Abstract
Phytohormone auxin plays an important role in fruit development and is perceived by the TIR1/AFB family of F-box proteins as auxin receptors involved in auxin signal pathway. Cucumber (Cucumis sativus L.) fruit development is either parthenocarpic or non-parthenocarpic. However, little is known on TIR1 and AFB participation in the early stage of cucumber fruit development. In present study, TIR1 and AFB2 were isolated from cucumber. CsTIR1 and CsAFB2 were highly expressed in leaves and ovaries. Their transcript levels decreased in parthenocarpic and pollinated fruits, but continuously up-regulated in aborted fruits, indicating that down-regulation of CsTIR1 and CsAFB2 may be in favor of cucumber fruit set and development. The transcript levels of CsTIR1 and CsAFB2 were significantly induced in leaves by NAA, 6-BA, GA3, ABA, and ethephon. The expression levels were up-regulated by ABA and ethephon treatments. This expression patterns was accordant with the aborted fruits. Thus, CsTIR1 and CsAFB2 may be important regulators during cucumber fruit development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cucumber (Cucumis sativus L.) belongs to the Cucurbitaceae family and is an important vegetable crop (Jeffrey 2008). After tomato (Solanum lycopersicum), the recently completed cucumber genome has been added to the existing crop database and a new model is being used for fruit development (Huang et al. 2009). Cucumber is an excellent plant to study fruit set and development, as its genotypes have different parthenocarpic capacities. Fruit development can be uncoupled from fertilization and seed development, and parthenocarpic fruits are seedless (Fos and Nuez 1996; Talon et al. 1992; Varoquaux et al. 2000). Parthenocarpy is a potentially desirable trait for many commercially grown fruits to avoid undesirable changes in structure, flavor, or nutrition proprieties (Martinelli et al. 2009).
Applying auxin and cytokinin to unpollinated cucumber ovaries (Kim et al. 1992; Fu et al. 2010), or overexpression of auxin biosynthesis gene in ovaries can promote the formation of fertilization-independent or parthenocarpic fruit (Yin et al. 2006). The increase in IAA content in cucumber ovaries is closely related to parthenocarpic fruit set (Kim et al. 1992). In addition, Auxin transport inhibitors induce parthenocarpy in cucumber by blocking the normal movement of auxin out of the ovary, thereby causing auxin accumulation within the ovary, which is sufficient to trigger parthenocarpy (Beyer and Quebedeaux 1974). Parthenocarpy has been staged by altering auxin signaling at different steps, indicating that auxin controls fruit development.
Auxin plays a key role in plant fruit development. Since decades, auxin signaling is known to be involved in transcriptional regulation by members of the Aux/IAA and ARF protein families, which are key regulators of auxin-modulated gene expression (Guilfoyle et al. 1998a, b; Leyser 2002; Walker and Estelle 1998). A significant advance in our understanding of the auxin-dependent mechanism underlying fruit set came from transgenic manipulation of Aux/IAA and ARF genes leading to parthenocarpic fruit in both tomato and Arabidopsis. Transgenic tomato lines with down-regulated SlIAA9 and decreased SlARF7 expression result in parthenocarpic fruit development before fertilization (Wang et al. 2005; De Jong et al. 2009b). In addition, inactivation of ARF8 causes parthenocarpic fruit development in tomato (Goetz et al. 2006, 2007). TIR1/AFB F-box proteins were later reported as auxin receptors that facilitate interaction with Aux/IAA proteins in an auxin-dependent manner (Dharmasiri et al. 2005b; Mockaitis and Estelle 2008). Aux/IAAs are targeted for degradation by ubiquitination and catalyzed by an SCF-type ubiquitin-protein ligase (Gray et al. 2001). TIR1 and its paralogs, AFB1, 2, 3, and 4 are the F-box subunits of the SCF E3-ubiquitin ligase complex and function as auxin receptors. The TIR1/AFB F-box protein acts as an auxin receptor and directly links auxin detection of binding to SCFTIR1/AFB to the degradation of the Aux/IAA proteins (Dharmasiri et al. 2005a, b; Kepinski and Leyser 2005b). Recently, overexpression of SlTIR1 results in parthenocarpic fruit formation in tomato (Ren et al. 2011). These findings suggest that auxin regulates most aspects of plant growth and development by TIR1-dependent degradation of Aux/IAA proteins. However, the auxin receptor molecular mechanism has remained elusive in cucumber fruit development.
