Abstract
Uremia Salt Lake, in North West Iran, has a hyper-saline water. A rare highly salinity-tolerant grass species, Aegilops cylindrica grows along its shores. Salinity tolerance of 44 genotypes of Ae. cylindrica, mainly collected from the Lake, was evaluated under control and 400 mM NaCl conditions using the physiological traits of plant height, dry weight, proline content, Na+ and K+ concentrations as well as K+/Na+ ratio. To evaluate the association between microsatellite (EST-SSR and SSR) markers and salinity tolerance, 35 primer pairs were used. Results showed a significant variation in the 44 genotypes studied in terms of their traits except for proline content. Ten most salinity-tolerant genotypes were identified based on their ability to survive, to produce the highest dry weight, and to sustain the least leaf Na+ concentration under salinity stress. The very high negative correlation found between Na+ concentration and salinity tolerance revealed the importance of individual or a combination of Na+ exclusion and excretion mechanisms contributing to the hyper-salinity tolerance of these genotypes. Clustering analysis based on marker data divided the 44 studied genotypes into two groups that were consistent with their saline and non-saline geographical areas. Results of molecular markers showed that four microsatellite markers (Xgwm312, Xwmc170, Xgwm291 and Xgwm410) generated a distinguished banding pattern in ten most salinity-tolerant genotypes. These results supported previous reports on their linkage with Na+ exclusion genes (HKT1;5 and HKT1;4) in wheat, which provided further evidence of usefulness of both genes and the linked markers to the salinity tolerance of the halophytic grass family species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil salinity is one of the most significant abiotic stresses that dramatically threatens crop production worldwide. According to FAO’s Land and Plant Nutrition Management Service, over 800 million hectares of land throughout the world are salt-affected (Munns and Tester 2008), that account for over 6 % of the world’s land area. Increased salinization of arable land is anticipated because of human activities and climate change. In Iran, saline soils comprise about 23.8 million hectares of total land area that accounts for about 30 % of the irrigated, arable land available (Ghassemi et al. 1995).
Salinity tolerance is a complex quantitative trait that is controlled by the interaction of several genes in many pathways of the physiological mechanisms such as Na+ and K+ transport in plants. Sodium exclusion is an important component of salinity tolerance that does not allow Na+ to accumulate to toxic concentrations within leaves and result in high K+/Na+ discrimination. Researchers have investigated the salinity tolerance of several accessions of Aegilops tauschii, and found that the accessions studied, similar to bread wheat, had much lower Na+ concentrations but higher K+/Na+ ratios in their leaves than did durum wheat (Gorham et al. 1987, 1990). The authors proposed that it was the D genome that imparted the property of Na+ exclusion to bread wheat and enhanced its K+/Na+ discrimination. Others have reported the existence of a Na+ exclusion locus in bread wheat that is located on the distal part of chromosome 4D (Dubcovsky et al. 1996).
Plants able to grow and complete their life cycle in habitats with soil salinity levels higher than 200 mM NaCl are known as halophytes (Flowers and Colmer 2008). Wild relatives of bread wheat are potential sources of valuable genetic materials such as salinity tolerance for wheat improvement. The use of wild relatives of Triticum species may offer an opportunity to increase salinity tolerance by providing availability to more variable germplasm (Shavrukov et al. 2009).
Jointed goat grass, Aegilops cylindrica host. (2n = 4x = 28; CCDD) species, is one of the wild relatives of wheat formed through amphidiploidization of a hybrid or hybrids between Ae. tauschii Coss. (2n = 2x = 14; DD) and Ae. markgrafii (Greuter) Hammer (2n = 2x = 14; CC).This species is known as a valuable source of genes related to biotic and abiotic stress tolerance. The screening results of Ae. cylindrica accessions collected from inland Pakistan and overseas revealed that about three quarters of the plants from 11 salinity-tolerant accessions survived at 300 mM NaCl for 7 weeks, and about 15 % survived at 400 mM NaCl in treatments using the mixture of Na2SO4:CaCl2:MgCl2:NaCl salts in a weight ratio of 10:5:1:4 using Hoagland solution (Farooq et al. 1989). Nevertheless, it is noted that the efficiency of pre-breeding via the formation of core collections can be increased by selecting representative subsets from large natural saline habitats. Iranian natural resources provide exceptional genetic resources for investigating halophytic adaption strategies in grass plant species naturally grown in high saline soils, particularly at the molecular level.
