Abstract
Accumulation of reactive oxygen species (ROS) causes oxidative stress under adverse environmental conditions, such as salinity. Ethylene decreases accumulation of ROS induced by salinity, but the mechanism is still unclear. To examine the interactions between salinity and ROS accumulation and the possible role of ethylene metabolism in regulation, we used mutant ein2-5 in Arabidopsis with loss of function in EIN2. The mutant is compared to the wild-type Col-0, completely insensitivity to ethylene at the morphological, physiological and molecular levels. The oxidative responses of the wild type and mutant to salinity were compared. Loss-of-function of EIN2 enhanced sensitivity to salinity, implying that EIN2 is required for plant response to salinity. Furthermore, salinity resulted in accumulation of large amounts of ROS in ein2-5 seedlings when compared with Col-0, suggesting that the loss-of-function of EIN2 exaggerates oxidative stress induced by salinity. Activities of the antioxidant enzymes SOD, POD and CAT decreased significantly in ein2-5 under salinity when compared with Col-0 plants. The expression profiles of the genes Fe-SOD, PODs and CAT1, which code for ROS scavenging enzymes were severely decreased in ein2-5 under salinity compared with Col-0, suggesting that EIN2 was involved in regulating expression of these genes. Taken together, our results demonstrate that loss-of-function of EIN2 increased oxidative stress induced by salinity and that EIN2 is involved in modulating ROS accumulation, at least in part, by decreasing activities of ROS-scavenging enzymes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity decreases plant growth and production, but the mechanisms of tolerance to salinity are poorly understood (Läuchli and Grattan 2007). If mechanisms were better understood, it might be possible to improve tolerance of plants to salt, particularly crop species. One mechanism by which salt is known to damage plants is the production of reactive oxygen species (ROS), such as O ·−2 , H2O2 and HO2 ·. These are generated as by-products of the essential energy-generating processes in photosynthesis and respiration (Van Breusegem and Dat 2006). In the case of photosynthesis, unfavorable environmental conditions, such as salt stress, especially under high light intensity or in combination with other stresses, disrupt photosynthesis and increase photorespiration, altering the normal homeostasis of cells and causing increased production of ROS (Miller et al. 2010). ROS is also produced by the action of light on protochlorophyllide, the immediate precursor of chlorophyll. If protochlorophyllide cannot be converted to chlorophyll, it accumulates and produces large amounts of ROS (Zhong et al. 2009). Because they are highly reactive, ROS cause irreversible damage to major cellular components, such as lipids, proteins, and nucleic acids (Apel and Hirt 2004; Miller et al. 2010). Although ROS have the potential to cause oxidative damage to cells during environmental stresses, recent studies have shown that low concentrations of ROS play a key role in signal transduction involved in mediating responses to various abiotic and biotic stresses. These two, somewhat opposing, facets of ROS underscore the need to control the steady-state concentration of ROS in cells during normal metabolism, as well as in response to different stresses (Miller et al. 2010). ROS accumulation during stress greatly depends on the balance between ROS production and ROS scavenging (Miller et al. 2010; Mittler et al. 2004). Therefore, ROS-scavenging mechanisms have an important role in protecting plants against stress conditions (Miller et al. 2010). ROS-scavenging enzymes, such as superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) that can eventually break down ROS to nontoxic molecules are essential for normal metabolism. Elucidating the mechanisms that control ROS accumulation during salt stress is important for enhancing plant tolerance to adverse environments.
