Abstract
Methanogens are a diverse group of organisms that can live in a wide range of environments. Herein, cobalt and tungsten assimilation pathways have proposed to be established in the genomes of Methanococcus maripaludies C5 and Methanosarcina mazei Go1, respectively. All of the proteins involved in the proposed pathways were identified from public domain databases and then complied manually to reconstruct the pathways. The function of proteins with unknown function was assigned by a combined prediction approach. Totally, 17 proteins were identified to cobalt transport and assimilation processes whereas 7 proteins reported to tungsten assimilation system. Phylogenetic analysis of this study revealed that heavy metal transporter of methanogens could be evolved from closely related members in the different genera of methanogens. Nevertheless, genes encoding for metal resistance proteins could be originated from thermophilic and sulfur reducing bacteria. Many metalloenzymes in methanogens were very unique to the species of methanogens. It implied that these metal ions were utilized to produce the precursors for energy driven processes of methanogens. This study suggested that in combination of systems models and evolutionary inference can only correlate metabolic fluxes and physiological changes in methanogens. In silico models of this study will provide insights to design experiments for heavy metal assimilation processes of methanogens growing under heavy metal-rich environments and or in a laboratory condition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A huge quantity of heavy metal ions have been deposited in the environment due to a general global increase in industrial activity over the past few decades (Silver and Phung 1996). Although transition metals cobalt and tungsten, essential components of many enzymes, are taken up by specific transport systems of several different types. All metals are toxic at higher concentrations, because they cause oxidative stress by formation of free radicals (Berti and Jacobs 1996). The transport of ions into the cytoplasm is generally a tightly controlled process mediated by membrane transport proteins. It is more regulated due to the selectively permeable plasma membrane of the cells, which usually has a large negative resting potential. This membrane potential provides a strong electrochemical gradient for the inward movement of metal ions. Most metal ions enter cells by an energy dependent saturable process via specific or generic metal ion carriers or channels (Bubb and Lester 1991). Metal chelate complexes may also be transported across the plasma membrane via specialized carriers (Cunningham and Berti 1993; Chellapandi 2011).
The transition metals cobalt and tungsten are essential cofactors for a number of prokaryotic enzymes involved in a variety of metabolic processes. Methanogens are of particular interest for bioremediation purpose since they can exist under heavy metals contaminated ecosystems. Florencio et al. (1994) studied the effect of cobalt on the growth rate and activity of different microorganisms involved in the anaerobic degradation of methanol. They reported that only methylotrophic methanogens and acetogens are stimulated by cobalt. Park et al. (2010) emphasized the importance of supplementing trace metals such as iron, cobalt, and nickel for maximum methanogenic activity, there is no evidence whether such supplements, even at relatively low concentration, could perturb anaerobic biomass. It strongly suggested that acetoclastic methanogens stressed due to reduced hydraulic/solids retention time may be susceptible to trace metal addition. Formate dehydrogenase, formyl methanufuran dehydrogenase, acetylene hydratase, and a class of phylogenetically related oxidoreductases are tungstoenzymes that catalyze the reversible oxidation of aldehydes (Kletzin and Adams 1996). Kessler et al. (1997) reported that tungsten has specifically to inhibit diazotrophic growth of Methanococcus maripaludis in the presence of molybdenum. The mechanism of nickel and cobalt uptake in many bacteria and most archaea is not known, although, for instance in methanogenes, Ni- and Co-containing enzymes are essential for energy metabolism and anabolism (Rodionov et al. 2006). Moreover, a little effort has been made on investigating the cellular systems of methanogens for controlling uptake and distribution of metal ions prevent toxic accumulation and subsequent oxidative damage of macromolecules.
Combining the relevant metabolic and genomic information of an organism allows for metabolic comparisons to be performed between various species of the same organisms as well as between different organisms (Francke et al. 2005). It also manifests the additional information of cellular and physiological processes of organisms (Vothknecht and Tumbula 1999). Proteomic investigations of methanogens in their native environments provide the most realistic information about their function but also pose the greatest experimental and bioinformatics challenges (Lacerda and Reardon 2009). Systems biology approaches have often been used to gain insights into the physiological responses of microorganisms to heavy metal stresses (Mergeay et al. 2003; Marrero et al. 2004; Baker-Austin et al. 2005; Rohlin and Gunsalus 2010). In this perspective, the present study was aimed to elucidate the cell function and physiology of methanogens in response to transport and assimilation processes of cobalt and tungsten ions using computational systems biology approach.
Materials and methods
Identification of known and missing proteins
A simple text mining approach was carried out to retrieve the genomic, proteomic and metabolic information of methanogens from public domain databases. A methanogenic bacterium which has shown more entries for protein sequences of heavy metal assimilation systems was selected as a model genome in this study. A protein sequence entry in NCBI and KEGG databases that was not yet reported for the present systems of a model genome has been searched by BLASTp tools (Altschul et al. 1997). The obtained sequence similarity hits with low e-values and high identity scores have been chosen for further functional assignments.
