchical feed-forward loop regulatory structure (Zhong and Ye 2014; Taylor-Teeples et al. 2015). On the top level, secondary wall NAC (NAM, ATAF1/2 and CUC2) master switches directly activate the expression of second level secondary wall MYB master switches and, together, both activate the bottom level of TFs (most of them from the MYB family) that are responsible for the coordinated expression of biosynthetic genes for SCW components. In addition, some biosynthetic genes are also directly activated by the NAC and MYB master switches, which bind to their target gene promoters via SNBE (Secondary Wall NAC Binding Element) and SMRE (Secondary Wall MYB-Responsive Element) sites, respectively (Zhong et al. 2010; Zhong and Ye 2012). In Arabidopsis, different members of NAC master switches perform the same function triggering SCW formation but in different SCW-containing cell types. VASCULAR-RELATED NAC-DOMAIN (VND) genes, VND1 to VND7, play a redundant role in regulating SCW biosynthesis in vessels whereas NAC SECONDARY WALL THICKENING PROMOTING FACTOR (NST) genes regulate SCW in xylary and interfascicular fibers (NST1, NST2 and NST3) and in the anther endothecium (NST1 and NST2 only). Importantly, despite their tissue-specific expression pattern, they activate similar sets of downstream genes (Zhong et al. 2006; Mitsuda et al. 2007; Yamaguchi et al. 2008; Zhou et al. 2014).
The functional redundant pair composed of MYB46 and MYB83 is a direct target of secondary wall NACs and constitute the second level of molecular switches that trigger the entire SCW biosynthetic program (Zhong et al. 2007; McCarthy et al. 2009). Both levels of master switches (NACs and MYBs) activate a battery of downstream TFs, including lignin-specific transcriptional activators and repressors (Zhong et al. 2008; Yamaguchi et al. 2011; Zhong and Ye 2012, 2014). However, very few TFs have been characterized to specifically regulate the biosynthesis of one of the SCW components. For instance, only three TFs have been characterized as lignin biosynthesis activators in Arabidopsis, namely MYB58, MYB63 and MYB85, while MYB4 is a repressor of C4H, inhibiting the biosynthesis of phenylpropanoid-derived compounds (Jin 2000; Zhong et al. 2008; Zhou et al. 2009). Functional characterization of their corresponding orthologues in other plant species such as poplar (Zhong et al. 2011b), pine (Bomal et al. 2008), maize (Fornalé et al. 2010) and switchgrass (Shen et al. 2012) suggest that their role in lignin biosynthesis regulation is conserved among different taxa. Finally, other downstream TFs such as KNAT7, SND2, SND3, MYB52, MYB54, MYB69 and MYB103 are likely involved in fine-tuning the transcriptional regulation of SCW deposition (Zhong and Ye 2015), working either as repressors (KNAT7) or as activators (all the others), but their putative orthologues in grass species have not been identified or functionally characterized so far.
Although extensively characterized over the past few years, the current model of the regulatory network controlling SCW deposition, and consequently also lignin deposition, is established predominantly for the eudicot model plant Arabidopsis (Fig. 2). Grasses have distinct vasculature patterning and composition of SCW from that of eudicots, suggesting a different transcriptional regulation (Handakumbura and Hazen 2012; Gray et al. 2012). Putative orthologues of several eudicot SCW regulators have been identified in the genome of grasses and gene expression profiling has helped identifying putative candidate regulators implicated in SCW deposition in some grasses. The most recent and significant results in the field are summarized here (Fig. 2).
Transcriptional regulatory networks controlling secondary cell wall (and lignin) deposition in Arabidopsis and grasses. Hierarchical levels are highlighted with different colors. Orthologous transcription factors between Arabidopsis and grasses are denoted by the same color. In Arabidopsis, NAC (NAM, ATAF1/2 and CUC2) master switches activate the entire secondary cell wall formation program in a cell-type-specific manner, while in grasses cell-type specificity of NAC activity is unknown. Regulatory factors upstream of NAC master switches in grasses remain elusive
Until recently, no transcription factor has been functionally characterized as a master switch able to activate the biosynthesis of all major components of SCW in grasses. In this regard, a first attempt was to use a heterologous system to evaluate the function of a group of rice and maize NAC and MYB transcription factors in the regulation of SCW deposition (Zhong et al. 2011a). The rice and maize NACs, named Secondary Wall-associated NACs OsSWNs and ZmSWNs, successfully complemented the Arabidopsis snd1 nst1 double mutant, which is defective in SCW thickening, and led to the ectopic deposition of cellulose, xylan and lignin when overexpressed in the wild-type background. In addition, overexpression of OsMYB46 and ZmMYB46 in Arabidopsis also led to the activation of the entire SCW biosynthetic program, suggesting that they are functional orthologues of Arabidopsis MYB46/MYB83. Furthermore, it was shown that OsSWNs and ZmSWNs bind to SNBEs present in the promoters of OsMYB46 and ZmMYB46 to activate their expression and, consequently, the expression of downstream genes involved in the SCW deposition (Zhong et al. 2011a). The authors concluded that the rice and maize SWNs and MYB46 s are master transcriptional activators involved in the regulation of SCW biosynthesis (Zhong et al. 2011a). Recently, the same research group reported the identification and functional characterization of a group of switchgrass master switches, including several NACs (PvSWNs) and one MYB (PvMYB46A), for their involvement in regulating SCW biosynthesis (Zhong et al. 2015).