Hyper-salinity tolerant Ae. cylindrica genotypes were collected from Uremia Salt Lake shore and used in this study. Uremia Lake has a hyper-saline (>200 g l−1) water and situated in a subsiding tectonic basin in North West Iran. It is the largest lake in the Middle East and the third largest salt water lake on earth. The Na+ and Cl− concentration in Uremia Lake is approximately four times the concentration of natural seawater and wetlands near the lake comprise a variety of salinity-tolerant plant species (Eimanifar and Mohebbi 2007).
While a limited number of studies have been reported to date on the physiological characterization of highly salinity-tolerant species, no adequate study has been conducted to determine the molecular markers associated with salinity tolerance at hyper-saline level. Therefore, the objectives of the present study were to assess the physiological response of the highly salinity-tolerant Ae. cylindrica genotypes and to assess the SSR and EST-SSR markers associated with the salinity tolerance.
Materials and methods
Plant materials
Forty-four genotypes of Ae. cylindrica were used in this study that included 20 collected from inside Uremia Lake (due to the shrinking of the Lake in recent years, according to GPS information), 20 from the surrounding areas, and 4 from non-saline areas (Sanandaj, Kordestan Province), (Fig. 1). The lake collection-sites were located at distances of around 20 km from each other. Individual plants were taken from each collection site, and the seeds were collected from each plant. To increase seeds for each pure line (hereafter termed “genotype”), the seeds thus collected were subsequently grown in separate rows under field conditions in the growing season in 2011–2012.
Growth conditions and salinity treatment
Seeds were germinated for 4 days at 4 °C on moist filter paper in Petri dishes. Germinated seeds were planted into 60 × 40 cm plastic trays containing gravel under greenhouse conditions. The excess solution passing through the drainage holes at the bottom of each tray was collected by an underneath plate. Seedlings were watered initially with tap water, and then irrigated with half-strength Hoagland nutrient solution (Hoagland and Arnon 1950) 3 days after emergence. At second leaf emergence, the nutrient solution was replaced with full strength Hoagland nutrient solution to which was added NaCl in three gradual steps of irrigation until final concentrations reached 400 mM (EC ≈ 40 ds m−1). Greenhouse air temperature ranged from 25 °C (day) to 17 °C (night).
A split plot design with randomized complete block arrangement was used with three replications. Salinity treatments (400 and 0 mM NaCl) were arranged as main plots and plant genotypes as subplot. After 4 weeks of salinity treatment, plant height, dry weight and survival rate were recorded. Leaf concentrations of Na+ and K+ were determined by flame photometry (Jenway PFP7, UK).
Proline content
Leaf tissue, 0.5 g, collected from salt-treated and control plants was quickly frozen and ground in liquid nitrogen. The tissue was extracted with 3 % (w v−1) sulfosalicylic acid and, after centrifugation (1 min, 7,500 ×g) the supernatant was used to determine proline content according to the method described by Bates et al. (1973).
Molecular markers
The DNA was extracted from fresh leaves following the protocol described by Doyle and Doyle (1990). 35 primer pairs including 10 EST-SSR pairs related to salinity tolerance and 25 SSR primer pairs (five of which are associated to salinity tolerance) were used in this study (Table 1). Polymerase chain reactions (PCRs) were carried out in 15 µl reactions comprising 40 ng of genomic DNA, 1.5 mM MgCl2, 0.3 µM of each primer, 200 µM of dNTPs and one unit Taq DNA polymerase (Fermentas, USA). The PCR profile consisted of denaturation at 95° for 5 min, followed by 32 cycles of 95° for 1 min, 50°–60° (depending on primers) for 1 min, and 72° for 1 min, with a final extension at 72° for 10 min. Amplified PCR products were separated on 12 % denaturing polyacrylamide gels using a vertical electrophoresis device followed by silver staining.