In this study, we isolated CsTIR1 and CsAFB2 mRNA sequences from cucumber fruit and the characteristics of the two genes were analyzed using a bioinformatics program. The expression levels of CsTIR1 and CsAFB2 in different tissues and early fruit developmental stages under different treatments indicated that these two genes are important regulators during fruit development. In particular, the declining levels of CsTIR1 and CsAFB2 played an important role in fruit initiation in natural parthenocarpic fruit. The expression levels in leaves using NAA, 6-BA, GA3, ABA and ethephon treatments were analyzed. The results indicated that the transcript levels of CsTIR1 and CsAFB2 were regulated by these hormones.
Materials and methods
Plant materials
Five cucumber cultivars, EC1, L8, S12533, and S12535 (strong parthenocarpic lines) and 8419s-1 (non-parthenocarpic line) were used in the experiments. Seedlings were grown in a greenhouse [12-h photoperiod, mean daily air temperatures, 29/17 °C (day/night), relative humidity, 85 %, photosynthetic photo flux density 800 μmol m−2 s−1] at Nanjing Agricultural University. All tissues (including root, stem, leaf, flower, and fruit) were collected from 10-week-old cucumber plants. The ovaries at the 12–15th node of the main stem were isolated to prevent pollen contamination on the day before anthesis. Ovaries of the parthenocarpy cultivars were isolated only for sampling. The experiments with cultivar 8419s-1 included three treatments: non-pollination, pollination, and 100 mg L−1 CPPU treatment of unpollinated ovaries according to Fu et al. (2010). Other cultivars were not pollinated. Except unpollinated ovaries of L8, S12533, and S12535 were sampled at 2 days after anthesis (DAA), and other samples were harvested at 0, 2, 4, 6, and 8 DAA (8419s-1 unpollinated fruits were not obtained at 8 DAA). Samples were frozen in liquid nitrogen and stored at −80 °C prior to RNA extraction.
Plant hormone treatment
8419s-1 seedlings were treated with different exogenous hormones. Germinated seeds were planted in nutrition pots, and 3-week-old seedlings were subjected to hormone treatments. Hormones treatments were carried out by spraying the leaves of the seedling with NAA (5, 10, and 50 μM), 6-BA (10 μM), GA3 (10 μM), ABA (10 μM), or ethephon (10 μM) and the plants were sampled at 0, 3, 6, and 9 h after spraying. Leaf samples were frozen in liquid nitrogen and stored at −80 °C.
Cloning the full-length cDNA of CsTIR1 and CsAFB2 and sequence analysis
The CsTIR1 and CsAFB2 CDS sequences in the cucumber genome database were Blast-searched with the SlTIR1 (GQ370812.1) and AtTIR1 (AAF78487) sequences, and genes ID of the homologous fragments were obtained. Csa01802 and Csa015043 have the highest identity with SlTIR1 and AtAFB2, respectively. We designed two primer pairs to amplify the full-length cDNA sequences of Csa01802 and Csa015043 (Table 1). The PCR products were directly sequenced (Invitrogen).
Searches for nucleotide and protein sequence similarities were completed with the BLAST algorithm at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al. 1997). Sequences were aligned using the DNAMAN program, v6.0. Structural domains were annotated using Smart (http://smart.embl-heidelberg.de/; Letunic et al. 2012). The phylogenetic tree, based on homology, was constructed with the MEGA program v4.0. The TIR1/AFB protein sequences used in the phylogenetic tree can be found at GenBank under the following accession numbers: Populus trichocarpa PtFBL1 (XP_002123035.1), PtFBL2 (XP_002321336.1), PtFBL3 (XP_002300140.1), PtFBL4 (XP_002328871.1), Ricinus communis RcTIR1 (XP_002520681.1), Vitis vinifera VvTIR1 (XP_002269127.1), Dimocarpus longan DiTIR1(ACX31301.2), Gossypium hirsutum GhTIR1 (ABG46343.1), Nicotiana tabacum NtTIR1 (ACT53268.1), S.lycopersicum SlTIR1 (GQ370812.1), Arabidopsis thaliana AtTIR1 (NP_567135.1), AtAFB2 (NP_566800.1), AtAFB3 (NP_563915.1), AtAFB5 (NP_568718.1), AtAFB18 (AAK76473.1), AtGRH1 (NP_567255.1), AtCOl1 (NP_565919.1), Oryza sativa OsTIR1 (AAV32196.1), Zea mays ZmTIR1 (NP_001148131.1), Arabidopsis lyrata subsp. lyrata AIF-box2 (XP_002875343.1), AIF-box3 (XP_002892715.1), AlAFB5 (XP_002865768.1), S. lycopersium SlCOl1 (AAR82926.1).