Ethylene is the only known gaseous plant hormone and classically recognized to affect plant developmental processes and fitness responses such as germination, flower and leaf senescence, fruit ripening and leaf abscission (Bleecker and Kende 2000; Johnson and Ecker 1998; Pierik et al. 2006). In addition, the involvement of ethylene in plant stress responses has been highlighted (Achard et al. 2006; Cao et al. 2007; Jung et al. 2009; Wang et al. 2007). In the past two decades, the linear signal pathway of ethylene has been established based on the identification of ethylene-response mutants in Arabidopsis (Cao et al. 2008). Ethylene is perceived by a five-member family of receptors (Dong et al. 2010; Vandenbussche et al. 2012). These receptors then transmit the signal to the downstream components constitutive triple response 1 (CTR1), ethylene insensitive 2 (EIN2) and ethylene insensitive 3 (EIN3), and coordinately and negatively regulate the responses to ethylene (Alonso et al. 1999; Alonso and Stepanova 2004; Bleecker and Kende 2000; Chao et al. 1997; Chen et al. 2005). The membrane-localized EIN2 protein is a central component in the ethylene signal transduction pathway, as it is the first positive regulator in the pathway. Hence, the EIN2 deficient loss of function mutant ein2-5 displays complete ethylene insensitivity (Alonso et al. 1999; Chen et al. 2005). Greening in the cotyledons of this mutant was more inhibited by NaCl than that of Col-0, but root length was similarly affected in both (Lin et al. 2012).
Accumulating evidence shows a connection between ethylene and cellular ROS production in many species (de Jong et al. 2002; Jung et al. 2009). For example, the overexpression of transcription factor JERF3 enhanced the expression of genes for ROS-scavenging enzymes and tolerance to salt (Wu et al. 2008). EIN2, as a bifunctional transducer of ethylene and stress responses (Alonso et al. 1999), plays important roles in mediating ozone stress, salt stress, ultraviolet-B (UV-B) stress, nutrient deficiency and lead stress (Alonso et al. 1999; Cao et al. 2009; Jung et al. 2009; Lei et al. 2011; Sun et al. 2011; Wang et al. 2007). However, it is unclear how EIN2 regulates salinity-induced production and accumulation of types of ROS, which are regarded as good candidates in mediating hormonal interaction (Ye et al. 2012).
Our previous work with Arabidopsis has demonstrated that disruption of EIN2 exaggerated ROS (H2O2) production and ultimately increased the sensitivity of plants to salinity during seed germination and seedling development (Lin et al. 2012; Yang et al. 2010). In the literature (Ellouzi et al. 2011), this is associated with accumulation of MDA, indicating the damaging effects of ROS.
The interactions between ROS accumulation, the effects of NaCl and role of ethylene are described in Fig. 1. Ethylene concentration, determined either endogenously or exogenously, is detected by the ethylene receptor (ETR), which then activates CTR1. This, in turn, activates EIN2 which interacts with EIN3/EIL1 to give the response of growth to ethylene. We hypothesize that EIN2 also regulates expression of genes for ROS scavenging enzymes, gene expression would be increased by salinity, ultimately leading to tolerance of salinity. Mutation in EIN2 (ein2-5) would decrease the ability of ein2-5 to stimulate gene expression, thus decreasing enzyme production and so allowing ROS to accumulate.
In this study, we investigated the underlying mechanisms involving ethylene and ROS under salinity. We hypothesize that EIN2 modulates ROS accumulation under salinity, at least in part, through regulating antioxidant systems as well as expression of genes such as Fe-SOD, PODs and CAT1.
Our results show that the ethylene signaling pathway component EIN2 is involved in regulating plant ROS accumulation under salinity, and that it may act as a limiting factor in the process, because loss-of-function of EIN2 exaggerates oxidative stress induced by salinity. These results have significance in understanding the function of ethylene signaling and EIN2 during plant response to salinity stress.
Materials and methods
Plant materials and growth conditions
Arabidopsis Columbia ecotype Col-0, and the ethylene-insensitive mutant ein2-5 (Alonso et al. 1999) were used in experiments. Seeds were sterilized in 70 % (v/v) ethanol containing 1 % (v/v) Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA), then sown on agar plates including MS basal salt mixture (Sigma-Aldrich, St Louis, MO, USA), 1 % sucrose, pH 5.7, 0.8 % agar. The plates were kept at 4 °C in the dark for 2 days and then transferred to light at 23 °C with a 16/8 h light/dark regime.