A phylogenetic analysis of NAC transcription factors in Brachypodium revealed the presence of eight SWN genes in the genome: six orthologues of Arabidopsis VND genes and two orthologues of SND/NST genes (Valdivia et al. 2013). All eight Brachypodium SWN genes were able to induce ectopic SCW deposition when transiently expressed in tobacco leaves, but only the transient expression of VND orthologues resulted in extensive cell death. Although it has been shown that a number of programmed cell death-related genes are direct targets of both VNDs and SNDs/NSTs (Zhong et al. 2010), these transcription factors control significantly different types of cell death: while VNDs trigger a fast cell death program in vessels, a much slower process regulated by SNDs/NSTs occurs in fibers (Courtois-Moreau et al. 2009; Zhong et al. 2010). Overexpression of BdSWN5, the orthologue of Arabidopsis VND7, in Brachypodium using an oestradiol-inducible system resulted in ectopic SCW formation in both roots and shoots as well as in the up-regulation of SCW-related genes such as cellulose synthase (BdCESA4), a xylem-specific protease (BdXCP1) and an orthologue of AtMYB46 (BdMYB1). Moreover, it has been shown that BdSWN5 is capable of transactivating the BdXCP1 promoter in a process mediated by SNBEs (Valdivia et al. 2013). Altogether, these results suggest that the SCW biosynthetic program in grasses is also controlled by NAC master switches acting as transcriptional activators that bind to conserved SNBEs on the promoter of downstream target genes.
In comparison to NAC and MYB master switches, a higher number of downstream transcription factors that directly target the expression of SCW biosynthetic genes have been characterized in different species of grasses. The role of maize R2R3-MYB transcription factor ZmMYB31 in the regulation of the phenylpropanoid pathway was investigated through overexpression in Arabidopsis (Fornalé et al. 2010). Several genes involved in monolignol biosynthesis were down-regulated and transgenic plants showed a severe growth phenotype and reduced lignin content with unaltered polymer composition. ZmMYB31 also repressed the synthesis of sinapoylmalate and indirectly redirected the carbon flux towards the production of anthocyanins, since it also represses lignin biosynthesis (Fornalé et al. 2010). In addition, chromatin immunoprecipitation (ChIP) demonstrated that ZmMYB31 interacts with the promoters of two lignin genes in vivo, ZmCOMT and ZmF5H, while the consensus DNA-binding sequence was determined in vitro as ACCT/AACC. Therefore, ZmMYB31 seems to be a transcriptional repressor of the phenylpropanoid pathway and is likely to play an important role in carbon partitioning along this pathway. Noteworthy, the ZmMYB31 paralogue in maize, ZmMYB42, was shown to also repress the phenylpropanoid pathway and affect the cell wall structure and composition when overexpressed in Arabidopsis (Sonbol et al. 2009). Moreover, the regulatory function of MYB31 and MYB42 as negative regulators of the phenylpropanoid pathway is conserved also in sorghum and rice (Agarwal et al. 2016). However, the patterns of TF binding for both regulatory (i.e., binding to the promoter of downstream target genes) and autoregulatory (i.e., binding to their own promoters) roles were shown to be species-specific, indicating a potential subfunctionalization following divergence of maize, sorghum and rice (Agarwal et al. 2016). These findings suggest that the molecular mechanisms controlling lignin deposition might significantly vary even among closely related plant species.
The Arabidopsis R2R3 MYB transcription factor AtMYB4 was shown to be a repressor of C4H and consequently to inhibit the biosynthesis of phenylpropanoid-derived compounds (Jin 2000). Recent data on the characterization of switchgrass PvMYB4 suggests that it is a functional orthologue of the Arabidopsis AtMYB4 gene (Shen et al. 2012). PvMYB4 was shown to bind to AC elements present in the promoter region of lignin biosynthetic genes leading to transcriptional repression of such targets. Overexpression of PvMYB4 in tobacco and switchgrass led to similar phenotypes, including reduced stature, reduced total phenolic content, lower abundance of ester-linked wall-bound p-coumaric acid and lower overall lignin content. Furthermore, PvMYB4 overexpression in switchgrass resulted in a strong down-regulation of several monolignol biosynthetic genes without affecting the expression levels of genes involved in flavonoid biosynthesis (Shen et al. 2012). These results, together with the fact that PvMYB4 is highly expressed in vascular bundles, suggest that PvMYB4 functions as a repressor of the lignin biosynthetic pathway in switchgrass.