Functional assignment for proteins with unknown function
The function of the missing proteins was predicted by combined functional annotation approach in which selected protein sequences subjected to domain and motif searches from KEGG motif server (http://www.motif.genome.jp/). Conserved domain (CD) of query sequence was searched from NCBI-CD database (Marchler-Bauer et al. 2005) using CD search tool (Marchler-Bauer and Bryant 2004) with expected threshold 0.01 and low complexity filter. ModWeb server (http://www.modbase.compbio.ucsf.edu/ModWeb20-html/modweb.html) was used to build 3D structures from the sequences and then protein models validated by structure analysis and verification server (http://www.nihserver.mbi.ucla.edu/SAVES/) using Prove and ProCheck algorithms. ProFunc server (Laskowski et al. 2005) was used to predict the protein function from the homology models by uploading PDB file.
Reconstruction of proposed heavy metal assimilation pathways
KEGG Automatic Annotation Server (KAAS) is useful as a rapid and high performance tool with high precision (98.5%) and sensitivity (97.4%) for bacterial genome annotation (Moriya et al. 2007). The known and missing protein sequences in dataset were used for automatic pathway reconstruction by KASS 1.6a server using bi-directional best hit of BLAST. Automatically reconstructed pathway of selected genomes was manually inspected and then verified with available metabolic information.
Modeling and simulation of heavy metal assimilation systems
CellDesigner 4.0 software (Funahashi et al. 2008) was used to draw the proposed pathway with all species in SBML level 1 script. SBML ODE (ordinary differential equations) Solver library (SOSlib) was used to similate the metabolic fluxus of metal-dependent enzymatic reactions and transport systems in time period (minutes). Chemical reaction network, arbitrary kinetic functions, initial concentrations of compartments, enzymes and metabolites have been assigned according to physiological hypotheses of metal transport phenomena. A time course simulation was carried out for enzymatic reactions by irreversible Michaelis–Menten kinetic function with enzyme kinetic data retrieved from Brenda database. Mass action kinetic function was used to simulate metal transport process (import, export and internal fluxes).
Phylogenetic tree construction
The sequences of protein family (transporter and metalloenzymes) in the proposed pathways were clustered with a complete deletion of gaps and corrected in multiple substitutions by multiple sequence alignment method implemented in ClustalX 2.0 software (Thompson et al. 1997). The parameter was: gap opening 10, gap extension 0.20, delay divergent sequence 30%, DNA transition weight 0.50 and Gonnet series protein weight matrix. Neighbor Joining (NJ) tree for every protein family was searched homogeneous patterns among all lineages using MEGA 4.0 software (Tamura et al. 2007) with 1,000 bootstraps values.
Results
The text mining results implied that M. maripaludies C5 genome has more number of protein sequence entries for cobalt utilization whereas Methanosarcina mazei Go1genome possessed a good established system to tungsten assimilation. Other methanogens have only limited number of protein entries in the database.
Cobalt assimilation system in M. maripaludies
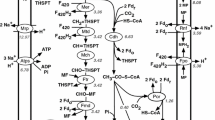
The genome of M. maripaludies is comprised of 10 protein coding genes (cbiS, cbiQ, cbiT, cbiP, cbiO1, cbiN, cbiM, corA, cobW, czcD) for cobalt transport and export processes, and 7 protein coding genes (cbiA, cbiN, cbiD, cbiB, cbiE, coxA, aroB) for cobalt-dependent biochemical reactions involved in the different metabolic pathways (Table 1). Among these, the function of proteins cbiP and cobW were predicted in this study. Cobalt ABC transporter, inner membrane subunit and predicted cobalt ABC transporter, permease have similar conserved domain (cbiQ; e-value 3.70 × 10−34), showing the functional resemblance for transporting cobalt ions across the cell membrane. Apart from cation diffusion facilitator family protein, this genome has a protein cobW that is functionally corresponded to Ni/Co exporter as the results of the conserved domain similarity (Ras-like GTPase superfamily; e-value 1.98 × 10−19) and other sequence and structural likeliness. This study shows that it has four ABC transporters (cbiPQST) and in addition to that, four transporter proteins (cbiMNO1 and corA) existed to increase the cobalt transport rate of this organism. Cation facilitator family protein (czcD) along with predicted Ni/Co exporter can regulate the cobalt ion export from cytoplasm of the cell. Cobalt assimilation process in the cell may be raised by the incorporation of cobalt ions into the enzymes, such as cbiA, cbiN aoxA and aroB. Cobalt ions can also contribute in photorespiration process indirectly by synthesizing precursors as the result of occurring enzymes cbiBDE. As represented in Fig. 1, cobalt ion export rate mediated by a protein complex (cobW-czcD) is lower than cobalt transport rate carried out by a transporter assembly (cbiAMNOQS). A transporter assembly cbiO1PT-corA is supported to increase the cobalt transport rate, which is rather than that of enzymatic detoxification processes.