Preliminary experiments with seedling grown with various NaCl concentrations showed that 100 mM NaCl decreased root growth by about 40 % and so this concentration was used to compare the effects of salinity on the mutant. Similarly, the response to ACC was tested and 50 μM adopted for the study. For different treatments, NaCl and ACC were added to the medium singly or coordinately as indicated. All the experiments were repeated three times with three replicates of each treatment.
Determination of chlorophyll and protochlorophyllide
Chlorophyll in seedlings was extracted and assayed according to Hiscox and Israelstam (1979). Protochlorophyllide was extracted as described by Shin et al. (2009) and determined from the relative fluorescence emission spectra of the samples obtained with a F-7500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan), with an excitation wavelength of 440 nm. Spectra were recorded between 600 and 700 nm with a bandwidth of 5 nm. Experiments were performed in triplicates for each treatment.
Microscopy and analysis
ROS accumulation in cotyledons was detected by the fluorescence probe H2DCF-DA (Sigma, St. Louis, MO, USA) according to Schopfer et al. (2001). Fluorescence microscopic images were acquired using a DM 4000B stereomicroscope (Leica, Germany). For measurements of ROS concentration in seedling roots, seedlings were incubated in the H2DCF-DA (10 μM) for 5 min and then washed with distilled water. After that the roots were observed and imaged with a laser scanning fluorescence microscope (Nikon C1 Plus, Japan). Dye excitation was at 488 nm; emitted light was detected at 522 nm. ROS fluorescence was quantified by the NIH ImageJ software program (Jung et al. 2009). For detection of superoxide anion (O ·−2 ) within the root tip, roots of intact plants were stained according to Dunand et al. (2007) for 15 min in a solution of 2 mM nitroblue tetrazolium (NBT) in 20 mM phosphate buffer (pH 6.1). The reaction was stopped by transferring the seedlings to distilled water. Then, the roots were observed and photographed with a DM 4000B stereomicroscope (Leica, Germany). Assessment of the staining intensity in the elongation zone was done with NIH ImageJ software. Both the experiments were repeated three times. At least 20 individual roots were analyzed for each genotype and treatment, and one typical image was selected for the figure.
Assay of antioxidant enzyme activity
For the detection of antioxidant enzyme activities, seedlings (0.5 g) were ground in liquid nitrogen and extracted in 5 mL of Tris–HCl buffer (50 mM, pH 7.0) containing 1 mM EDTA-Na2 and 1 % (w/v) soluble polyvinylpyrrolidone. The homogenates were centrifuged (10,000g, 30 min, 4 °C) and the supernatant was collected and used for the assay of enzyme activities. SOD (E.C. 1.15.1.1) activity was determined by measuring its ability to inhibit photochemical reduction of NBT following Beauchamp and Fridovich (1971) with modifications. The reaction mixture was 50 mM sodium phosphate buffer (pH 7.3), methionine (13 mM), NBT (75 mM), EDTA (0.1 mM), riboflavin (4 mM) and 0.1 ml of the extract. The reaction was started by the addition of riboflavin, and carried out for 20 min under 170 μmol photons m−2 s−1 provided by fluorescent lamps. The absorbance at 560 nm was determined. One unit of SOD was defined as the amount of enzyme that produced 50 % inhibition of NBT reduction.
POD (E.C. 1.11.1.7) was assayed according to Rathmell and Sequeira (1974) as follows: The reaction mixture contained 1.8 ml of 100 mM sodium phosphate buffer (pH 6.0), 0.1 ml guaiacol, 0.1 ml of 12 mM H2O2 and 0.1 ml of leaf extract. The formation of the conjugate product of guaiacol was measured at 460 nm. The increase in A436 was measured using an extinction coefficient of 26.6 mM−1 cm−1 for the conjugate as it was formed.
CAT (E.C. 1.11.1.6) activity was determined according to Aebi (1984). The decomposition of H2O2 was followed at 240 nm in a quartz cuvette (extinction coefficient of H2O2 was 0.04 mM−1 cm−1). The reaction mix consisted of 2.7 ml 0.1 M sodium phosphate buffer (pH 7.0), 0.1 ml of 300 mM H2O2 solution and 0.2 ml of the extract. All experiments were performed in triplicate for each treatment.