RNA extraction and qRT-PCR for gene expression analysis
Total RNA was extracted from different cucumber tissues using Trizol reagent (Invitrogen), according to manufacturer’s instructions. The RNA was treated with DNase I (Fermentas, UK) for 30 min at 25 °C and purified according to manufacturer’s instructions. The first-strand cDNA synthesis was performed using 2 μg of total RNA and a Fermentas Reverse Transcription Kit. The expression levels of CsTIR1 and CsAFB2 were determined by real-time RT-PCR. The primer sequences used are listed in Table 1. For the quantification of the PCR products, Cs-Actin was used as control (GenBank Accession No. AB010922). Relative fold differences were calculated based on the comparative Ct method. The formula is calculated as follows: 2−∆∆Ct according to the previous report by Audran–Delalande et al. (2012).
Determination of IAA and ZR concentration
The extraction and purification method for IAA and ZR were modified from those described by Yang et al. (2001). Samples of 8419s-1 fruits were ground in an ice-cooled mortar in 8 mL of 80 % (v/v) methanol extraction medium containing 1 mM butylated hydroxytoluene as an antioxidant. The extracts were incubated at 4 °C for 4 h and centrifuged at 3,500 rpm for 8 min at 4 °C. The supernatants were combined and passed through Chromosep C18 columns (C18 Sep-Park Cartridge, Waters Corp., Milford, MA, USA), prewashed with 10 mL of 100 % and 5 mL of 80 % (v/v) methanol, respectively. The hormone fractions were eluted with 10 mL of 100 % (v/v) methanol and 10 mL of ether and collected and dried under N2 gas. The residues were dissolved in 2 mL phosphate buffer saline (PBS) containing 0.1 % (v/v) Tween 20 and 0.1 % (w/v) gelatin (pH 7.5) for ELISA.
The antibodies against IAA and ZR horseradish peroxidase used in ELISA were produced at the Phytohormones Research Institute (China Agricultural University). ELISA was performed in a 96-well microtitration plate. Each well was coated with 100 μL coating buffer (1.5 g L−1 Na2CO3, 2.93 g L−1 NaHCO3, and 0.02 g L−1 NaN3, pH 9.6) containing 0.25 μg mL−1 antigen against the hormone. The coated plates were incubated for 4 h at 37 °C and overnight at 4 °C for IAA and ZR, and then kept at room temperature for 30 min. After washing four times with PBS + Tween 20 [0.1 % (v/v)] buffer (pH 7.4), each well was filled with 50 μL of either extract IAA or ZR standards (0–200 ng L−1 dilution range) and 50 μL of 20 μg mL−1 antibody against IAA or ZR. The plates were incubated at 37 °C for 30 min, and then washed as described above. In each well, 100 μL of 1.25 μg mL−1 IgG-horseradish peroxidase substrate was added and incubated for 30 min at 37 °C. The plates were rinsed four times with PBS + Tween 20 buffer, and 100 μL color-appearing solution containing 1.5 mg mL−1 0-phenylenediamine and 0.008 % (v/v) H2O2 was added to each well. The reaction was stopped by adding 50 μL 6 N H2SO4 per well when the 200 ng mL−1 standard developed a pale color, and the 0 ng mL−1 standard had a deep color. Color development in each well was detected using an ELISA Reader (Model EL301, Bio-TEK, Winooski, VT, USA) at optical density of A490. IAA and ZR contents were calculated following the report by Weiler et al. (1981). The results are presented as mean ± SE of three replicates.
Statistics
CsTIR1 and CsAFB2 mRNA levels were determined as mean ± SE. Differences among treatments were analyzed by one-way ANOVA. P values ≤0.05 and ≤0.01 was considered significant according to Tukey’s multiple range tests.
Results
Isolation of the full-length CsTIR1 and CsAFB2 cDNA and genome sequence
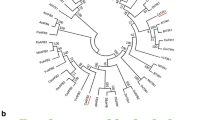
A 2,243 bp full-length cDNA of the putative SlTIR1-like gene and a 2,479 bp full-length cDNA of the putative AtAFB2-like gene (CsTIR1 and CsAFB2, respectively) were obtained by sequencing and were deposited in GenBank under accession numbers JX901282 and JX901283, respectively. The CsTIR1 and CsAFB2 cDNAs contained 1,755- and 1,764-bp open reading frames and encoded proteins of 587 and 620 amino acid residues, respectively. The genomic organizations of CsTIR1 and CsAFB2 were analyzed (Figs. 1a and 2a). The result showed that CsTIR1 contained an F-box region and six leucine-rich repeat (LRR) domains (Fig. 1b), while CsAFB2 contained an F-box and seven LRR domains (Fig. 2b). A phylogenetic tree comprising 25 TIR1 sequences from different species was constructed (Fig. 3). Phylogenetic analysis indicated that CsTIR1 and CsAFB2 were the most similar to SlTIR1 and PtFBL3, closely followed by AtTIR1 and AtAFB2, respectively.