Tungsten assimilation system
In this study, two tungsten-specific transporter proteins, such as torB and torP are appeared in the inner membrane of M. mazei Go1, but no tungsten-specific exporter (Table 1). Despite of this, it has a multidrug efflux pump protein (ebrA1) to expel out the excessive tungsten ions from cell. Concerning tungsten-dependent enzymes, tungsten ion is directly incorporated into formylmethanofuran dehydrogenase (fwdD) and indirectly required for the catalytic activity of NADH/F420 H2 dehydrogenase (fpoI) and aldehyde ferredoxin oxidoreductase (aor2). The results obtained for the metabolic behavior of tungsten assimilation system indicated that the rates of tungsten transport, detoxification and assimilation are regulated one by one depending on concentrations of metabolites produced after importing it (Fig. 2). The biochemical process regulated by tungsten ions is observed to end within 90–100 min. Among tungsten-dependent enzymes, enzymatic rate of formylmethanofuran dehydrogenase is effectively dependent on tungsten ion concentration in the cytoplasm. Furthermore, the reaction rate of NADH/F420 H2 dehydrogenase is higher than the reaction rate of aldehyde ferredoxin oxidoreductase.
Phylogenetic analysis of metal transporter proteins and metalloenzymes
Phylogenetic analysis revealed that transporter families of methanogens are grouped separately in response to metals which are highly assimilated by individual organism (data not shown). FecC and cbiTO in methanogens are shared their phylogeny within closely related members in methanogens while cbiN resembled with Thermosipho africanus and Arthrospira maxima. CbiS in methanogens is closely related to Clostridium perfringens. ABC trasporter in Methanosarcina genus shows phylogenetic relationship with Dehalogenimonas kykanthroporepellens, Rhodomicrobium vannielii and Thermoanaerobacter tengcongensis. A phylogenetic relationship is occurred between methanogens and Clostridium botulinum for czcD. Tungsten transporter (torP) in methanogens is evolutionally relatedness to Dehalogenimonas kykanthroporepellens whereas cbiQ similar to Thermicola sp.
The genera of Methanosarcina and Methanococcus are clustered separately for cbiX family in the phylogenetic tree and then with thermophilic archaea (Fig. 3). CbiX family of these organisms are again clustered with cbiN family of Methanosarcina genus and Geobater uraniireducens and Pelobacter propionicus. Phylogenetic analysis of cbiDBE family showed a relatedness between methanogens and acetogenic bacteria as shown in Fig. 4. A clade of methanogens for aroB family is evolutionarly related to the same family in the members of sulfur utilizing archaea and of haloarchaea. Aor2 family of M. mazei is phylogenetically corresponded with tungsten containing aor2 and then with members in the groups of thermophiles and of proteobacteria. A phylogenetic relatedness is noticed between methanogens and Desulfobacterium autotrophicum for cdh2 family; methanogens and Ferroglobus placidus and Archaeoglobus fulgidus for cdh2/coxA family (Fig. 5).
Discussion
Heavy metal toxicity on cellular metabolism of methanogens is still being a major problem for commodity production using industrial effluents containing heavy metals incorporation (Pedone et al. 2004). Since, in silico description has revealed on understanding cobalt and tungsten assimilation behaviors of methanogens. The rate evolution of a gene remains the same as long as the biological function does not change (Kimura 1983). The identification of orthologous genes among many species is the shortest way to predict functions of sequenced genomes (Razia et al. 2010). Initially, orthologs of known nickel and cobalt transporter genes in available prokaryotic genomes were identified by similarity search. A combined functional prediction (sequence similarity search, phylogenetic interface, structure-function relationship) was employed for assigning function of missing proteins with unknown function. Perhaps, such approach can provide more biological reliability, when the predicted proteins incorporated in the proposed systems.