In contrast to ZmMYB31/ZmMYB42 and PvMYB4, which are transcriptional repressors of the phenylpropanoid pathway, SbMYB60 was recently identified as a positive regulator of lignin biosynthesis in Sorghum bicolor (Scully et al. 2016). This R2R3 MYB transcription factor is the co-orthologue of AtMYB58 and AtMYB63, a functional redundant pair of transcriptional activators of the lignin biosynthetic pathway in Arabidopsis (Zhou et al. 2009). Overexpression of SbMYB60 in sorghum resulted in the up-regulation of genes involved in monolignol biosynthesis and accumulation of monolignol biosynthetic proteins, which consequently led to increased lignin content without affecting the levels of cell wall polysaccharides. Moreover, higher abundance of soluble monolignol intermediates such as hydroxycinnamates was found in both stalks and leaves of transgenic plants. Constitutive expression of SbMYB60 also resulted in ectopic lignification in leaf midribs (Scully et al. 2016). Thus, despite the low amino acid identity (41%) between SbMYB60 and its Arabidopsis co-orthologues, these regulators of the monolignol biosynthetic pathway are conserved between monocots and eudicots, although some differences might be expected due to differences in vascular patterning and cell wall type between these groups of plants. Interestingly, a rice MYB transcription factor, OsMYB58/63 was shown to up-regulate the expression of the SCW-related cellulose synthase gene, cellulose synthase A7 (OsCesA7) and specifically controls the deposition of cellulose, in contrast to its putative orthologues in Arabidopsis (AtMYB58 and AtMYB63) and sorghum (SbMYB60) (Noda et al. 2015). This observation highlights the importance of further evaluation of the contribution and specific role of distinct transcription factors in the complex transcriptional network controlling SCW deposition in grasses.
Future perspectives
Secondary cell walls account for the majority of plant biomass, which constitute a renewable resource for the production of chemicals, materials and fuels. Lignin is considered the most recalcitrant factor affecting plant biomass digestibility, due to its ability to immobilizing hydrolytic enzymes and blocking their access to cell wall polysaccharides. Hence, it is essential to unravel the molecular mechanisms underlying lignin metabolism in order to better exploit the potential of lignocellulosic biomass. Noteworthy, lignin itself is a target product in biorefineries as there is an increasing interest in converting lignin into high-value products such as carbon fiber, plastics, polymeric foams and a variety of fuels and chemicals (Ragauskas et al. 2014). Despite the recent advances in our understanding of the molecular mechanisms involved in the regulation and biosynthesis of lignin in grasses, this knowledge is still fragmentary, especially when compared to that of eudicot plants. Noteworthy, grasses differ considerably from eudicots in vascular patterning and cell wall composition, suggesting the presence of many grass-specific molecular mechanisms that are not found in eudicots and whose knowledge cannot be extrapolated from data obtained with eudicot model plants. However, a major technical bottleneck still hampers the proper characterization of molecular aspects of SCW deposition and lignin metabolism in grasses: efficient genetic transformation protocols for a number of grass species. Although protocols for stable genetic transformation of major grass crops are available, the process is highly genotype-specific and typically presents very low transformation efficiency. In this regard, a recent report described a new approach to boost monocot transformation rates in a broad range of genotypes from different grass species. By using the overexpression of maize morphogenic genes Baby boom (Bbm) and Wuschel2 (Wus2) in the transformation construct, Lowe et al. (2016) were able to create a biological context conducive to transformation efficiency, enhancing the recovery of transgenic calli in recalcitrant varieties of maize, sorghum, rice and sugarcane. Until recently, there was no model system with an efficient, reproducible genetic transformation protocol to validate gene function in vivo within C4 grasses, which include the major bioenergy crops such as maize, sugarcane, sorghum, switchgrass and Miscanthus. In this regard, Setaria viridis has been proposed as an alternative C4 grass model, with a highly efficient Agrobacterium-mediated transformation protocol recently published, with transformation efficiency of up to 29% (Martins et al. 2015b). Moreover, cost-effective and time-saving spike-dip methods that bypass the laborious tissue culture procedures were also developed (Martins et al. 2015a; Saha and Blumwald 2016). Altogether, these technical advances open new opportunities to further evaluate the function of candidate genes in many aspects of SCW and lignin metabolism in C4 grasses.