Sequence analysis of cucumber CsTIR1. a Genomic structure of CsTIR1. Black lines represent introns, gray boxes are exons, and white boxes are untranslated regions (UTRs). b Alignment of predicted amino acid sequences of AtTIR1, SlTIR1, and CsTIR1. Numbers at the right indicate the positions of the amino acid residues. Shaded black regions indicate the identical amino acids. The putative F-box motif and conserved LRR domains are indicated by thin lines
Sequence analysis of cucumber CsAFB2. a Genomic structure of CsAFB2. Black lines represent introns, gray boxes are exons, and white boxes are UTRs. b Alignment of predicted amino acid sequences of AtAFB2, AtAFB3, and CsAFB2. Numbers at the right indicate the positions of the amino acid residues. Black shows the identical amino acids. The putative F-box motif and conserved LRR domains are indicated by thin lines
CsTIR1 belongs to a distinct subfamily of the auxin receptor TIR1 protein family. The phylogenetic tree was created using the neighbor-joining approach and MEGA4 software. The phylogenetic tree was constructed with protein sequences from the following species: Populus trichocarpa PtFBL1, PtFBL2, PtFBL3, and PtFBL4; Ricinus communis RcTIR1; Vitis vinifera VvTIR1; Dimocarpus longan DiTIR1; Gossypium hirsutum GhTIR1; Nicotiana tabacum NtTIR1; S. lycopersicum SlTIR1; Arabidopsis thaliana AtTIR1; AtAFB2, AtAFB3, AtAFB5, AtAFB18, AtGRH1, and AtCOl1; Oryza sativa OsTIR1; Zea mays. L. ZmTIR1; Arabidopsis lyrata subsp. Lyrata AIF-box2, AIF-box2, AlAFB5; and S. lycopersicum SlCOl1
The expression patterns of CsTIR1 and CsAFB2 in different cucumber organs
CsTIR1 and CsAFB2 transcript levels were assessed in different cucumber organs to explore their potential role during plant development. The expression analysis performed by qRT-PCR showed that CsTIR1 and CsAFB2 were expressed in all organs (Fig. 4), but differences among organs were observed. CsTIR1 was expressed at its highest levels in leaves, followed by ovaries, anthers, and roots. The expression level was similar in the young fruit (2 days after pollination) and root. CsTIR1 expression in the stem was very low (Fig. 4a). Although CsAFB2 is an F-box protein, its expression pattern was different with that of CsTIR1. The highest transcript level of CsAFB2 was observed in the ovaries, followed by the leaves, anthers, roots, stems, and young fruits (Fig. 4b).
Expression analysis of CsTIR1 and CsAFB2 in different cucumber organs. a, b Expression levels of CsTIR1 and CsAFB2 in roots, stems, leaves, anthers, ovaries, and young fruits. qRT-PCR analyses of total RNA isolated from roots, stems, leaves, anthers, ovaries, and young fruit pollinated after 2 days, which was used to assess CsTIR1 and CsAFB2 transcript levels. Data are mean ± SD normalized relative to Cs-Actin gene transcript level. All samples were run in triplicate. Asterisks indicate significant differences (t test; *P ≤ 0.05; **P ≤ 0.01)
The expression analysis of CsTIR1 and CsAFB2 in early developmental fruit stages
We further investigated the expression levels of the two genes during early fruit development under different treatments (Fig. 5). First of all, the CsTIR1 transcript level was analyzed at different fruit developmental stages (Fig. 5a–d). The expression level was gradually declined during natural parthenocarpic fruit development (EC1 unpollinated) from 0 to 8 DAA (Fig. 5a). This result was consistent with CsTIR1 expression in induced parthenocarpic and pollinated fruits (Fig. 5b and c). In contrast, CsTIR1 transcript level was increased in aborted fruits, which were non-parthenocarpic fruits under apoptosis (Fig. 5d). Then, expression level of CsAFB2 during early fruit development was performed. It was agreed with expression level CsTIR1 (Fig. 5a–d), except for the highest expression level during pollinated fruit development occurred 4 days after pollination (Fig. 5c). The expression levels of CsTIR1 and CsAFB2 declined significantly from the ovarian stage to fruit formation, but increased in aborted fruit. This dynamic expression pattern suggests that CsTIR1 and CsAFB2 might play important roles in cucumber fruit development and that pollination might stimulate its quick expression followed by a decrease when the fruit has successfully set.