Expression analysis
Quantitative real-time PCR (qRT-PCR) was performed in a quantitative PCR instrument (Bole, USA). Total RNA was extracted from seedlings 6 h before or after treatment using an RNAiso Plus kit (Takara Inc., Japan) according to the manufacturer’s instructions. Total RNA (5 μg) was used in reverse transcription with a RevertAid Reverse Transcriptase and an oligo d(T)primers (TaKaRa, Japan). qRT-PCR was performed using a RealMasterMix kit (Tiangen, China) with 40 cycles as follows: 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 10 min. All quantifications were normalized to the amplification of ACTIN2 gene (locus number At3g18780). Primer sequences of all genes used for qRT-PCR are listed in Table 1. Experiments were performed in triplicate for each treatment.
Statistical analysis
Statistical analyses were carried out using the SPSS-17 statistical software package. The results are represented as the mean ± standard error (SE). Differences between treatments were separated by the least significant difference (LSD) test at 0.05 probability.
Results
Sensitivity of seedlings of ein2-5 and Col-0 plants to salt stress
Without application of NaCl, under optimal culture conditions, there were no significant differences between ein2-5 and wild-type plants. However, when cultured on media with NaCl, ein2-5 was more sensitive than Col-0 (Fig. 2a). Root elongation of ein2-5 was significantly decreased by salt when compared with the wild type (Fig. 2b). The increased sensitivity of ein2-5 to NaCl was also shown by the larger reduction in total chlorophyll content in ein2-5 compared with the wild type (Fig. 2c). Furthermore, the results also showed that ein2-5 seedlings accumulated more protochlorophyllide than Col-0 under NaCl (Fig. 2d).
The ein2-5 mutant plants were more sensitive to salinity stress than Col-0 plants. a Representative photographs of Col-0 and ein2-5 plants germinated and grown on MS agar plates with or without 100 mM NaCl as indicated for 6 days. b Primary root length of Col-0 and ein2-5 plants is shown in a. c, d Total chlorophyll content and the amount of protochlorophyllide of Col-0 and ein2-5 plants is shown in a, respectively. Values shown are mean ± SE, bars indicate standard errors. The letters indicate statistically significant differences (P < 0.05)
Effects of salinity on ROS accumulation in ein2-5 and Col-0
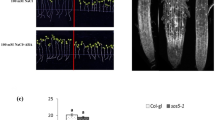
We first investigated the accumulation of ROS in cotyledons of seedlings treated with NaCl. As the results (Fig. 3a) indicate NaCl treatment increased ROS accumulation drastically in the ein2-5 mutant, but little in the wild-type plants. In addition, we also measured ROS accumulation in seedling root tips. Larger amounts of ROS were found in NaCl-treated ein2-5 root tips than in Col-0 plants (Fig. 3b). Moreover, the increased ROS accumulation induced by salinity was alleviated in Col-0, but not in ein2-5 by the ethylene biosynthesis precursor, ACC (Fig. 3b).
Accumulation of ROS in Col-0 and ein2-5 plants germinated and grown on MS agar plates with or without 100 mM NaCl as indicated for 6 days. a, b Representative images for ROS accumulation of cotyledons and roots indicated by H2DCF-DA in Col-0 and ein2-5 plants, respectively. c NBT (nitroblue tetrazolium) staining to show the presence of superoxide anions in Col-0 and ein2-5 roots. Values shown are mean ± SE. The letters indicate statistically significant differences (P < 0.05)
Accumulation of superoxide anion (O ·−2 ), a main form of ROS, displayed the similar pattern as ROS accumulation in roots of both mutant and Col-0 seedling (Fig. 3c). Salinity significantly stimulated O ·−2 accumulation, especially in ein2-5 seedlings which increased fourfold compared with control under NaCl. Similarly, application of ACC significantly inhibited ROS accumulation in Col-0 but not in ein2-5.
