Abstract
The adipose tissue is a highly regulated endocrine and paracrine organ that secretes a wide variety of biologically active molecules involved in the control of energy balance and the regulation of body weight. Our work aimed to analyze the dysregulation of the adipocyte metabolism and compare the gene expression patterns between omental (OM) and subcutaneous (SC) adipose tissue from obese and lean subjects by using whole-genome DNA microarrays. OM and SC adipose tissues were obtained from 43 obese subjects undergoing bariatric surgery and from six lean individuals. Gene expression analysis was performed by whole-genome microarrays and Taqman RT-PCR. The analysis of microarrays showed upregulation of 545 genes in OM and 47 in SC adipose tissue, whereas 723 and 27 genes were downregulated in OM and SC tissue, respectively, in obese patients. Significantly altered genes showed at least a twofold change of p < 0.05. Validation of the arrays with 28 genes was carried out by using low density microfluidic cards which confirmed the changes found in most genes. We focused on the altered expression of gene coding for enzymes and transcription factors involved in lipid metabolism. Interestingly, some of these genes have not been previously described in obesity. Our results show that adipose tissue from obese subjects entails defense mechanisms against an excessive expansion and fat accumulation, repressing both lipogenesis and lipolysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Obesity is a multifactorial illness with severe associated metabolic comorbidities such as diabetes, cardiovascular damage, hyperlipemia, or cancer whose prevalence is increasing throughout the world alarmingly. Obesity is characterized by the accumulation of body fat produced by an imbalance between caloric intake and energy expenditure.
The anatomical distribution of adipose tissue plays an important role in the development of these metabolic disorders. Subcutaneous (SC) and omental (OM) fat depots are distinct and produce different metabolic effects: Omentectomy leads to a decrease of glucose and insulin levels [1], while the elimination of SC adipose tissue by liposuction does not improve the metabolic syndrome [2]. Recently, transplantation of SC adipose tissue into the OM depot and vice versa has been performed successfully in mice [3, 4]; however, only the transplantation of SC adipose tissue into the abdominal cavity had beneficial effects, improving insulin sensitivity, glucose tolerance, and reducing body weight. Furthermore, there is an evidence for a role of developmental genes in the origin of obesity and body fat distribution, suggesting that genetically programmed developmental differences in adipocytes and their precursors in different regions of the body play an important role in obesity, body fat distribution, and potential functional differences between visceral and SC adipose tissue.
The visceral adipose tissue (OM included) is known to exhibit morphological and functional differences in comparison with SC fat, including differences of insulin action on regulation of lipolysis. The visceral adipose tissue is more abundant in men and is composed by the mesenteric fat and the epiplon or omentum. The amount of visceral fat is about 21% of the total body fat in men and only 8% in females; however, while there is a continuous increase of absolute visceral fat volume with increasing obesity in men, women do not seem to increase their visceral fat substantially until they have accumulated some 30 L of total fat volume. Above this level, the ratio of visceral to total fat storage is similar in both genders [5]. On the contrary, the SC adipose tissue is more abundant in females and is associated to an improved sensitivity to insulin and a lower risk to develop obesity-associated disorders.
So far, no whole-genome studies have been conducted to characterize possible changes in the gene expression profile comparing both OM and SC adipose tissue from obese versus lean subjects, as only partial array experiments have been conducted previously involving partial genome, a unique localization, or a unique condition (obese or lean).
In the present study, we analyzed the effect of obesity on gene expression profile of obese human SC and OM adipose tissue versus lean patients by using DNA microarrays and real-time PCR. Differences between both fat depots indicate new target genes not previously found in obesity.
Methods
Subjects
Patients were classified as obese or lean according to their body mass index (BMI), following the World Health Organization criteria, BMI ≥30 and <25 kg/m2, respectively. Surgeries were performed at the Hospital Universitario La Paz (Madrid), Hospital Clínico S. Carlos (Madrid), and Clínica Universitaria de Navarra (Pamplona). Approval of the protocols was obtained from the Ethical Committees of the Hospitals and our institution, in agreement with the European laws on biomedical research and with the principles of the Declaration of Helsinki.