Statistical analysis
The data on physiological traits were subjected to analysis of variance using PROC GLM of SAS Ver. 9.1 (SAS Institute 2002). Mean comparisons were conducted using Fisher’s lease significant differences LSD0.05 test.
For molecular data, allelic polymorphism information content values were calculated according to the formula developed by Nei (1973):
where, P 2 ij is the frequency of the jth allele for the ith locus summed across all alleles for the locus. Binary data were used to compute a pairwise similarity matrix using the Dice similarity index. Pairwise similarities were used to construct a dendrogram by using UPGMA (unweighted pair group method with arithmetic averages) employing SAHN (sequential, agglomerative, hierarchical and nested) clustering in NTSYS-pc version 2.11a (Rohlf 2002).
Results
Physiological traits
Analyses of variance showed significant differences among Ae. cylindrica genotypes for dry weight, plant height, Na+ and K+ concentrations as well as K+/Na+ discrimination when exposed to saline conditions compared with the control (Table 2). Salinity stress significantly affected all traits except proline content. Moreover, a significant genotype × salinity interaction was observed for all the traits.
Shoot dry weight of the genotypes decreased in the salinity treatment (400 mM NaCl) compared with that of the control, ranging from 7 to 37 % reduction (data not shown). According to these results, 10 out of the 44 genotypes were identified as the highest (<20 % dry weight loss) salt-tolerant ones. These superior salinity-tolerant genotypes also acquired the highest survival rates under 400 mM NaCl treatment (data not shown). On the other hand, four genotypes collected from non-saline region did not survive at salinity treatment (0 % survival rate).
Considerable variation among genotypes in response to salinity was also observed for plant height. Plant height in all the genotypes decreased significantly as salinity increased from 0 to 400 mM NaCl (Table 3). Plant height reduction of the genotypes ranged from 1 to 22 % (data not shown).
A significant variation in Na+ and K+ accumulation was observed in the genotypes when exposed to salinity stress. Na+ concentration was sixfold lower in genotype number 26 than the mean value for all the genotypes in 400 mM NaCl. This genotype not only possessed the lowest amount of Na+ concentration (69.6 mg g DW−1), but had also the highest K+/Na+ discrimination (10.24) among the 44 genotypes studied (Table 3).
Proline content was not significantly affected by salinity stress. The highest proline content was observed in genotype number 20 (3.04 µmol mg FW−1) for 400 mM salinity while the lowest was recorded for genotype number 16 (0.11 µmol mg FW−1) under control conditions (Table 3).
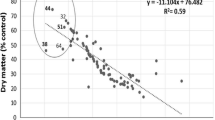
The relationship between trait and salinity tolerance represented by biomass production under 400 mM NaCl indicated that Na+ and K+ concentrations, K+/Na+ discrimination and plant height correlated significantly with salinity tolerance (Table 4). On the other hand, proline content did not correlate with salinity tolerance in Ae. cylindrica genotypes. Na+ concentration (r = 0.94) and plant height (r = 0.30) showed the highest and the lowest correlations with salinity tolerance, respectively. Results of linear regression analysis showed that the ten most salinity-tolerant genotypes (n26-n43-n36-n15-n42-n41-n39-n29-n35-n30) were superior for salinity tolerance where they possessed the lowest Na+ concentrations under saline conditions among the 44 genotypes studied (Fig. 2). The regression coefficient calculated between Na+ content and salinity tolerance (R 2 = 0.69) was greater than that between K+ content and salinity tolerance (R 2 = 0.38), and resulted, in turn, in a smaller regression coefficient between K+/Na+ and salinity tolerance (R 2 = 0.47).