Activities of ROS-scavenging enzymes SOD, POD and CAT under salinity
The antioxidant systems of the ethylene response mutant ein2-5 with Col-0 plants were studied (Fig. 4). SOD activity was the same in both Col-0 and the mutant without NaCl, whereas it was much larger with NaCl in both plants, particularly in Col-0. POD activity was higher in ein2-5 than that in Col-0 without NaCl was unaffected by NaCl in Col-0, but great decreased in ein2-5. Under optimal condition, without NaCl, the ein2-5 mutant had lower activity of CAT than that Col-0 plants.
Activities of ROS-scavenging enzymes SOD (a), POD (b) and CAT (c) in Col-0 and ein2-5 plants germinated and grown on MS agar plates with or without 100 mM NaCl as indicated for 6 days. Values shown are mean ± SE, bars indicate standard errors. The letters indicate statistically significant differences (P < 0.05)
Expression profiling of ROS-related genes in seedlings under salt stress
Under salt stress, the expression of DEFL, a well-established ROS marker gene, was up-regulated in both Col-0 and ein2-5 seedlings. Generally, the expression was higher in ein2-5 than in Col-0 particularly with NaCl (Fig. 5a). Furthermore, in the Col-0 seedlings, a significant up-regulation of the genes FeSOD, PODs and CAT1 under salt stressed conditions was observed compared to ein2-5 seedlings in which no change occurred with NaCl (Fig. 5b–d).
Discussion
Under stress conditions, such as salinity, the rate of ROS production was dramatically elevated as already shown in the literature (Miller et al. 2010). As these radicals are capable of causing oxidative damage to proteins, DNA and lipids, and are therefore toxic, strict control of ROS concentration is essential to prevent damage to metabolism and to ensure accurate execution of their signaling functions. In the present study, we highlight the role of EIN2 in modulating ROS accumulation under salt stress. The results also demonstrate that EIN2 modulates salinity-induced ROS accumulation, at least in part, through regulating antioxidant activities, and we show that this is probably the consequence of regulating expression of genes for ROS-scavenging enzymes.
EIN2 is a central component of the ethylene signaling transduction pathway in plants, and its null mutant ein2 in Arabidopsis is completely insensitive to ethylene (Alonso et al. 1999; Cao et al. 2008). Mutations in EIN2 delayed leaf senescence (Grbić and Bleecker 1995) and resulted in constitutive activation of several antioxidant enzymes that conferred enhanced resistance to oxidative stress (Alonso et al. 1999). However, our results suggest that EIN2 play a different role in plant response to oxidative stress induced by salinity.
Under salt stress seed germination and seedling growth of plants with ein2-1, an allele of ein2-5, were much more severely inhibited by salinity than the wild type, suggesting that EIN2 is important for salt tolerance (Cao et al. 2007; Wang et al. 2007). However, the underlying mechanisms which determine the activity of antioxidant enzymes and thereby ROS accumulation have been rarely studied. Our new results show that sensitivity of EIN2 mutants is due to decreased gene expression and thus to less antioxidant enzyme activity which lead to ROS accumulation and inhibition of function.
In cotyledons such as we studied, overaccumulation of protochlorophyllid, the precursor of chlorophyll, would produce large amounts of ROS and cause oxidative damage to ein2-5 seedlings (Fig. 2d). This showed that the increased sensitivity to salinity of ein2-5 was correlated with the increased ROS production induced by salinity (Yang et al. 2010), and this may due to the mutants inability to scavenge ROS.
Ethylene acts upstream of ROS in Arabidopsis roots under adverse environments (Jung et al. 2009), and our findings were consistent with this (Fig. 3b, c). The EIN2 gene encodes a master positive regulator in the ethylene signaling pathway and is involved in response to oxidative stress (Alonso et al. 1999). It is now possible to interpret the results of our study, based on the important roles of EIN2 in regulating ROS accumulation in salinity stressed seedlings. Before salinity treatment, there were no significant differences with respect to ROS accumulation in cotyledons or roots as indicated in situ by molecular probes (Fig. 3). However, salinity significantly enhanced ROS accumulation in ein2-5 as compared to Col-0 plants. It appears that loss-of-function of EIN2 exaggerates salinity induced ROS accumulation in Arabidopsis.