For microarrays experiments, human adipose tissue was obtained from the OM and SC depots from 11 obese patients (seven females and four males) undergoing bariatric surgery and from six lean subjects (three males and three females) by Nissen fundoplication, after giving their informed consent. Four lean patients presented gastroesophageal reflux or hiatus hernia, but no other pathology. SC samples from two of the lean females were obtained by liposuction. For the validation experiments, further 32 obese patients (17 females and 15 males) providing 30 samples from each depot because some of them were not paired were included in the analysis maintaining the samples of the lean subjects used in the arrays. Most of the lean and obese patients were included in a previous study to address a completely different question [6].
RNA Extraction and Microarray Hybridization
RNA of each adipose sample was extracted with the RNeasy lipid tissue kit (Qiagen Madrid, Spain) following the manufacturer’s protocol. Integrity and concentration of RNA were checked with the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
RNA was amplified into digoxigenin-labeled cRNA using Applied Biosystem Chemiluminiscent RT-ITV Labeling KIT (Foster City, CA, USA) for hybridization at 55°C for 16 h to “Human Genome Survey Microarray V2.0” from Applied Biosystems (Foster City, CA, USA), containing 60 bp oligonucleotides specific for 32,878 probes corresponding to 29,098 genes. These microarrays contain an additional 2,180 annotated genes to a total of 29,098 genes identified, based on a combination of the public and Celera Genomics databases, computational annotation, and manual curation. Ten arrays were utilized for OM adipose tissue and 14 for SC adipose tissue from men and women.
Detection at 458 nm was performed with the Chemiluminiscent Detection kit (Applied Biosystems, Foster City, CA, USA), and images were collected using the 1700 Chemiluminiscent Microarray Analyzer. Chemiluminiscent signals were quantified, corrected for background and spot, and spatially normalized, according to the platform manufacturer’s recommendation with the Applied Biosystems software (Foster City, CA, USA). Thus, probes having a S/N ratio below 3 and a Flag value above 5,000 in all samples were discarded. Before being compiled into an expression profile data matrix, all arrays were quantile-normalized to be comparable. This quantile normalization procedure transforms the data into log2 scale.
Genes differentially expressed were selected by using the lineal models of the LIMMA package [7] from the Bioconductor project (http://www.bioconductor.org) [8] contained in the Spotfire Integromics software. p values were calculated by two sample t test and used to calculate adjusted p value as false discovery rate (FDR; or expected proportion of false positives among the rejected hypotheses) using the Benjamini and Hochberg [9] procedure also included in the Bioconductor package.
The genes differentially expressed (obese versus lean) were classified by Protein Analysis Through Evolutionary Relationships (PANTHER) which is a freely available, comprehensive online software system for relating gene sequence to specific molecular functions, biological processes, and pathways (http://www.pantherdb.org/)
Real-Time PCR Validation
To validate the data obtained from the arrays, we used Taqman low density arrays (LDAs) also called microfluidic cards with specific Taqman probes. Based on quantitative real-time PCR, LDAs maintain the technical sensitivity and allow to quantify simultaneously a maximal number of 384 genes per card. In our study, 18S and IPO8 served as endogenous control genes, but data were referred to IPO8 as we have recently reported IPO8 as an outstanding gene for normalization of gene expression studies in human adipose tissue in contrast to 18S [6]. For validation experiments, we included genes previously described to vary in obesity that served us as positive controls; however, most of the genes to validate the arrays were chosen on the basis of their differential expression obtained in the arrays and their relationship with processes related to obesity, but not previously described in obesity itself. Thus, genes selected by data mining were hybridized to cDNA from 32 additional obese patients and four lean subjects. The cDNA was synthesized from approximately 50 ng of each RNA sample with the High Capacity RNA-to-cDNA Kit (Applied Biosystems).
PCRs were run in an ABI7900 HT Fast Real Time PCR System. Differences in gene expression were measured by the 2−ΔΔCt method, which calculates the mRNA abundance in relation to a control reference gene. Statistically significant differences were determined by the Student t test (p < 0.05).
Results and Discussion
Study Population
The biochemical profile of the lean and obese patients appears in Table 1. As expected, BMI differed significantly between lean and obese subjects. Age and total cholesterol did not show statistical differences. Although significant changes were present in the plasma concentrations of basal glucose and triglycerides between lean and obese patients, they all were within the normal ranges. No differences between the two groups of obese patients (arrays and LDAs) were found.