The expression levels of CsTIR1 and CsAFB2 during early cucumber fruit development stages. CsTIR1 and CsAFB2 expression patterns in EC1 unpollinated fruits (a). 8419s-1 fruits treated with CPPU (b). 8419s-1 pollinated fruits (c). 8419s-1 non-pollination fruits (d). QRT-PCR analyses were preformed using RNA generated from differently treated cucumber ovaries at 0, 2, 4, 6, and 8 days after anthesis (the 8419s-1 non-pollination ovary at 8 days was not obtained). All samples were run in triplicate. Asterisks indicate significant differences (t test; *P ≤ 0.05; **P ≤ 0.01)
We next evaluated whether CsTIR1 and CsAFB2 expression levels were similar to those in other developing parthenocarpic fruit which were sampled at 2 DAA. CsTIR1 and CsAFB2 mRNA levels in parthenocarpic ovaries were significantly lower compared with aborted ovaries (Fig. 6). The results indicated that the expression levels of CsTIR1 and CsAFB2 were down-regulated in early developing cucumber fruit.
Transceipt levels of CsTIR1 and CsAFB2 in different parthenocarpy cultivars. a, b CsTIR1 and CsAFB2 expression levels in different parthenocarpic fruits and pollinated fruit. Unpollinated fruit of 8419s-1 was the control, indicating that fruit set was not successful. Ovaries that were pollinated and treated with CPPU began to format fruits. EC1, L8, S12533, and S12535 are the natural parthenocarpic cucumber lines. Data are expressed as relative values. Each value represents mean ± SE of three replicates
Changes in IAA and cytokinin content during cucumber fruit development
IAA content was measured in developing cucumber ovaries and young fruits to investigate the relationship between endogenous IAA and expression levels of CsTIR1 and CsAFB2 (Fig. 7a). IAA concentration decreased in pollinated and induced parthenocarpic fruits (8419s-1) at 2 days after pollination (DAP), then increased at 4 DAP. However, IAA concentration increased in aborted fruit at 2 DAP, after that decreased. These results were in contrast with CsTIR1 and CsAFB2 expression levels. This showed that increased endogenous IAA can not up-regulate the transcript levels of both genes in early stage of cucumber fruit development.
Analysis of endogenous hormone IAA and ZR in cucumber young fruit. Analysis of endogenous IAA concentration in cucumber young fruits (a). Analysis of endogenous ZR concentration in cucumber young fruits (b). Treatments included the pollinated, non-pollination and treated with CPPU of 8419s-1 fruits. Samples were collected at 0, 2, 4 and 6 days after anthesis. Data are mean ± 1 SE of three replicates
ZR content was detected in 8419s-1 young fruits and aborted fruits for exploring the roles of CsTIR1 and CsAFB2 in fruit development (Fig. 7b). ZR concentration decreased in pollinated and induced parthenocarpic fruits (8419s-1) at 2 DAP, then increased in pollinated fruits at 4 DAP and in induced parthenocarpic fruits at 6 days after CPPU treatment. ZR content has been declining in aborted fruits. However, ZR concentration was the highest at 4 DAP, and the expression level of CsAFB2 in pollinated at 4DAA was also the highest. The ZR level in aborted fruits was decreased from 0 to 6 days after anthesis. However, the expression patterns of CsTIR1 and CsAFB2 appeared the opposite situation. The result indicated that cytokine level may affect the transcript of CsAFB2 in fruit development.