Molecular markers
Out of the 35 EST-SSR and SSR primer pairs used, 26 primers generated clear patterns with distinctive polymorphism. Of the nine non-productive primers, six were monomorphic (Xcfd183, Xgwm131, Xgwm3, Xbe240888, Xec906203 and Xec906222), and three were not amplified (Xtxp-7, Xtxp-8 and Xtxp-19).
A total of 128 alleles were detected for the 26 loci. The number of alleles per locus ranged from 2 for Xcfd18 to 8 for Xcinua178 with an average number of 4.4 per locus (Table 1). The PIC-values ranged from 0.10 for Xgwm186 to 0.41 for Xgwm165, with an average value of 0.23 for all primers (Table 1).
The UPGMA dendrogram based on SSRs and EST-SSR markers divided the genotypes into two main groups (Fig. 3). The first included 40 genotypes originating from the hyper-saline shore area of Uremia Lake (North West Iran), and the second group comprised four genotypes originating from the non-saline area in the west of Iran.
Discussion
The physiological response of Ae. cylindrica genotypes, most of which were collected from the hyper-saline area, to salinity stress and the association of SSR markers with the salinity tolerance were investigated. The results revealed the significant effect of salinity on physiological traits with the exception of leaf proline content. Moreover, proline content did not correlate with salinity tolerance criteria such as survival rate (r = 0.21) and dry matter production (r = 0.13) under the 400 mM NaCl treatment (Table 4). This result is in agreement with that of Colmer et al. (1995) who observed a non-significant role of proline in salinity tolerance of wheat × Lophopyrum elongatum amphiploid plants. On the other hand, overexpression of the gene responsible for proline biosynthesis (P5CS) in wheat led to salinity tolerance enhancement (Sawahel and Hassan 2002). The results of the present study and the one mentioned above on the interspecific salinity-tolerant hybrids may indicate the non-significant role of proline in salinity tolerance of the grass species with the highest salinity tolerance. These results may shed light on the contradictory reports on the role of proline in salinity tolerance indicating that the level of proline contribution depends on the genetic background of the plant species (Ashraf and Harris 2004). Complementarily, it could be hypothesized that the role of proline content may be conceived to be very limited or even irrelevant in such hyper-salinity-tolerant grasses as Ae. cylindrica.
Ten most salinity-tolerant genotypes of Ae. cylindrica were identified based on their ability to survive the whole experimental period and to produce the highest dry weight under 400 mM NaCl salinity stress. Survival rate and biomass production are considered as the most important criteria to discriminate salinity tolerance in crop plants (Munns and James 2003). Only 10 out of the 20 genotypes collected from inside the Salt Lake were found to be most salinity tolerant. This may suggest that the combined effects of waterlogging and salinity under such conditions may exclude some of the genotypes from being able to tolerate the hyper-saline conditions. Although no direct comparison can be made between our results and those of other workers due to environmental differences, the level of salinity tolerance in the Ae. cylindrica genotypes originating from the hyper-saline shore areas of Uremia Lake exceeded that previously reported by Farooq et al. (1989). They reported that 15 % of the accessions, belonging to the Ae. cylindrica species obtained overseas, survived at an EC level of 30 dS m−1 (≈300 mM NaCl). These results highlight the impact of natural selection on the adaptation of such important key grass species to its natural hyper-saline habitat and on the enrichment of salinity-tolerance gene pool of this wild relative of wheat.