Global View on Gene Expression Transcripts by Microarrays
The microarray data were quantified, filtered, and normalized as described in the “Methods” section. After filtering, from the original 29,098 genes, 14,564 genes (50%) in the OM depot and 19,183 genes (66%) in the SC depot were expressed with a good quality signal. From the filtered genes, 545 (3.7%) and 47 genes (0.25%) were upregulated in OM and SC tissue, respectively, and 723 (5%) and 27 (0.14%) genes were downregulated in OM and SC fat with a p value <0.05. These figures represent a 43% and 64% of the overexpressed genes in the OM and SC tissues, respectively, and a 57% and 36% of the repressed genes in those tissues. A list of genes significantly altered in both depots is shown in Table 2. Only the upregulated genes with a fold change above 6 and the downregulated genes with a fold change ≤0.25 are shown. Many of the altered genes have not been previously described to be involved in obesity. As shown in Table 2, calgranulin B (S100A9) was a highly induced gene in both OM and SC adipose tissues from obese patients versus leans. S100A9 is a member of the S100 family of calcium-modulated proteins that control vascular inflammation and atherothrombosis, and it has been suggested as a cardiovascular marker in type 1 diabetes [10]. S100A9 complexes with another member of the family (S100A8) and the presence of this complex, called calprotectin, has been shown very recently to be elevated in plasma of obese patients independently of classical inflammation markers [11]. Among the downregulated genes, one of the most repressed in both OM and SC adipose depots was NDRG4 which is involved in response to stress and has been previously reported to be decreased in SC adipose tissue from obese men [12].
Although in the past 10 years numerous studies using microarrays in human adipose tissue have been published, all of them show partial results because they only used tissue from a single depot (OM or SC, but not both) [12–19], or a single group of individuals (lean or obese, but not both) [14, 18–22], or because the collection of the genes used did not include the complete genome [13–15, 20, 21, 23]. This makes the comparison of our results with already published studies difficult to interpret, and some discrepancies may be due to differences in the experimental design.
For example, our arrays differ from other reports performed in SC adipose tissue which show substantially more altered genes than those obtained in our analysis (even with less genes in the arrays). The discrepancy could be due to the use of samples of one single gender; conversely, our study included both men and women adipose tissues samples. To our knowledge, the work by Lee and coworkers [16] is the only one including both genders, but the studied population was slightly different to the general population, as Pima Indians show the highest prevalence rates of obesity and type 2 diabetes.
Biological Processes Associated with Differentially Expressed Genes
One of the aims of the studies with arrays is the interpretation of the altered genes within a biological context. By using the PANTHER® database, we studied the biological processes which are predicted to be significantly altered in both adipose depots by the changes observed in the gene expression profile between obese and lean subjects.
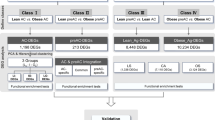
In both OM and SC depots, the genes included in unclassified biological processes were overrepresented among the up- and downregulated genes, exactly 18% and 36% were increased in OM and SC fat, respectively, while 26% and 20% were decreased in those adipose tissues depots (Fig. 1). Excluding this group, the induced genes in OM adipose tissue were found to be grouped mainly in processes of immunity and defense, protein metabolism, and signal transduction, while the repressed genes were associated with the two latter and also with nucleic acid metabolism (Fig. 1). In SC adipose tissue, less biological processes were represented as compared to OM tissue. Immunity and defense and signal transduction were more represented among the upregulated genes. Developmental processes, signal transduction, intracellular protein traffic, and those related to nucleic acid metabolism were predominant among the downregulated genes (Fig. 1).
We next focused our analysis on the genes coding for proteins involved in lipid fatty acid and steroid metabolism, specifically in the lipogenesis and lipolysis pathways. In this context, the proportion of up- and downregulated genes in this pathway was 2.5% and 2.2%, respectively, in OM tissue, while in SC tissue, only repressed genes were found (6%).