The expression of CsTIR1 and CsAFB2 in responses to hormones
To investigate the CsTIR1 and CsAFB2 expression in responses to hormones, the expression levels were further analyzed in leaves treated with different concentrations of NAA. CsTIR1 and CsAFB2 were induced slowly following auxin treatment, and transcript levels were highest at 6 h after treatment, following declined at 9 h (Fig. 8), indicating that CsTIR1 and CsAFB2 expression induced by exogenous auxin and their expression patterns were similar in response to 5–50 μM NAA. In addition, we further carried out real-time quantitative PCR analyses. The result showed that CsTIR1 and CsAFB2 were induced by other hormones, including: 6-BA, GA3, ABA, and ethephon (Fig. 9). CsTIR1 was just up-regulated in the leaves by 6-BA at 3 h, then declined (Fig. 9a). Exogenous GA3 application caused a rapid up-regulation expression of CsTIR1, the highest level appeared at 6 h. Figure 9a shows CsTIR1 expression in response to ABA and ethephon, which were identical and significantly higher than those of the control. The CsAFB2 transcript increased significantly at 3 and 6 h after 6-BA treatment, and then declined (Fig. 9b). The CsAFB2 expression level in response to GA3 increased by 3 h and remained high 9 h after treatment. CsAFB2 expression in response to ABA was similar to that of CsTIR1 (Fig. 9b). Taken together, these results indicated that CsTIR1 and CsAFB2 were induced by exogenous hormones except 6-BA, which down-regulated expression. This was consistent with those of CPPU-induced parthenocarpic fruits.
Quantitative RT-PCR analysis of CsTIR1 and CsAFB2 in response to the different concentrations of NAA. a, b CsTIR1 and CsAFB2 expression patterns in leaves of 8419s-1 treated with different concentrations of NAA (5, 10, and 50 μM). QRT-PCR analyses were performed using RNA generated from cucumber leaves after treatments at different times (0, 3, 6, and 9 h). The control was 0 h. Data are mean ± SD normalized relative to Cs-Actin gene transcript level. All samples were run in triplicate. Asterisks indicate significant differences (t test; *P ≤ 0.05; **P ≤ 0.01)
The expression analysis of CsTIR1 and CsAFB2 in cucumber leaves under the treatments of exogenous hormones or hormone analogs a, b Expression levels of CsTIR1 and CsAFB2 under different treatments, including: 6-BA, GA3, ABA, and ethephon. The control was 0 h. Data are mean ± SD normalized relative to Cs-Actin gene transcript level. All samples were run in triplicate. Asterisks indicate significant differences (*P ≤ 0.05; **P ≤ 0.01)
Discussion
Fruit development is closely related to auxin (de Jong et al. 2009a; Pandolfini et al. 2007; Serrani et al. 2008). TIR1 and AFB2 play important roles as auxin receptors in regulating fruit development. Cucumber is an important economic crop with important research value. In this study, CsTIR1 and CsAFB2 genes were isolated from cucumber. Sequence analysis of the CsTIR1 protein indicated that it contained one conserved F-box and six LRR domains (Figs. 1a and 2a), which was the same as that of the SlTIR1 structural domain (Ren et al. 2011). Protein interactions were predicted from the Arabidopsis protein interaction database (http://arabidopsis.org/). The prediction showed that CsTIR1 interacts with SKP1, Cullin, SAPK7, and auxin-induced protein 22D in cucumber. Pull-down assay had shown that SKP1 links the F-box protein to Cullin to form SCF-ubiquitin ligase complexes (Bai et al. 1996; Dharmasiri et al. 2005a; Kepinski and Leyser 2005a), indicating that TIR1 may be a conserved gene in plants.
In tomato, the expression level of SlTIR1 was the highest in flower, down-regulated in immature green fruit (Ren et al. 2011). Our results also showed that the expression levels of CsTIR1 and CsAFB2 were high at flower day and decreased during the early stage of natural parthenocarpic cucumber fruit (2–8 DAA) (Fig. 5). We further analyzed the expression patterns of CsTIR1 and CsAFB2 in induced parthenocarpic fruit. The CsTIR1 and CsAFB2 transcripts were also down-regulated in the induced parthenocarpic fruit of cucumber, which uncoupled fruit initiation without pollination and fertilization and gave rise to parthenocarpic fruits (Fig. 5b). The result was consistent with decline in the expression levels of CsTIR1 and CsAFB2 in pollinated fruit (Fig. 5c). This suggested that the fruit set and development may be accompanied by down-regulated CsTIR1 and CsAFB2 expression. However, Ren et al. (2011) reported that overexpression SlTIR1 resulted in parthenocarpic fruit formation in tomato. This is in contradiction with our study that expression levels of CsTIR1 and CsAFB2 were down-regulated in parthenocarpic fruit. The reason may be that TIR1 gene plays an important role at the stages of flower-to-fruit transition; we just analyzed the expression levels of CsTIR1 and CsAFB2 in parthenocarpic fruit after anthesis.