Regarding the biology of grass lignin, many questions remain unanswered. For instance, with the recent identification of CSE as a central enzyme to the lignin biosynthetic pathway in Arabidopsis, it would be interesting to determine how caffeate esters are channeled into the production of G and S lignin units in grasses given that CSE orthologues were mainly not found in the majority of grass genomes. Moreover, the recent observation that the flavonoid tricin is an authentic lignin monomer in grasses brings the question of whether the regulation of tricin biosynthesis follows the same genetic program employed for the biosynthesis of the canonical monolignols. In addition, the transport of monolignols and other minor or alternative lignin monomers to the apoplast remains a poorly understood aspect of lignin metabolism, even in dicots. So far, only one monolignol transporter belonging to the ABCG subfamily of ATP-binding cassette transporters was characterized through reverse genetics and shown to act as a p-coumaroyl alcohol-specific transporter in Arabidopsis (Alejandro et al. 2012), while the mechanism by which the major lignin monomers coniferyl alcohol and sinapyl alcohol are transported remains elusive. Unraveling the mechanism of monolignols transport is of special interest in grasses, a group of plant whose lignin incorporate a wide range of alternative monomers such as the flavonoid tricin and different preacylated monolignols. Finally, loss-of-function mutants with altered lignification have helped the characterization of both class III peroxidases (Lee et al. 2013; Shigeto et al. 2013; Fernández-Pérez et al. 2015) and laccases (Berthet et al. 2011; Zhao et al. 2013) involved in lignin polymerization in Arabidopsis. Noteworthy, very specific functions were revealed for class III peroxidases, such as the involvement of Prx64 in a mechanism of localized lignin deposition in the endodermis of Arabidopsis plants (Lee et al. 2013). Conversely, only a few laccase genes involved in lignification were functionally characterized in grasses. Advances in plant systems biology and genetic transformation will certainly allow further identification and characterization of class III peroxidases with a role in lignin polymerization in grasses.
Regarding the transcriptional network controlling SCW deposition, it would be interesting to evaluate whether any transcriptional regulator in grasses shows cell-type specificity as some master regulators in Arabidopsis. The identification of cell-type-specific master regulators in grasses might be important for the engineering of plant biomass by targeting SCW deposition in specific cell types (such as fibers) without detrimental effects on plant growth and development. Moreover, despite the identification of orthologues of SCW- and lignin-related transcription factors in grass genomes, it is not clear whether the nature of the regulatory network characterized for Arabidopsis is completely conserved in grasses. Given the differences in vasculature patterning and cell wall composition, it is expected that grass-specific features will be found among the mechanisms controlling SCW deposition in this large group of plants. Answering these questions might be very useful for the understanding of the biology of SCW and lignin in grasses and for the rational engineering of grass crops for downstream applications.
Author contribution statement
IC wrote the manuscript and prepared the figures; MSS, MSB, AF, TFS and ER wrote the manuscript. All authors read the final version of the manuscript and agreed with the submission.
References
Agarwal T, Grotewold E, Doseff AI, Gray J (2016) MYB31/MYB42 Syntelogs exhibit divergent regulation of phenylpropanoid genes in maize, sorghum and rice. Sci Rep 6:28502
Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, Martinoia E (2012) AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol CB 22:1207–1212. doi:10.1016/j.cub.2012.04.064
Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115:1053–1074. doi:10.1093/aob/mcv046
Barros J, Serrani-Yarce JC, Chen F, Baxter D, Venables BJ, Dixon RA (2016) Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat Plants 2:16050. doi:10.1038/nplants.2016.50
Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, Lapierre C, Jouanin L (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23:1124–1137. doi:10.1105/tpc.110.082792
Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JEK, Blais S, Tremblay L, Morency M-J, Pavy N, Grima-Pettenati J, Séguin A, MacKay J (2008) Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot 59:3925–3939. doi:10.1093/jxb/ern234
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–363. doi:10.1146/annurev-genet-102209-163508