Validation of Selected Genes with Microfluidic Cards
In order to validate the microarray data, an independent technique was used with a group of genes. Twenty-eight up- and downregulated genes were further analyzed by quantitative real-time PCR in 30 additional OM and SC adipose samples from obese patients, whose adipose tissues were not included in the microarrays. Most of the chosen genes belonged to the lipid fatty acid and steroid metabolism pathway to allow us to have more confident data in this functional network of huge relevance in bioenergetics and obesity. Some of these genes are known to change in obesity and others correspond to new genes not previously reported to be altered in this pathology and whose expression changes between groups. We have validated genes involved in the molecular mechanisms that regulate the energy balance, such as transcription factors, enzymes of energy metabolism, fatty acid metabolism and cholesterol, and proteins with a role in signaling cascades. We have made two comparisons: obese versus lean patients and OM versus SC location in obese patients.
The quantitative PCR was performed by using Taqman LDAs. The LDAs included 18S and importin-8 (IPO8) as endogenous control genes, but the expression was finally referred to IPO8 as it has been shown to be the most suitable gene in OM and SC depots from obese and lean subjects, as well as in cultured differentiated preadipocytes [6]. The validation of these genes in independent adipose samples reinforces the results from the arrays. Moreover, regarding many of those genes, our data are in agreement with the expression studies recently published by other groups (see below), which validates the general performance and provides confidence in our overall data.
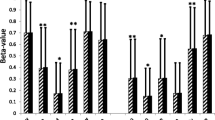
When analyzed by LDAs, the selected genes showed concordant results with those from the arrays, although not all the changes were always significantly different. Results appear in Table 3, which shows the differential expression in obese versus lean subjects in OM and SC adipose tissue in both LDAs and arrays and the ratio OM/SC in obese. Table 3 also shows correlation analysis of the validated genes in the OM and SC depots with the biochemical parameters (BMI, glucose, cholesterol, and triglycerides) of the subjects included in the LDA study.
The most striking result in this expression analysis is the repression of lipid metabolism in general, suggesting a defensive mechanism against an excess of fatty acids. As shown in Table 3, OM adipose tissue is more affected than SC tissue in obesity, confirming the results from the arrays and suggesting the relationship between visceral obesity and other pathologies such as insulin resistance, hyperlipemias, and cardiovascular disease. In addition, most of the significant correlations were found in genes from this depot with the plasmatic triglycerides levels.
Two essential and antagonic pathways, lipolysis and lipogenesis, play a crucial role in energy balance. Alterations in these pathways are associated with obesity [24]. Both are repressed in our study in a significant manner in OM and SC adipose tissue, though repression is higher in the OM depot as determined from our results with the arrays confirmed by microfluidic cards. The more likely cause is the higher sensitivity of OM tissue to catecholamines stimulated lipolysis, whereas the SC tissue is more sensitive to the antilipolytic effect of insulin [24].
The fatty acid synthase (FASN) and acetyl-CoA-carboxylase 2 (ACACB) are essential for lipogenesis. These enzymes play an important role in the development of obesity and regulation of body weight [25]. Their repression blocks the synthesis of triglycerides, which suggests it can be a defensive mechanism against an excess of fatty acids. The repression of FASN decreases the body weight in rodents, increases the β-oxidation of fatty acids, and decreases the appetite suggesting a role as a regulator of energy homeostasis [25–27]. Our array data show a decrease in FASN in both OM and SC adipose tissues, in agreement with previous studies [14, 28]. ACACB was decreased in our study in the obese group. This is in contrast with the knockout mice that showed a lean phenotype [29], but in agreement with a study with obese Zucker rats and their lean counterparts, which in addition showed major species differences in adipose SC and visceral depots in humans and rats [30]. Two transcription factors, sterol regulatory element binding transcription factor-1 (SREBF1) and MLX interacting protein-like (MLXIPL), which regulate the transcription of genes involved in the lipogenic pathway [31], are repressed in OM and SC adipose tissue. In addition, MLXIPL induces the glycolytic pathway, in response to insulin and glucose, and reduces it in the presence of polyunsaturated fatty acids [32]. Though SREBF1 has been associated to obesity in humans [33], to our knowledge, it is the first time that MLXIPL is described as a downregulated gene in human adipose tissue obtained from obese patients. The decrease in lipogenic genes, more acute in the OM depot, could also be associated to an inflammation enhancement. As already seen in Fig. 1, genes involved in immunity and defense processes were highly represented, mostly those which showed upregulation. This is in agreement with Poulain-Godefroy and coworkers who found an association between an increased inflammation and a decrease of lipogenic markers in omental fat women [34].