The ten most salinity-tolerant genotypes also possessed the least leaf Na+ concentration under salinity stress among the 44 genotypes used in this study. This result is not surprising as these most salinity-tolerant genotypes had been collected from inside Uremia Lake according to GPS information. Uremia Lake has been shrinking in recent years. Thus, it can be justified that natural selection readily produces genotypes with increased NaCl tolerance in Ae. cylindrica, the species having been recorded from a number of saline habitats which support NaCl-tolerant genotypes. Given the consistency of results presented here, it can also be explained that most salinity-tolerant genotypes have well developed salinity tolerance mechanisms to cope with saline environments. The tolerance mechanisms to salinity stress include: (1) salt reduction within the plant such as exclusion (from root and leaf), excretion (salt glands, bladder hairs and re-translocation); (2) impeding the harmful effects of salt within plant cells via compartmentalization, and succulence; and (3) osmotic adjustment. It is interesting to note that the excretion of salt crystal on the leaves of the above-mentioned genotypes were observed in the 400 Mm NaCl treatment (data not shown). In the most salinity-tolerant genotypes grown under high salinity (400 mM NaCl), salt excretion, which was recognizable by the salt crystal deposits on stem hairs and leaf surfaces, could be observed with the naked eye. Further botanical work on the salt excretion and glandular structure of these genotypes is underway in our laboratory. Within a species, Na+ concentration in leaves has been known as an index for estimating Na+ exclusion (Colmer et al. 2006). Plants with the ability to maintain low Na+ and high K+ concentrations (high K+/Na+ ratio) in leaves have reportedly exhibited a greater salinity tolerance within the species studied (Garthwaite et al. 2005; Poustini and Siosemardeh 2004). Despite the existing reports on Na+ exclusion in wheat and some of its wild relative as well as the halophytic species of Triticeae (Colmer et al. 2006), the present study is the first to report both salt excretion and exclusion in Ae. cylindrica.
Analysis of microsatellite markers detected relatively low PIC that referred to small genetic diversity in the 44 genotypes of Ae. cylindrica studied. Bordbar et al. (2011) reported that Ae. cylindrica had the smallest PIC value (0.24) among the eight Aegilops and Triticum species, which is consistent with our results. Using C-banding and FISH analysis, Badaeva et al. (2002) hypothesized that the small intraspecific polymorphism of Ae. cylindrica might represent a recent origin of this species. Furthermore, using chloroplast and nuclear microsatellites, Gandhi et al. (2005) suggested that Ae. cylindrica displayed very low levels of genetic diversity and, further, that this allotetraploid must have originated only recently. Results of clustering analysis of Ae. cylindrica genotypes using 26 microsatellite markers divided the genotypes into two groups which precisely correspond to their saline and non-saline geographical locations.
Molecular markers associated with salinity tolerance
Results of cluster analysis based on microsatellite data showed that the ten most salinity-tolerant genotypes were located in close proximity to each other in the same cluster but not clearly separated from the remaining 30 genotypes, all of which had been collected from the saline-habitat (Fig. 3, highlighted with a circle). Therefore, additional markers associated with salinity tolerance need to be used to discriminate clearly the ten most salinity-tolerant genotypes from the remaining ones. The most salinity-tolerant group of genotypes shared the same haplotype at the four markers Xwmc170, Xgwm291, Xgwm312 and Xgwm410. Two of these markers (Xgwm291 and Xgwm410) have been previously reported to be co-segregated with Nax2, which removes Na+ from the xylem in roots, and mapped to the distal end of chromosome 5AL in wheat (Byrt et al. 2007). The Nax2 locus on 5AL corresponds to the locus for a putative Na+ transporter, HKT1;5. The Nax2 region on 5AL chromosome is homoeologous to the region on chromosome 4DL containing the major gene for Na+ exclusion in common wheat, Kna1 (Byrt et al. 2007; James et al. 2011).The Ae. cylindrica species share a common genome with wheat (D genome) and this supports the presence of the Na+ exclusion gene to which the Xgwm291 and Xgwm410 microsatellite markers are linked. Na+ exclusion in the most salinity-tolerant grass, Ae. cylindrica, is not only associated with the D genome but should have also be enhanced by the C genome as suggested by Colmer et al. (2006).
The other two markers (Xgwm312 and Xwmc170) associated with salinity tolerance in the present study was found to be located on 2AL and closely linked to the Nax1 locus (HKT1;4 gene) (Lindsay et al. 2004). The HKT1;4 gene confers a reduced Na+ transport rate from roots to shoots and a reduced Na+ retention in leaf sheaths, which leads to a higher sheath-to-blade Na+ concentration ratio (Byrt et al. 2007). It may be speculated that the CC and or DD genome of Ae. cylindrica possess a region which is orthologous to the 2AL mapped Nax1 locus in wheat.