Bout S, Vermerris W (2003) A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol Genet Genomics 269:205–214. doi:10.1007/s00438-003-0824-4
Carpita NC, McCann MC (2008) Maize and sorghum: genetic resources for bioenergy grasses. Trends Plant Sci 13:415–420. doi:10.1016/j.tplants.2008.06.002
Cass CL, Peraldi A, Dowd PF, Mottiar Y, Santoro N, Karlen SD, Bukhman YV, Foster CE, Thrower N, Bruno LC, Moskvin OV, Johnson ET, Willhoit ME, Phutane M, Ralph J, Mansfield SD, Nicholson P, Sedbrook JC (2015) Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J Exp Bot 66:4317–4335. doi:10.1093/jxb/erv269
Cesarino I, Araujo P, Domingues Junior AP, Mazzafera P (2012a) An overview of lignin metabolism and its effect on biomass recalcitrance. Braz J Bot 35:303–311
Cesarino I, Araujo P, Sampaio Mayer JL, Paes Leme AF, Mazzafera P (2012b) Enzymatic activity and proteomic profile of class III peroxidases during sugarcane stem development. Plant Physiol Biochem 55:66–76. doi:10.1016/j.plaphy.2012.03.014
Cesarino I, Araujo P, Sampaio Mayer JL, Vicentini R, Berthet S, Demedts B, Vanholme B, Boerjan W, Mazzafera P (2013) Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S: G ratio of Arabidopsis lac17 mutant. J Exp Bot 64:1769–1781. doi:10.1093/jxb/ert045
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761. doi:10.1038/nbt1316
Courtois-Moreau CL, Pesquet E, Sjödin A, Muñiz L, Bollhöner B, Kaneda M, Samuels L, Jansson S, Tuominen H (2009) A unique program for cell death in xylem fibers of Populus stem. Plant J 58:260–274. doi:10.1111/j.1365-313X.2008.03777.x
Del Río JC, Marques G, Rencoret J, Martínez ÁT, Gutiérrez A (2007) Occurrence of naturally acetylated lignin units. J Agric Food Chem 55:5461–5468. doi:10.1021/jf0705264
Del Río JC, Prinsen P, Rencoret J, Nieto L, Jiménez-Barbero J, Ralph J, Martínez ÁT, Gutiérrez A (2012a) Structural characterization of the lignin in the cortex and pith of elephant grass (Pennisetum purpureum) stems. J Agric Food Chem 60:3619–3634. doi:10.1021/jf300099g
Del Río JC, Rencoret J, Prinsen P, Martinez AT, Ralph J, Gutierrez A (2012b) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60:5922–5935. doi:10.1021/jf301002n
Del Río JC, Lino AG, Colodette JL, Lima CF, Gutiérrez A, Martínez ÁT, Lu F, Ralph J, Rencoret J (2015) Differences in the chemical structure of the lignins from sugarcane bagasse and straw. Biomass Bioenergy 81:322–338. doi:10.1016/j.biombioe.2015.07.006
Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W (2015) Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27:695–710. doi:10.1105/tpc.114.134643
Escamilla-Treviño LL, Shen H, Hernandez T, Yin Y, Xu Y, Dixon RA (2013) Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass (Panicum virgatum L.). Plant Mol Biol 84:565–576. doi:10.1007/s11103-013-0152-y
Fernández-Pérez F, Pomar F, Pedreño MA, Novo-Uzal E (2015) The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol Plant 154:395–406. doi:10.1111/ppl.12310
Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, Fuguet E, Torres J-L, Rovira P, Puigdomènech P, Rigau J, Grotewold E, Gray J, Caparrós-Ruiz D (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64:633–644. doi:10.1111/j.1365-313X.2010.04363.x
Gray J, Caparrós-Ruiz D, Grotewold E (2012) Grass phenylpropanoids: regulate before using! Plant Sci 184:112–120. doi:10.1016/j.plantsci.2011.12.008
Ha CM, Escamilla-Trevino L, Yarce JCS, Kim H, Ralph J, Chen F, Dixon RA (2016) An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J 86:363–375. doi:10.1111/tpj.13177
Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1)—a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14:545–553. doi:10.1046/j.1365-313X.1998.00153.x
Handakumbura PP, Hazen SP (2012) Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Plant Physiol 3:74. doi:10.3389/fpls.2012.00074
Hatfield R, Ralph J, Grabber JH (2008) A potential role for sinapyl p-coumarate as a radical transfer mechanism in grass lignin formation. Planta 228:919–928. doi:10.1007/s00425-008-0791-4
Jin H (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161. doi:10.1093/emboj/19.22.6150
Lam PY, Zhu F-Y, Chan WL, Liu H, Lo C (2014) Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol 165:1315–1327. doi:10.1104/pp.114.239723
Lam PY, Liu H, Lo C (2015) Completion of tricin biosynthesis pathway in rice: cytochrome P450 75B4 is a unique chrysoeriol 5′-hydroxylase. Plant Physiol 168:1527–1536. doi:10.1104/pp.15.00566
Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167:1284–1295. doi:10.1104/pp.114.253757
Lan W, Morreel K, Lu F, Rencoret J, Del Río JC, Voorend W, Vermerris W, Boerjan WA, Ralph J (2016a) Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol. doi:10.1104/pp.16.02012