Regarding the lipolysis, it would be expected to be increased in response to a fat excess, but not all the enzymes participating in this pathway are induced. Thus, enzymes as acyl-coenzyme A oxidase 2 (ACOX2) that initiates the β-oxidation are repressed in the OM depot as a likely effect against an induction of other enzymes in the ulterior phase of dehydrogenation of fatty acids. Mitchell et al. found a linkage of serum insulin to chromosome 3p in the region that contains the ACOX2 gene [35]. Recently, it has been reported that cell death-inducing DFFA-like effector A (CIDEA) is involved in the regulation of lipid storage in human adipocytes [36], regulating lipolysis. When CIDEA is overexpressed, lipogenesis increases and lipolysis decreases. CIDEA is repressed in our study in both OM and SC depot from obese people, but more in SC tissue, where repression of lipogenesis is not as pronounced as in OM depot. Dahlman et al [14] found an upregulation of CIDEA in the SC fat of obese women submitted to energy-restricted diets.
Among the genes involved in lipolysis, adenylyl cyclases (ADCY) are clearly repressed, mostly in OM tissue. It is important to note that four out of the six ADCYs downregulated in the arrays were confirmed by the LDA validation. Thus, ADCY1, ADCY2, ADCY3, and ADCY4 were decreased in the OM tissue of obese patients, among all the ADCYs, and ADCY3 was also repressed in the SC depot. The ADCY catalyzes the conversion of ATP into cyclic adenosine-3′,5′-monophosphate (cAMP), which regulates several physiological functions in mammals including lipolysis. cAMP is generated by two families of enzymes from the ADCY superfamily: One comprises of transmembrane ADCY encoded by nine distinct genes (ADCY1 to ADCY9) with key roles in cellular responses. Until now, the isoforms of the human fat were unclear; however, rat preadipocytes and adipocytes from epididymal white fat also express types 2, 4, 5, and 6 ADCY isoforms [37]. Recently, it has been shown that a polymorphism in the ADCY3 gene is associated with obesity in a population of Swedish men [38]. Moreover, adult type 3 ADCY-deficient mice are obese [39]. Surprisingly, no previous reported microarray studies found alterations of the expression of these proteins.
This ADCY repression could be due to the sympathetic tone that is known to influence weight gain. Adenosine is considered a regulatory metabolite and signals via binding to the four adenosine receptors (AR) classified by their ability to inhibit or stimulate ADCY. The most prominent target of the A1AR is ADCY, and because it couples to Gi proteins, its agonistic binding leads to inhibition of the ADCY enzymes. In this way, agonists of A1AR inhibit ADCY in BAT [40]. A1AR are predominant in white fat and measurement of ADCY activity and regulation in isolated adipocytes and isolated plasma membranes suggest that A1AR is unusually and tonically active in obese rats, lowering ADCY levels, through the Gi proteins, and thereby lowering cAMP levels in these tissues and inhibiting lipolysis [41].
Although A1AR was filtered in our arrays and we cannot be certain that a high A1AR expression occurs in human obesity, our results show arguments in favor of an abnormally active inhibitory branch of the ADCY system that could influence and decrease the lipolytic pathway in these subjects confirming the low lipolysis observed in obese patients in vivo [42, 43].
Another antilipolytic regulator is NPY. Its receptors NPY1R and NPY5R are involved in the regulation of body weight and are overexpressed in OM adipose tissue, in agreement with previous results [13]. In our study, NPY5R was also altered in SC adipose tissue.
In this balance between synthesis and degradation of triglycerides, another anabolic pathway is affected in our results: gluconeogenesis. The study of this pathway has been focused in liver or muscle in obese individuals but not in adipose tissue. The synthesis of glycogen is the result of glycogen synthase 1 (GYS1) and glycogen phosphorylase (PYGM) activity [44]. Both enzymes are repressed in OM and SC adipose tissue of obese patients and also forkhead box O1 (FOXO1A), the transcription factor that activates the process of gluconeogenesis [45]. The repression of the synthesis of glycogen is due to the insulin resistance state that is characterized by a decrease in insulin action, implying an alteration in the actions of the hormone on carbohydrates, lipids, and protein metabolism.