Marker-assisted selection (MAS) is the process of using morphological, biochemical, or DNA markers as indirect selection criteria for selecting agriculturally important traits such as salinity tolerance in crop breeding. This process is used to improve the effectiveness or efficiency of selection for the traits of interest in breeding programs. Nevertheless, the four SSR markers strongly associated with Na+ exclusion have been further verified by their association with salinity tolerance in the Ae. cylindrica species. However, the efficiency of the three SSR markers Xgwm410, Xgwm291 and Xgwm312 has been previously confirmed in MAS for salinity tolerance in hexaploid wheat (James et al. 2011).
Conclusions
Our observations revealed the importance of saline-habitat species collection as an objective measure in sampling genetic resources to improve crops for withstanding salinity tolerance. The results of the present study indicate that Na+ exclusion is one of the most important physiological attributes of Ae. cylindrica with respect to salinity tolerance. Microsatellite markers used in the present study differentiated the genotypes originating from saline and non-saline geographical areas. The microsatellite markers (Xgwm312, Xwmc170, Xgwm291 and Xgwm410), associated with salinity tolerance, supported previous reports on their linkage with Na+ exclusion genes (HKT1;5 and HKT1;4) in wheat, and hence provided further evidence of usefulness of both the genes and the markers in the salt tolerance trait of the most salinity-tolerant grass family species. Taken overall, the results suggest that the species possessing the ability to colonize natural saline-habitat and containing variability in salinity tolerance in their normal populations are always to be found exploiting saline habitats by forming salinity-tolerant ecotypes. It is also important to indicate that variability in salinity tolerance is generally available in plant species that could be selected for in plant breeding programs.
Author contribution
AA and FH collected the seeds and contributed significantly to the study design. The experiment was conducted by MA and RK with the supervision of AA, MMM and BEST. The data were analyzed by MA, who also wrote the paper, with significant inputs from AA.
Abbreviations
- EC:
-
Electrical conductivity
- DW:
-
Dry weight
- FW:
-
Fresh weight
References
Ahmad M, Shahzad A, Iqbal M, Asif M, Hiran AH (2013) Morphological and molecular genetic variation in wheat for salinity tolerance at germination and early seedling stage. Aust J Crop Sci 7:66–74
Ashraf M, Harris PJC (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166:3–16
Badaeva ED, Amosova AV, Muvavenko OV, Samatadze TE (2002) Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Syst Evol 231:163–190
Bates LS, Waldern RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Bordbar F, Rahiminejad MR, Saeidi H, Blattner FR (2011) Phylogeny and genetic diversity of D-genome species of Aegilops and Triticum (Triticeae, Poaceae) from Iran based on microsatellites, ITS, and trnL-F. Plant Syst Evol 291:117–131
Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143:1918–1928
Colmer TD, Epstein E, Dvorak J (1995) Differential solute regulation in leaf blades of various ages in salt sensitive wheat and a salt-tolerant wheat × Lophopyrum elongatum (Host) A. Love amphiploid. Plant Physiol 108:1715–1724
Colmer TD, Flowers TJ, Munns R (2006) Use of wild relatives to improve salt tolerance in wheat. J Exp Bot 57:1059–1078
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dubcovsky J, Maria GS, Epstein E, Luo MC, Dvorak J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92:448–454
Eimanifar A, Mohebbi F (2007) Urmia Lake (Northwest Iran): a brief review. Saline Syst 3:1–8
Farooq S, Niazi M, Iqbal N, Shah TM (1989) Salt tolerance potential of wild resources of the tribe Triticeae II. Screening of species of the genus Aegilops. Plant Soil 119:255–260