For detoxification of ROS, particularly during stress condition, ROS-scavenging enzymes such as SOD, POD and CAT are essential (Gechev et al. 2006; Miller et al. 2010). SOD scavenges O ·−2 generated during metabolism, and is thus a key enzyme in cellular defense, generating H2O2 (Gechev et al. 2006). Increased production of O ·−2 caused by salinity induces gene expression and increases the activity of SOD in both the wild type and mutant plants. However, they were changed less in ein2-5 than in Col-0 suggesting that EIN2 is involved in regulating SOD activity. This is achieved, at least in part, through modulation of the expression of Fe-SOD under salinity. The activity of CAT in salt stressed ein2-5 plants was increased significantly compared with control plants, indicating that there is a high concentration of H2O2 in NaCl-stressed plants, as CAT is active only at relatively high H2O2 concentrations (Gechev et al. 2006). Taken together, these results highlight that loss-of-function of EIN2 exaggerates oxidative stress caused by salinity and rules out that EIN2 is involved in the process by modulating activities of ROS-scavenging enzymes, such as SOD, POD and CAT (Fig. 4).
Stress acclimation and adaptation in plants depend largely on changes in gene expression that initiate remodeling in metabolism and physiology in response to adverse environmental cues (Wang et al. 2007). After treatment with salinity, ein2-5 showed a greater expression of a well-established ROS marker gene DEFL (At2g43510), previously shown to be ubiquitously induced by various ROS (Gadjev et al. 2006), but not of the genes for SOD, POD or CAT, showing why ein2-5 was particularly sensitive to oxidative stress induced by increased ROS accumulation. It is known that SOD has three different isoenzymes distributed in different organelles. The more important isoenzyme, Fe-SOD, is found in chloroplasts (Alscher et al. 2002). The increase of Fe-SOD expression under salinity implies that chloroplasts are more sensitive to ROS than other cellular organelles (McKersie et al. 2000; Wu et al. 1999), probably because of generation of ROS by light acting on protochlorophyllide and chlorophyll. Chloroplasts may be less able to scavenge abiotic induced ROS, and decrease it is accumulation. However, disruption of EIN2 decreases gene expression and production of antioxidant enzymes, making the chloroplasts even more sensitive to salinity. Measurements of transcript levels of other ROS-scavenging genes, including PODs and CAT1, showed that their expression was also disrupted in ein2-5 compared with Col-0 under salinity (Fig. 4). This underscores the involvement of EIN2 in regulating expression of ROS-scavenging genes. Taken together, we conclude that EIN2 modulates salinity induced ROS accumulation through the ROS-scavenging pathway.
It has been proposed that EIN2 acts as a cross-talk point for multiple signaling pathways involving hormones and stresses (Alonso et al. 1999; Bouchez et al. 2007; Cao et al. 2008; Wang et al. 2007). In parallel, small concentrations of ROS are involved in mediating plant responses to abiotic and biotic environmental stimuli as signal transduction molecules (Miller et al. 2010; Mittler et al. 2004; Torres and Dangl 2005). EIN2 may integrate stress signaling and hormone interactions to regulate gene expression and metabolic adjustments, and thus determine the equilibrium during plant development in the presence of abiotic stress such as salinity. Being located upstream of transcription factors in abscisic acid (ABA) induced gene expression, EIN2 regulates plant developmental responses through an ABA responsive pathway under salinity stress (Wang et al. 2007). Loss-of-function mutation in EIN2 increased ABA content, and ROS accumulation increased under stress conditions (Gechev et al. 2006; Jiang and Zhang 2001; Zhang et al. 2001). Therefore, we cannot exclude the possibility that EIN2 also modulates ROS accumulation under salinity through an ABA pathway.