Another signaling pathway in obesity is the one related to leptin because obesity is characterized by high leptin levels and leptin resistance. Leptin signaling is mediated by its receptors and the JAK/STAT pathway, and the resistance decreases the activation of this pathway [46]. STAT3 is regulated by PIAS3 [47] whose expression is depressed in our study. Though we have not found previous results regarding PIAS3 in human adipose tissue, Deng et al. [48] showed an increased expression in ob/ob mice which is in contrast with our results; however, ob/ob mice are leptin deficient and are not comparable to human obesity in which leptin is high. In fact, these authors suggest that PIAS3 increase could be a consequence of the absence of leptin because high fat diet-induced obese rats had unchanged PIAS3 mRNA levels [49].
Another gene involved in the neovascularization is tenomodulin (TNMD). This protein expression correlates with leptin levels and is associated with insulin resistance [50]. Though TNMD has a general role against angiogenesis, its specific function in adipose tissue is not known. In our study, TNMD is induced in both depots from obese individuals in agreement with very recently published results [51].
Our study, in addition, has allowed to find a variety of transcription factors that might have a role in the development of obesity. Of those, we have validated a few like Hoxa7, a gene involved in development that has been related to obesity development and fat distribution [52]; Foxoa1 (already mentioned above) because it promotes the transcription of genes that increase glucose production together with PGC1a [53]; NCOA6, is a key coactivator of multiple nuclear receptors and transcription factors that include the adipogenic PPARG and CEBPA [54]; and ZBTB16, members of the Krüppel-like factor family, with a repressor function. Among the genes of this family, klf2 has been described as a negative regulator of the adipocyte differentiation [55]. ZBTB16 could be a candidate involved in metabolic syndrome as it is found in a genomic region that has been associated with higher body weight, higher lipid profile, and insulin resistance in rats [56]. Figure 2 shows a scheme of some of the altered genes in these pathways.
One limitation of our study is the use of whole adipose tissue which includes other cell types different to adipocytes such as the stromal–vascular fraction. At this respect, an increased inflammatory response and macrophage infiltration of the adipose tissue have been shown in severely obese patients [57].
The use of microarrays for the study of obesity represents a powerful tool to determine differences in the expression profile of obese patients. These differences reveal potential pathways involved in the origin and development of this pathology that might be used to develop new therapeutic strategies. This genome-wide study reveals that obese subjects entail defense mechanisms against an excessive expansion and fat accumulation, as they present decreased lipolysis and lipogenesis in both fat depots. Our study also reveals a large number of genes not previously classified in functional processes due to the lack of knowledge about the function of many genes and the pathway involved. Fortunately, the functional databases are being regularly updated and the interpretation of our data can be continuously re-evaluated.
References
Thorne A, Lonnqvist F, Apelman J, et al. A pilot study of long-term effects of a novel obesity treatment: omentectomy in connection with adjustable gastric banding. Int J Obes Relat Metab Disord. 2002;26:193–9.
Klein S, Fontana L, Young VL, et al. Absence of an effect of liposuction on insulin action and risk factors for coronary heart disease. N Engl J Med. 2004;350:2549–57.
Hocking SL, Chisholm DJ, James DE. Studies of regional adipose transplantation reveal a unique and beneficial interaction between subcutaneous adipose tissue and the intra-abdominal compartment. Diabetologia. 2008;51:900–2.
Tran TT, Yamamoto Y, Gesta S, et al. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410–20.
Kvist H, Chowdhury B, Grangard U, et al. Total and visceral adipose-tissue volumes derived from measurements with computed tomography in adult men and women: predictive equations. Am J Clin Nutr. 1988;48:1351–61.
Hurtado del Pozo C, Calvo RM, Vesperinas-Garcia G, et al. IPO8 and FBXL10: new reference genes for gene expression studies in human adipose tissue. Obesity (Silver Spring). 2010;18:897–903.
Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3.
Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;51:289–300.
Johansson F, Kramer F, Barnhart S, et al. Type 1 diabetes promotes disruption of advanced atherosclerotic lesions in LDL receptor-deficient mice. Proc Natl Acad Sci USA. 2008;105:2082–7.
Mortensen OH, Nielsen AR, Erikstrup C, et al. Calprotectin—a novel marker of obesity. PLoS ONE. 2009;4:e7419.
Shea J, French CR, Bishop J, et al. Changes in the transcriptome of abdominal subcutaneous adipose tissue in response to short-term overfeeding in lean and obese men. Am J Clin Nutr. 2009;89:407–15.