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Gandhi HT, Vales MI, Watson CJW, Mallory-Smith CA, Mori N, Rehman M, Zemetra RS, Riera-Lizarazu O (2005) Chloroplast and nuclear microsatellite analysis of Aegilops cylindrica. Theor Appl Genet 111:561–572
Gandhi HT, Vales MI, Mallory-Smith C, Riera-Lizarazu O (2009) Genetic structure of Aegilops cylindrica host in its native range and in the United States of America. Theor Appl Genet 119:1013–1025
Garthwaite AJ, Bothmer RV, Colmer TD (2005) Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl− into the shoots. J Exp Bot 56:2365–2378
Ghassemi F, Jakeman AJ, Nix HA (1995) Salinisation of land and water resources: human causes, extent and management and case studies. In: Centre for resource and environmental studies. The Australian National University, Canberra
Gorham J, Hardy C, Wyn Jones RG, Joppa LR, Law CN (1987) Chromosomal location of a K+/Na+ discrimination character in the D genome of wheat. Theor Appl Genet 74:584–588
Gorham J, Wyn Jones RG, Bristol A (1990) Partial characterization of the trait for enhanced K+/Na+ discrimination in the D genome of wheat. Planta 180:590–597
Hoagland DR, Arnon DI (1950) The water culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:32p
James RA, Blake C, Byrt CS, Munns R (2011) Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J Exp Bot 62:2939–2947
Leonova IN, Roder MS, Nasyrova F (2009) The application of wheat microsatellite markers for the detection of interspecific variation in tetraploid Aegilops species with C and U genomes. Cereal Res Commun 37:335–343
Li-Fang Z, Li-Xiao S, Yi-Gao F, Bao-Li Q, Hai-Bin X, Zi-You P, Zeng-Jun Q (2008) Development and chromosome mapping of new wheat EST-SSR markers and application for characterizing rye chromosomes added in wheat. Acta Agron Sin 34:926–933
Lindsay P, Lagudah S, Hare A, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol 31:1105–1114
Moghaieb EA, Abdel-Hadi A, Talaat B (2011) Molecular markers associated with salt tolerance in Egyptian wheats. Afr J Biotechnol 79:18092–18103
Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Poustini K, Siosemardeh A (2004) Ion distribution in wheat cultivars in response to salinity stress. Field Crops Res 85:125–133
Roder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rohlf FJ (2002) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.11a. Applied Biostatistics, New York
SAS Institute (2002) SAS version 9.1. SAS Institute. Cary, North Carolina
Sawahel WA, Hassan AH (2002) Generation of transgenic wheat plants producing high levels of the osmoprotectant proline. Biotechnol Lett 24:721–725
Shahzad A, Ahmad M, Iqbal M, Ahmed I, Ali GM (2012) Evaluation of wheat landrace genotypes for salinity tolerance at vegetative stage by using morphological and molecular markers. Genet Mol Res 11:679–692
Shavrukov Y, Langridge P, Tester M (2009) Salinity tolerance and sodium exclusion in genus Triticum. Breed Sci 59:671–678
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–537
Xu ZL, Ali Z, Yi JX, He XL, Zhang DY, Yu GH, Khan AA, Khan IA, Ma HX (2011) Expressed sequence tag-simple sequence repeat-based molecular variance in two Salicornia (Amaranthaceae) populations. Genet Mol Res 10:1262–1276
Xu SC, Gong YM, Mao WH, Hu QZ, Zhang GW, Fu W, Xian QQ (2012) Development and characterization of 41 novel EST-SSR markers for Pisum sativum (Leguminosea). Am J Bot 99:149–153
Acknowledgments
The authors would like to thank Professor Junhua Peng from Chinese Academy of Sciences for his onsite assistance and guidance in seed collection.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Zwiazek.
Rights and permissions
About this article
Cite this article
Arabbeigi, M., Arzani, A., Majidi, M.M. et al. Salinity tolerance of Aegilops cylindrica genotypes collected from hyper-saline shores of Uremia Salt Lake using physiological traits and SSR markers. Acta Physiol Plant 36, 2243–2251 (2014). https://doi.org/10.1007/s11738-014-1602-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-014-1602-0