In the introduction, we proposed a regulatory model for EIN2 in modulating ROS accumulation under salinity (Fig. 1) and our results support the hypothesis suggested by it. In the model, ethylene is perceived and transduced, affecting the transcription factors EIN3/EIL and initiating the ethylene response. Genetic data indicate that EIN2 mediates an essential step in the signal propagation between the CTR1 and EIN3/EIL. In the linear signal pathway, EIN2 could modulate ROS concentration by up-regulating expression of genes such as Fe-SOD, PODs and CAT1 at the transcriptional level. These genes encode for ROS-scavenging antioxidant enzymes such as SOD, POD and CAT and decrease salinity-induced ROS effectively, thus ultimately, decreasing oxidative stress induced by ROS and improving plant tolerance to salinity. In conclusion, we show that loss-of-function in EIN2, giving the ein2-5 plants, exaggerates oxidative stress induced by salinity, suggesting that EIN2 is a limiting factor in the regulation of metabolism and adaptation to salinity. The studies have emphasized that EIN2 disruption alters expression of Fe-SOD, PODs and CAT1 genes under salinity stress, and so decreases activities of antioxidant enzymes, such as SOD, POD and CAT in ein2-5 plants compared with Col-0, indicating that EIN2 is involved in regulating ROS-scavenging enzymes and thus ROS accumulation, at least in part, through regulating gene expression.
Author contribution
Y. L. and Z. T. designed the research, Y. L. and D. C. performed the research, Y. L., Y. Z. and Z. T. analyzed the data, and Y. L., M. P. and Z. T. wrote the paper.
Abbreviations
- ACC:
-
1-Aminocyclopropane-1-carboxylic acid
- CAT:
-
Catalase
- Col:
-
Columbia
- CTR1:
-
Constitutive triple response 1
- EIN2:
-
Ethylene insensitive 2
- EIN3:
-
Ethylene insensitive 3
- ETR:
-
Ethylene receptor
- H2DCF-DA:
-
2′,7′-Dichlorodihydrofluorescein diacetate
- H2O2 :
-
Hydrogen peroxide
- JERF3:
-
Jasmonate and ethylene-responsive factor 3
- MDA:
-
Malondialdehyde
- NBT:
-
Nitroblue tetrazolium
- POD:
-
Peroxidase
- qRT-PCR:
-
Quantitative real time-PCR
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
References
Achard P, Cheng H, Grauwe LD, Decat J, Schoutteten H, Moritz T, Straeten DVD, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311:91–94
Aebi H (1984) Catalase in vitro. Methods Enzymol 105:121–126
Alonso JM, Stepanova AN (2004) The ethylene signaling pathway. Science 306:1513–1515
Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284:2148–2152
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53:1331–1341
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Beauchamp C, Fridovic I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44:276–287
Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16:1–18
Bouchez O, Huard C, Lorrain S, Robby D, Balagué C (2007) Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol 145:465–477
Cao WH, Liu J, He XJ, Mu RL, Zhou HL, Chen SY, Zhang JS (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143:707–719
Cao YR, Chen SY, Zhang JS (2008) Ethylene signaling regulates salt stress response. Plant Signal Behav 3:761–763
Cao SQ, Chen ZY, Liu GQ, Jiang L, Yuan HB, Ren G, Bian XH, Jian HY, Ma XL (2009) The Arabidopsis ethylene-insensitive 2 gene is required for lead resistance. Plant Physiol Biochem 47:308–312
Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSI-TIVE3 and related proteins. Cell 89:1133–1144
Chen Y, Etheridge N, Schaller GE (2005) Ethylene signaling transduction. Ann Bot 95:901–915
de Jong AJ, Yakimova ET, Kapchina VM, Woltering EJ (2002) A critical role for ethylene in hydrogen peroxide release during programmed cell death in tomato suspension cells. Planta 214:537–545
Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C (2010) Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biochem 285:40706–40713
Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol 174:332–341
Ellouzi H, Hamed KB, Cela J, Munné-Bosch S, Abdelly C (2011) Early effects of salt stress on the physiological and oxidative status of Cakile maritima (halophyte) and Arabidopsis thaliana (glycophyte). Physiol Plant 142(2):128–143
Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141:436–445
Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28:1091–1101