Lan W, Rencoret J, Lu F, Karlen SD, Smith BG, Harris PJ, del Río JC, Ralph J (2016b) Tricin-Lignins: Occurrence and Quantitation of Tricin in Relation to Phylogeny. Plant J n/a–n/a. doi:10.1111/tpj.13315
Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153:402–412. doi:10.1016/j.cell.2013.02.045
Lewis N, Yamamoto E (1990) Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41:455–496. doi:10.1146/annurev.pp.41.060190.002323
Li L, Hill-Skinner S, Liu S, Beuchle D, Tang HM, Yeh C-T, Nettleton D, Schnable PS (2015) The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J 81:493–504. doi:10.1111/tpj.12745
Li M, Pu Y, Yoo CG, Ragauskas AJ (2016) The occurrence of tricin and its derivatives in plants. Green Chem 18:1439–1454. doi:10.1039/C5GC03062E
Liu C-J (2012) Deciphering the enigma of lignification: precursor transport, oxidation, and the topochemistry of lignin assembly. Mol Plant 5:304–317. doi:10.1093/mp/ssr121
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho M-J, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco SC, RegisterIII JC, Zhao Z-Y, Xu D, Jones TJ, Gordon-Kamm WJ (2016) Morphogenic regulators baby boom and wuschel improve monocot transformation. Plant Cell. doi:10.1105/tpc.16.00124
Lu F, Karlen SD, Regner M, Kim H, Ralph SA, Sun R-C, Kuroda K, Augustin MA, Mawson R, Sabarez H, Singh T, Jimenez-Monteon G, Zakaria S, Hill S, Harris PJ, Boerjan W, Wilkerson CG, Mansfield SD, Ralph J (2015) Naturally p-hydroxybenzoylated lignins in palms. BioEnergy Res 8:934–952. doi:10.1007/s12155-015-9583-4
Marita JM, Hatfield RD, Rancour DM, Frost KE (2014) Identification and suppression of the p-coumaroyl CoA:hydroxycinnamyl alcohol transferase in Zea mays L. Plant J 78:850–864. doi:10.1111/tpj.12510
Marriott PE, Gómez LD, McQueen-Mason SJ (2016) Unlocking the potential of lignocellulosic biomass through plant science. New Phytol 209:1366–1381. doi:10.1111/nph.13684
Martins PK, Nakayama TJ, Ribeiro AP, Cunha BADB, Nepomuceno AL, Harmon FG, Kobayashi AK, Molinari HBC (2015a) Setaria viridis floral-dip: a simple and rapid Agrobacterium-mediated transformation method. Biotechnol Rep 6:61–63. doi:10.1016/j.btre.2015.02.006
Martins PK, Ribeiro AP, Cunha BADB, Kobayashi AK, Molinari HBC (2015b) A simple and highly efficient Agrobacterium-mediated transformation protocol for Setaria viridis. Biotechnol Rep 6:41–44. doi:10.1016/j.btre.2015.02.002
McCarthy RL, Zhong R, Ye Z-H (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50:1950–1964. doi:10.1093/pcp/pcp139
Meyermans H, Morreel K, Lapierre C, Pollet B, Bruyn AD, Busson R, Herdewijn P, Devreese B, Beeumen JV, Marita JM, Ralph J, Chen C, Burggraeve B, Montagu MV, Messens E, Boerjan W (2000) Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme a O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275:36899–36909. doi:10.1074/jbc.M006915200
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280. doi:10.1105/tpc.106.047043
Moura JCMS, Bonine CAV, Viana JOF, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52:360–376. doi:10.1111/j.1744-7909.2010.00892.x
Noda S, Koshiba T, Hattori T, Yamaguchi M, Suzuki S, Umezawa T (2015) The expression of a rice secondary wall-specific cellulose synthase gene, OsCesA7, is directly regulated by a rice transcription factor, OsMYB58/63. Planta 242:589–600. doi:10.1007/s00425-015-2343-z
Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG, Sibout R, Lapierre C, Ralph J, Sedbrook JC (2014) p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J 77:713–726. doi:10.1111/tpj.12420
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344:709. doi:10.1126/science.1246843
Ralph J (2010) Hydroxycinnamates in lignification. Phytochem Rev 9:65–83. doi:10.1007/s11101-009-9141-9
Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H-JG (1994) Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc 116:9448–9456. doi:10.1021/ja00100a006
Ralph J, Grabber JH, Hatfield RD (1995) Lignin-ferulate cross-links in grasses: active incorporation of ferulate polysaccharide esters into ryegrass lignins. Carbohydr Res 275:167–178. doi:10.1016/0008-6215(95)00237-N
Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, Boerjan W (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3:29–60. doi:10.1023/B:PHYT.0000047809.65444.a4
Rencoret J, Ralph J, Marques G, Gutierrez A, Martinez AT, del Rio JC (2013) Structural characterization of lignin isolated from coconut (Cocos nucifera) coir fibers. J Agric Food Chem 61:2434–2445. doi:10.1021/jf304686x