Gomez-Ambrosi J, Catalan V, Diez-Caballero A, et al. Gene expression profile of omental adipose tissue in human obesity. FASEB J. 2004;18:215–7.
Dahlman I, Linder K, Arvidsson Nordstrom E, et al. Changes in adipose tissue gene expression with energy-restricted diets in obese women. Am J Clin Nutr. 2005;81:1275–85.
Maury E, Ehala-Aleksejev K, Guiot Y, et al. Adipokines oversecreted by omental adipose tissue in human obesity. Am J Physiol Endocrinol Metab. 2007;293:E656–65.
Lee YH, Nair S, Rousseau E, et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–83.
Meugnier E, Faraj M, Rome S, et al. Acute hyperglycemia induces a global downregulation of gene expression in adipose tissue and skeletal muscle of healthy subjects. Diabetes. 2007;56:992–9.
Wang S, Sparks LM, Xie H, et al. Subtyping obesity with microarrays: implications for the diagnosis and treatment of obesity. Int J Obes (Lond). 2009;33:481–9.
Capel F, Viguerie N, Vega N, et al. Contribution of energy restriction and macronutrient composition to changes in adipose tissue gene expression during dietary weight-loss programs in obese women. J Clin Endocrinol Metab. 2008;93:4315–22.
Gabrielsson BG, Johansson JM, Lonn M, et al. High expression of complement components in omental adipose tissue in obese men. Obes Res. 2003;11:699–708.
Gabrielsson BG, Johansson JM, Jennische E, et al. Depot-specific expression of fibroblast growth factors in human adipose tissue. Obes Res. 2002;10:608–16.
Vohl MC, Sladek R, Robitaille J, et al. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12:1217–22.
Gabrielsson BL, Carlsson B, Carlsson LM. Partial genome scale analysis of gene expression in human adipose tissue using DNA array. Obes Res. 2000;8:374–84.
Arner P. Differences in lipolysis between human subcutaneous and omental adipose tissues. Ann Med. 1995;27:435–8.
Mobbs CV, Makimura H. Block the FAS, lose the fat. Nat Med. 2002;8:335–6.
Kumar MV, Shimokawa T, Nagy TR, et al. Differential effects of a centrally acting fatty acid synthase inhibitor in lean and obese mice. Proc Natl Acad Sci USA. 2002;99:1921–5.
Loftus TM, Jaworsky DE, Frehywot GL, et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–81.
Ortega FJ, Mayas D, Moreno-Navarrete JM, et al. The gene expression of the main lipogenic enzymes is downregulated in visceral adipose tissue of obese subjects. Obesity (Silver Spring). 2010;18:13–20.
Choi CS, Savage DB, Abu-Elheiga L, et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci USA. 2007;104:16480–5.
Kreuz S, Schoelch C, Thomas L, et al. Acetyl-CoA carboxylases 1 and 2 show distinct expression patterns in rats and humans and alterations in obesity and diabetes. Diabetes Metab Res Rev. 2009;25:577–86.
Uyeda K, Yamashita H, Kawaguchi T. Carbohydrate responsive element-binding protein (ChREBP): a key regulator of glucose metabolism and fat storage. Biochem Pharmacol. 2002;63:2075–80.
Dentin R, Benhamed F, Pegorier JP, et al. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J Clin Invest. 2005;115:2843–54.
Oberkofler H, Fukushima N, Esterbauer H, et al. Sterol regulatory element binding proteins: relationship of adipose tissue gene expression with obesity in humans. Biochim Biophys Acta. 2002;1575:75–81.
Poulain-Godefroy O, Lecoeur C, Pattou F, et al. Inflammation is associated with a decrease of lipogenic factors in omental fat in women. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1–7.
Mitchell BD, Cole SA, Hsueh WC, et al. Linkage of serum insulin concentrations to chromosome 3p in Mexican Americans. Diabetes. 2000;49:513–6.
Misra A, Garg A, Abate N, et al. Relationship of anterior and posterior subcutaneous abdominal fat to insulin sensitivity in nondiabetic men. Obes Res. 1997;5:93–9.
Serazin-Leroy V, Morot M, de Mazancourt P, et al. Differences in type II, IV, V and VI adenylyl cyclase isoform expression between rat preadipocytes and adipocytes. Biochim Biophys Acta. 2001;1550:37–51.