Grbić V, Bleecker AB (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J 8:595–602
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Jiang M, Zhang J (2001) Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol 42:1265–1273
Johnson PR, Ecker JR (1998) The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet 32:227–254
Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21:607–621
Läuchli A, Grattan SR (2007) Plant growth and development under salinity stress. In: Jenks MA, Hasegawa P, Jain SM (eds) Advances in molecular breeding toward drought and salt tolerant crops. Springer, Netherlands, pp 1–32
Lei G, Shen M, Li ZG, Zhang B, Duan KX, Wang N, Cao YR, Zhang WK, Ma B, Ling HQ, Chen SY, Zhang JS (2011) EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ 34:1678–1692
Lin Y, Wang J, Zu Y, Tang Z (2012) Ethylene antagonizes the inhibition of germination in Arabidopsis induced by salinity by modulating the concentration of hydrogen peroxide. Acta Physiol Planta 34:1895–1904
McKersie BD, Murnaghan J, Jones KS, Bowley SR (2000) Iron-superoxide dismutase expression in transgenic alfalfa increases winter survival without a detectable increase in photosynthetic oxidative stress tolerance. Plant Physiol 122:1427–1438
Miller G, Suzuki N, Ciftci-Yilma S, Mittler R (2010) Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ 33:453–467
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498
Pierik P, Tholen D, Poorter H, Visser E, Voesenek L (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11:176–183
Rathmell WG, Sequeira L (1974) Soluble peroxidase in fluid from the intercellular spaces of tobacco leaves. Plant Physiol 53:317–318
Schopfer P, Plachy C, Frahry G (2001) Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol 125:1591–1602
Shin R, Kim K, Kang H, Zulfuarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negative-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106:7660–7665
Sun ZH, Chen W, Chen ZY, Zhang QA, Fang L, Li J, Jiang ST, Cao SQ (2011) A role for Ethylene-Insensitive 2 gene in the regulation of the ultraviolet-B response in Arabidopsis. Acta Physiol Planta 33:1025–1030
Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8:397–403
Van Breusegem F, Dat JF (2006) Reactive oxygen species in plant cell death. Plant Physiol 141:384–390
Vandenbussche F, Vaseva I, Vissenberg K, Van Der Straeten D (2012) Ethylene in vegetative development: a tale with a riddle. New Phytol 194:895–909
Wang YN, Liu C, Li K, Sun F, Hu H, Li X, Zhao Y, Han C, Zhang W, Duan Y, Liu M, Li X (2007) Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol Biol 64:633–644
Wu G, Wilen RW, Robertson AJ, Gusta LV (1999) Isolation, chromosomal localization, and differential expression of mitochondrial manganese superoxide dismutase and chloroplastic copper/zinc superoxide dismutase genes in wheat. Plant Physiol 120:513–520
Wu LJ, Zhang Z, Zhang H, Wang XC, Huang R (2008) Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol 148:1953–1963
Yang L, Zu YG, Tang ZH (2010) Ethylene improves Arabidopsis salt tolerance mainly via retaining K+ in shoots and roots rather than decreasing tissue Na+ content. Environ Exp Bot. doi:10.1016/j.envexpbot.2010.08.006
Ye N, Zhu G, Liu Y, Zhang A, Li Y, Liu R, Shi L, Jia L, Zhang J (2012) Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J Exp Bot 63:1809–1822
Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP (2001) Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol 126:1438–1448
Zhong S, Zhao M, Shi T, Shi H, An F, Zhao Q, Guo H (2009) EIN3EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA 106:21431–21436
Acknowledgments
We thank Prof. Hongwei Guo (Peking University) for providing Arabidopsis seeds and Dr. Zhong-hua Zhang for technical assistance. We are also grateful to the two anonymous reviewers for their valuable comments on the work. This study was supported by the National Natural Science Foundation of China (Grant No. 31000176).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Z.-L. Zhang.
Rights and permissions
About this article
Cite this article
Lin, Y., Chen, D., Paul, M. et al. Loss-of-function mutation of EIN2 in Arabidopsis exaggerates oxidative stress induced by salinity. Acta Physiol Plant 35, 1319–1328 (2013). https://doi.org/10.1007/s11738-012-1172-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-012-1172-y