Genetic studies have shown that the TIR1 and AFB functions were in a partially redundant manner to mediate the auxin response. A triple and quadruple tir/afb mutant analysis demonstrated that these genes have overlapping functions and are collectively essential for Arabidopsis growth and development (Dharmasiri et al. 2005b). This is possible reason that CsAFB2 and CsTIR1 have similar expression characteristics in different organs. However, the expression levels of CsTIR1 and CsAFB2 during early fruit development were different in pollinated fruits. The highest expression level of CsTIR1 was observed at 2 days earlier than that of CsAFB2, which appeared at 4 days after pollination. Thus, the result indicated that CsTIR1 and CsAFB2 may have different responses to pollination/fertilization.
In cucumber, pollination increases the levels of natural IAA and zeatin, which are essential to fruit development (Boonkorkaew et al. 2008). The increase in IAA concentration in cucumber young fruits is closely related to fruit development (Kim et al. 1992; Beyer and Quebedeaux 1974). However, the expression levels of CsTIR1 and CsAFB2 were down-regulated at the relevant fruit development stage when endogenous IAA level increased during fruit development (Fig. 7a). The result indicated that down-regulation expression levels of CsTIR1 and CsAFB2 and increase endogenous IAA content could help to fruit set and growth. The maximum content of IAA and ZR in pollinated fruits appeared at 4 DAP (Fig. 7), and the highest expression level of CsAFB2 was also at 4 DAP (Fig. 5c). This indicated that CsAFB2 may be induced by hormones from pollinated fruit. The hypothesis was verified in cucumber leaves treated by exogenous hormones.
Some studies have found that synthetic auxin analogs, such IAA, NAA, and 2, 4-dichlorophenoxyacetic acid could promote the binding of Aux/IAA proteins to the TIR1 F-box protein to enhance TIR1-Aux/IAA interaction (Dharmasiri et al. 2005a; Kepinski and Leyser 2005a; Tan et al. 2007). This is coincident with that CsTIR1 and CsAFB2 transcripts were accumulated in leaves following 5–50 μM NAA treatment (Fig. 8). Kepinski and Leyser (2005a, b) reported that 0.5 μM natural IAA resulted in a greatly enhanced co-purification of TIR1-Myc with the Aux/IAA domain II peptide, the synthetic auxin NAA was also able to promote the interaction, but with lower activity, it has a clear promotive effect at 10 μM. However, any changes in TIR1/AFB2 RNA levels were not detected even after treatment with high concentration of IAA (1 μM) in Arabidopsis root (Parry et al. 2009). In this study, the 5, 10 and 50 μM NAA were chose for detecting the CsTIR1 and CsAFB2 responses to exogenous auxin, and found that the expression levels of CsTIR1 and CsAFB2 in treated leaves at 6 h were gradually up-regulated with the increase of NAA concentration. The result indicated that exogenous auxin can regulate the transcript of CsTIR1 and CsAFB2 in cucumber leaves.
In conclusion, this study indicated that CsTIR1 and CsAFB2 may play an important role in cucumber fruit development. Down-regulated CsTIR1 and CsAFB2 transcripts may have promotive effect on cucumber parthenocarpic fruit development. The transcripts of CsTIR1 and CsAFB2 were induced by exogenous hormones. Future studies should utilize transgenic tools to verify CsTIR1 and CsAFB2 functions in cucumber.
Author contribution
Li Cui was responsible for experimental design and results interpretation, and wrote the paper. Tinglin Zhang and Ji Li performed cloning of CsTIR1 and CsAFB2 and qRT-PCR studies. Qunfeng Lou was involved in bioinformatic analysis. Jinfeng Chen helped revise the manuscript. Li Cui was responsible for experimental design and results interpretation, and wrote the paper. Tinglin Zhang and Ji Li performed cloning of CsTIR1 and CsAFB2 and qRT-PCR studies. Qunfeng Lou was involved in bioinformatic analysis. Then, Li Cui was responsible for revising manuscript. Jinfeng Chen helped revise the manuscript and approved manuscript final version.