Riboulet C, Guillaumie S, Mechin V, Bosio M, Pichon M, Goffner D, Lapierre C, Pollet B, Lefevre B, Martinant JP, Barriere Y (2009) Kinetics of phenylpropanoid gene expression in maize growing internodes: relationships with cell wall deposition. Crop Sci 49:211–223. doi:10.2135/cropsci2008.03.0130
Rosler J, Krekel F, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113:175–179. doi:10.1104/pp.113.1.175
Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181:783–795. doi:10.1534/genetics.108.098996
Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70:818–830. doi:10.1111/j.1365-313X.2012.04933.x
Saha P, Blumwald E (2016) Spike-dip transformation of Setaria viridis. Plant J 86:89–101. doi:10.1111/tpj.13148
Sattler SE, Funnell-Harris DL, Pedersen JF (2010) Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Sci 178:229–238. doi:10.1016/j.plantsci.2010.01.001
Scully ED, Gries T, Sarath G, Palmer NA, Baird L, Serapiglia MJ, Dien BS, Boateng AA, Ge Z, Funnell-Harris DL, Twigg P, Clemente TE, Sattler SE (2016) Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J 85:378–395. doi:10.1111/tpj.13112
Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DGJ, Wang H, Jackson L, Tang Y, Neal Stewart C, Chen F, Dixon RA (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193:121–136. doi:10.1111/j.1469-8137.2011.03922.x
Shen H, Mazarei M, Hisano H, Escamilla-Trevino L, Fu C, Pu Y, Rudis MR, Tang Y, Xiao X, Jackson L, Li G, Hernandez T, Chen F, Ragauskas AJ, Stewart CN, Wang Z-Y, Dixon RA (2013) A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell 25:4342–4361. doi:10.1105/tpc.113.118828
Shigeto J, Kiyonaga Y, Fujita K, Kondo R, Tsutsumi Y (2013) Putative cationic cell-wall-bound peroxidase homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J Agric Food Chem 61:3781–3788. doi:10.1021/jf400426g
Sonbol F-M, Fornalé S, Capellades M, Encina A, Touriño S, Torres J-L, Rovira P, Ruel K, Puigdomènech P, Rigau J, Caparrós-Ruiz D (2009) The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol Biol 70:283–296. doi:10.1007/s11103-009-9473-2
Tang HM, Liu S, Hill-Skinner S, Wu W, Reed D, Yeh C-T, Nettleton D, Schnable PS (2014) The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J 77:380–392. doi:10.1111/tpj.12394
Taylor-Teeples M, Lin L, de Lucas M, Turco G, Toal TW, Gaudinier A, Young NF, Trabucco GM, Veling MT, Lamothe R, Handakumbura PP, Xiong G, Wang C, Corwin J, Tsoukalas A, Zhang L, Ware D, Pauly M, Kliebenstein DJ, Dehesh K, Tagkopoulos I, Breton G, Pruneda-Paz JL, Ahnert SE, Kay SA, Hazen SP, Brady SM (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517:571–575. doi:10.1038/nature14099
Tohge T, Watanabe M, Hoefgen R, Fernie AR (2013) The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol Biol 48:123–152. doi:10.3109/10409238.2012.758083
Valdivia ER, Herrera MT, Gianzo C, Fidalgo J, Revilla G, Zarra I, Sampedro J (2013) Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J Exp Bot 64:1333–1343. doi:10.1093/jxb/ers394
Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6:46. doi:10.1186/1754-6834-6-46
van der Weijde T, Alvim Kamei CL, Torres AF, Vermerris W, Dolstra O, Visser RGF, Trindade LM (2013) The potential of C4 grasses for cellulosic biofuel production. Front Plant Sci 4:107. doi:10.3389/fpls.2013.00107
Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Plant Biol 11:278–285. doi:10.1016/j.pbi.2008.03.005
Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol 196:978–1000. doi:10.1111/j.1469-8137.2012.04337.x
Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, Welsh L, Haustraete J, McClellan C, Vanholme B, Ralph J, Simpson GG, Halpin C, Boerjan W (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341:1103–1106. doi:10.1126/science.1241602
Vargas L, Cesarino I, Vanholme R, Voorend W, de Lyra Soriano Saleme M, Morreel K, Boerjan W (2016) Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotechnol Biofuels 9:139. doi:10.1186/s13068-016-0551-9
Vicentini R, Bottcher A, dos Santos Brito M, dos Santos AB, Creste S, de Andrade Landell MG, Cesarino I, Mazzafera P (2015) Large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PLoS One 10:e0134909
Vignols F, Rigau J, Torres MA, Capellades M, Puigdomènech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7:407–416. doi:10.1105/tpc.7.4.407
Vogel J (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11:301–307. doi:10.1016/j.pbi.2008.03.002
Wang Y, Chantreau M, Sibout R, Hawkins S (2013) Plant cell wall lignification and monolignol metabolism. Front Plant Sci 4:220. doi:10.3389/fpls.2013.00220
Wang J, Feng J, Jia W, Chang S, Li S, Li Y (2015a) Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol Biofuels 8:145. doi:10.1186/s13068-015-0331-y