Nordman S, Abulaiti A, Hilding A, et al. Genetic variation of the adenylyl cyclase 3 (AC3) locus and its influence on type 2 diabetes and obesity susceptibility in Swedish men. Int J Obes (Lond). 2008;32:407–12.
Wang Z, Li V, Chan GC, et al. Adult type 3 adenylyl cyclase-deficient mice are obese. PLoS ONE. 2009;4:e6979.
Schimmel RJ, McCarthy L, Dzierzanowski D. Effects of pertussis toxin treatment on metabolism in hamster brown adipocytes. Am J Physiol. 1985;249:C456–63.
Dhalla AK, Chisholm JW, Reaven GM, et al. A1 adenosine receptor: role in diabetes and obesity. Handb Exp Pharmacol 2009;271–95.
Lillioja S, Foley J, Bogardus C, et al. Free fatty acid metabolism and obesity in man: in vivo in vitro comparisons. Metabolism. 1986;35:505–14.
Connacher AA, Bennet WM, Jung RT, et al. Effect of adrenaline infusion on fatty acid and glucose turnover in lean and obese human subjects in the post-absorptive and fed states. Clin Sci (Lond). 1991;81:635–44.
Taylor AJ, Ye JM, Schmitz-Peiffer C. Inhibition of glycogen synthesis by increased lipid availability is associated with subcellular redistribution of glycogen synthase. J Endocrinol. 2006;188:11–23.
Puigserver P, Rhee J, Donovan J, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–5.
Cheng A, Uetani N, Simoncic PD, et al. Attenuation of leptin action and regulation of obesity by protein tyrosine phosphatase 1B. Dev Cell. 2002;2:497–503.
Chung CD, Liao J, Liu B, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–5.
Deng J, Hua K, Caveney EJ, et al. Protein inhibitor of activated STAT3 inhibits adipogenic gene expression. Biochem Biophys Res Commun. 2006;339:923–31.
Peiser C, McGregor GP, Lang RE. Leptin receptor expression and suppressor of cytokine signaling transcript levels in high-fat-fed rats. Life Sci. 2000;67:2971–81.
Tolppanen AM, Pulkkinen L, Kolehmainen M, et al. Tenomodulin is associated with obesity and diabetes risk: the Finnish Diabetes Prevention Study. Obesity (Silver Spring). 2007;15:1082–8.
Saiki A, Olsson M, Jernas M, et al. Tenomodulin is highly expressed in adipose tissue, increased in obesity, and down-regulated during diet-induced weight loss. J Clin Endocrinol Metab. 2009;94:3987–94.
Gesta S, Bluher M, Yamamoto Y, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci USA. 2006;103:6676–81.
Puigserver P, Rodgers JT. Foxa2, a novel transcriptional regulator of insulin sensitivity. Nat Med. 2006;12:38–9.
Lee J, Saha PK, Yang QH, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA. 2008;105:19229–34.
Hackl H, Burkard TR, Sturn A, et al. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 2005;6:R108.
Seda O, Liska F, Sedova L, et al. A 14-gene region of rat chromosome 8 in SHR-derived polydactylous congenic substrain affects muscle-specific insulin resistance, dyslipidaemia and visceral adiposity. Folia Biol (Praha). 2005;51:53–61.
Weisberg SP, McCann D, Desai M, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808.
Acknowledgments
This work was supported by grants from Fundación de Investigación Médica Mutua Madrileña (FMM2005 and 2006) to RMC and MJO and SAF2001-2243 and SAF2006-01319 from Plan Nacional (MEC) to MJO. CIBEROBN is an initiative of Instituto de Salud Carlos III (ISCIII), Spain. We thank the Genomic Unit from CNIC (Madrid) for the microarray processing and Integromics (Granada, Spain) for their help processing and statistics of the microarray data. We thank Ricardo Escalante for critical reading of the manuscript.
Conflict of interest disclosure
The authors have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hurtado del Pozo, C., Calvo, R.M., Vesperinas-García, G. et al. Expression Profile in Omental and Subcutaneous Adipose Tissue from Lean and Obese Subjects. Repression of Lipolytic and Lipogenic Genes. OBES SURG 21, 633–643 (2011). https://doi.org/10.1007/s11695-010-0246-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11695-010-0246-4