Abbreviations
- CPPU:
-
N-(2-ehloro-4-Pyidyl)-N′-Phenylurea
- TIR1:
-
Transport inhibitor response 1
- AFB:
-
Auxin signaling F-box
- NAA:
-
a-Naphthalene acetic acid
- 6-BA:
-
6-Benzylaminopurine
- GA3 :
-
Gibberellin
- ABA:
-
Abscisic acid
- ORF:
-
Open reading frame
- UTR:
-
Untranslated region
- SKP1:
-
S-Phase kinase-associated protein 1
- SAPK7:
-
Serine/threonine-protein kinase 7
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Audran–Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012) Genome-wide identification, functional analysis and expression profiling of Aux/IAA gene family in tomato. Plant Cell Physiol 53(4):659–672
Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86(2):263–274
Beyer E, Quebedeaux B (1974) Parthenocarpy in cucumber: mechanism of action of auxin transport inhibitors. J Am Soc Hortic Sci 99(5):385–390
Boonkorkaew P, Hikosaka S, Sugiyama N (2008) Effect of pollination on cell division, cell enlargement, and endogenous hormones in fruit development in a gynoecious cucumber. Sci Hortic 116(1):1–7
de Jong M, Mariani C, Vriezen WH (2009a) The role of auxin and gibberellin in tomato fruit set. J Exp Bot 60(5):1523–1532
De Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009b) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57(1):160–170
Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435(7041):441–445
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell 9(1):109–119
Fos M, Nuez F (1996) Molecular expression of genes involved in parthenocarpic fruit set in tomato. Physiol Plant 98(1):165–171
Fu F, Mao W, Shi K, Zhou Y, Yu J (2010) Spatio-temporal changes in cell division, endoreduplication and expression of cell cycle-related genes in pollinated and plant growth substances-treated ovaries of cucumber. Plant Biology 12(1):98–107
Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell Online 18(8):1873–1886
Goetz M, Hooper LC, Johnson SD, Rodrigues JCM, Vivian–Smith A, Koltunow AM (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145(2):351–366
Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414(6861):271–276
Guilfoyle T, Hagen G, Ulmasov T, Murfett J (1998a) How does auxin turn on genes? Plant Physiol 118(2):341–347
Guilfoyle T, Ulmasov T, Hagen G (1998b) The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell Mol Life Sci 54(7):619–627
Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41(12):1275–1281
Jeffrey C (2008) A review of the Cucurbitaceae. Bot J Linn Soc 81(3):233–247
Kepinski S, Leyser O (2005a) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435(7041):446–451
Kepinski S, Leyser O (2005b) Plant development: auxin in loops. Curr Biol 15(6):208–210
Kim IS, Okubo H, Fujieda K (1992) Endogenous levels of IAA in relation to parthenocarpy in cucumber (Cucumis sativus L.). Sci Hortic 52(1–2):1–8
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40(D1):302–305
Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53(1):377–398
Martinelli F, Uratsu SL, Reagan RL, Chen Y, Tricoli D, Fiehn O, Rocke DM, Gasser CS, Dandekar AM (2009) Gene regulation in parthenocarpic tomato fruit. J Exp Bot 60(13):3873–3890
Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24:55–80
Pandolfini T, Molesini B, Spena A (2007) Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci 12(8):327–329
Parry G, Calderon–Villalobos L, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray W, Bennett M, Estelle M (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci 106(52):22540–22545
Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y (2011) The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J Exp Bot 62(8):2815–2826
Serrani JC, Ruiz–Rivero O, Fos M, García–Martínez JL (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J 56(6):922–934
Talon M, Zacarias L, Primo–Millo E (1992) Gibberellins and parthenocarpic ability in developing ovaries of seedless mandarins. Plant Physiol 99(4):1575–1581
Tan X, Calderon–Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446(7136):640–645
Varoquaux F, Blanvillain R, Delseny M, Gallois P (2000) Less is better: new approaches for seedless fruit production. Trends Biotechnol 18(6):233–242
Walker L, Estelle M (1998) Molecular mechanisms of auxin action. Curr Opin Plant Biol 1(5):434–439
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell Online 17(10):2676–2692
Weiler E, Jourdan P, Conrad W (1981) Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 153(6):561–571
Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127(1):315–323
Yin Z, Malinowski R, Ziółkowska A, Sommer H, Plcader W, Malepszy S (2006) The DefH9-iaaM-containing construct efficiently induces parthenocarpy in cucumber. Cell Mol Biol Lett 11(2):279–290
Acknowledgments
This work was supported by funds from the National Natural Science Foundation of China grant (The 973 Program: 2012CB3904), “Youth Science and Technology Innovation Fund” program of Nanjing agricultural university (No. KJ2012013), “Fundamental Research of Nanjing Agricultural University (Y0201100253)” and “Ph.D. Programs Foundation of Ministry of Education of China (20120097120037)”.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Wang.
Rights and permissions
About this article
Cite this article
Cui, L., Zhang, T., Li, J. et al. Cloning and expression analysis of Cs-TIR1/AFB2: the fruit development-related genes of cucumber (Cucumis sativus L.). Acta Physiol Plant 36, 139–149 (2014). https://doi.org/10.1007/s11738-013-1394-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-013-1394-7