Wang Y, Bouchabke-Coussa O, Lebris P, Antelme S, Soulhat C, Gineau E, Dalmais M, Bendahmane A, Morin H, Mouille G, Legée F, Cézard L, Lapierre C, Sibout R (2015b) LACCASE5 is required for lignification of the brachypodium distachyon culm. Plant Physiol 168:192–204. doi:10.1104/pp.114.255489
Watts KT, Mijts BN, Lee PC, Manning AJ, Schmidt-Dannert C (2006) Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem Biol 13:1317–1326. doi:10.1016/j.chembiol.2006.10.008
Wen J-L, Sun S-L, Xue B-L, Sun R-C (2013) Quantitative structural characterization of the lignins from the stem and pith of bamboo (Phyllostachys pubescens). Holzforschung 67:613–627. doi:10.1515/hf-2012-0162
Wilkerson CG, Mansfield SD, Lu F, Withers S, Park J-Y, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, Ralph J (2014) Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344:90–93. doi:10.1126/science.1250161
Withers S, Lu F, Kim H, Zhu Y, Ralph J, Wilkerson CG (2012) Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J Biol Chem 287:8347–8355. doi:10.1074/jbc.M111.284497
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55:652–664. doi:10.1111/j.1365-313X.2008.03533.x
Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T (2011) VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 66:579–590. doi:10.1111/j.1365-313X.2011.04514.x
You T-T, Mao J-Z, Yuan T-Q, Wen J-L, Xu F (2013) Structural elucidation of the lignins from stems and foliage of arundo donax linn. J Agric Food Chem 61:5361–5370. doi:10.1021/jf401277v
Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang Z-Y, Dixon RA (2013) LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25:3976–3987. doi:10.1105/tpc.113.117770
Zhong R, Ye Z-H (2012) MYB46 and MYB83 Bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53:368–380. doi:10.1093/pcp/pcr185
Zhong R, Ye Z-H (2014) Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci 229:193–207. doi:10.1016/j.plantsci.2014.09.009
Zhong R, Ye Z-H (2015) Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56:195–214. doi:10.1093/pcp/pcu140
Zhong R, Demura T, Ye Z-H (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18:3158–3170. doi:10.1105/tpc.106.047399
Zhong R, Richardson EA, Ye Z-H (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792. doi:10.1105/tpc.107.053678
Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20:2763–2782. doi:10.1105/tpc.108.061325
Zhong R, Lee C, Ye Z-H (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3:1087–1103. doi:10.1093/mp/ssq062
Zhong R, Lee C, McCarthy RL, Reeves CK, Jones EG, Ye Z-H (2011a) Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol 52:1856–1871. doi:10.1093/pcp/pcr123
Zhong R, McCarthy RL, Lee C, Ye Z-H (2011b) Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 157:1452–1468. doi:10.1104/pp.111.181354
Zhong R, Yuan Y, Spiekerman JJ, Guley JT, Egbosiuba JC, Ye Z-H (2015) Functional characterization of NAC and MYB transcription factors involved in regulation of biomass production in switchgrass (Panicum virgatum). PLoS One 10:e0134611
Zhou J-M, Gold ND, Martin VJJ, Wollenweber E, Ibrahim RK (2006) Sequential O-methylation of tricetin by a single gene product in wheat. Biochim Biophys Acta-Gen Subj 1760:1115–1124. doi:10.1016/j.bbagen.2006.02.008
Zhou J, Lee C, Zhong R, Ye Z-H (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21:248–266. doi:10.1105/tpc.108.063321
Zhou J, Zhong R, Ye Z-H (2014) Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9:e105726. doi:10.1371/journal.pone.0105726
Acknowledgements
Marvin was used for drawing the chemical structures shown in Fig. 1, Marvin 15.3.30 (version number), 2015, ChemAxon (http://www.chemaxon.com). Igor Cesarino acknowledges the Foundation for Research of the State of São Paulo (FAPESP) for the BIOEN Young Investigators Awards research fellowship (Grant 2015/02527-1). Marcella Siqueira Simões is indebted to FAPESP for a master fellowship (Grant 2015/18361-5). Michael dos Santos Brito thanks FAPESP for the financial support (Grant 2014/08468-4) and Young Investigators Awards research fellowship (Grant 2014/23017-9). Elisson Romanel acknowledges FAPESP for the financial support (Grants 2014/17486-6 and 2014/06923-6) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support (Grant 444912/2014-2). Tatiane da Franca Silva thanks CNPq for the financial support (Grant 448042/2014-2). Amanda Fanelli is indebted to CNPq for a doctorate fellowship (Grant 142474/2015-0).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L.A. Kleczkowski.
Rights and permissions
About this article
Cite this article
Cesarino, I., Simões, M.S., dos Santos Brito, M. et al. Building the wall: recent advances in understanding lignin metabolism in grasses. Acta Physiol Plant 38, 269 (2016). https://doi.org/10.1007/s11738-016-2293-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-016-2293-5