Cobalt ions are imported into the cell through a membrane complex formed by a protein assembly cbiAMNOQS located in the membrane of M. maripaludies. CobW is associated czcD to form the first efflux system while cbiO1PT with corA assembled to form second efflux system. Transport of cobalt ions against their concentration gradient through membrane may be derived by copper transporting P-type ATPase (copP). Apart from the metal transporter systems, it assumed five possible ways to utilize cobalt ions in the cytoplasm; (1) a cobalt ion incorporated with sirohydrochlorin can be converted cobalt-sirohydrochlorin reversibly by enzyme cbiX; (2) cob (II)yrinic acid a,c-diamide and hydrogen ion are produced from hydrogenobyrinic acid a,c-diamide and cobalt by enzyme cbiA; (3) cbiE, a cobalt-dependent enzyme, is used for synthesis of precorrin-8X and carbon dioxide; (4) carbon monoxide dehydrogenase is contributed to produce carbon dioxide from carbon monoxide that can be activated in presence of cobalt ion; (5) 3-dehydroquinate and phosphate are final products of enzyme reaction catalyzed by aorB. As the consequence of these reactions, hydrogen ions and CO2 are produced that can be served as substrates for methane biosynthesis. It suggested that cobalt ions can take an account to indirectly regulate the methanogenesis of methanogens, particularly hydrogen utilizing methanogens. Cobalt ion is required for nitrogen metabolism because of 3-dehydroquinate synthase activity dependent on the concentration of cobalt ions. The proposed cobalt assimilation systems is accorded with earlier investigations (Lorowitz et al. 1992; Bhattacharya et al. 1995; Brindley et al. 2003).
Tungsten is the heaviest atom and the only third-row transition element that exhibits biological activity in enzymes (Bevers et al. 2008). Tungsten-specific transporter (torBP) is occurred in the membrane of M. maripaludies, which is functionally identical to tungsten transport protein A (wtpA) of hyperthermophilic archaeon Pyrococcus furiosus (Bevers et al. 2006). The presence of tungsten-dependent enzymes (fwdD, fpoI, aor2) in this genome suggested that methanogenic archaea are utilizing tungsten as one of the key metals for their energy-driven process. Concentration gradient energy driven process can be activated on pumping tungsten ions across the cell membrane and excessive or accumulated ions can be indirectly utilized for catalytic activities of these enzymes. As similar to cobalt assimilation systems, carbon dioxide is evolved as end-products that may be served as a substrate of the methane biosynthesis. Thus, it revealed the physiological importance of tungsten ions in methanogens for their energetic metabolisms.
ABC transporter plays a crucial role in substrate uptake, export, and osmoregulation in methanogens (Nies 2003). Many bacterial species possess transporters from only one family, and few species have a redundant set of nickel/cobalt transporters from different families (Rodionov et al. 2006). Methanogens have typical transporter for capable and selective transport of these metal ions. Even if metal transporter can be evolved from closely related members in the different genera of methanogens, metal resistance proteins may be originated from thermophilic and sulfur reducing bacteria. Protein superfamilies of methanogens for metal transport process have a reasonable sequence similarity to closely related protein families of proteobacteria. It may be reasonable due to unique genomic and evolutionary features and ancient metabolic adaptation. Nevertheless, these protein families can be evolved into present bacterial lineages at a slow evolution rate (Ranea et al. 2004; Dupont et al. 2010). It suggests that methanogens have typical metal transporter as compared to present bacterial system.
Metalloenzymes are very unique to the species of methanogens. CbiX/cbiN, coxA, cdh2, and aor2 families show a phylogenetic proximity between methanogens and thermophilic archaea in accordance to earlier report (Itoh 2003). A close evolutionary resemblance is noted between methanogens and acidogenic bacteria for cbix/cbiN. It may possibly occur when such proteins of these organisms shared for energetic metabolisms in a microbial consortium (Lee et al. 2008; Henderson et al. 2010). AroB family of methanogens has phylogenetically corresponded with sulfur utilizing archaea as a result of sulfur cycling and methanogenesis are primarily drive microbial colonization (Borin et al. 2009). Hence, it revealed the phylogenetic and metabolic relatedness for heavy metal assimilation pathways can likely be established among methanogens, sulfur utilizing archaea and methylotrophs, which is agreed with earlier works (Boetius et al. 2000; Thomsen et al. 2001; Orphan et al. 2001, 2003; Teske et al. 2003; Caldwell et al. 2008). Nonetheless, in silico representation should couple with an experimental program that may provide valuable information including genetic and metabolic regulatory systems of methanogens. The present pathway models will thus provide a simple idea to understand the heavy metal assimilation behaviors of methanogens.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST, a new generation of protein database search program. Nucleic Acids Res 25:3389–3402
Baker-Austin C, Dopson M, Wexler M (2005) Molecular insight into extreme copper resistance in the extremophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Microbiology 151:2637–2646
Berti WR, Jacobs LW (1996) Chemistry and phytotoxicity of soil trace elements from repeated sewage sludge applications. J Environ Qual 25:1025–1032
Bevers LE, Hagedoorn PL, Krijger GC, Hagen WR (2006) Tungsten transport protein A (WtpA) in Pyrococcus furiosus: the first member of a new class of tungstate and molybdate transporters. J Bacteriol 188:6498–6505
Bevers LE, Hagedoorna PL, Hagen WR (2008) The bioinorganic chemistry of tungsten. Coord Chem Rev 253:269–290
Bhattacharya SK, Uberoi V, Madura RL, Haghighi-Podeh MR (1995) Effect of cobalt on methanogenesis. Environ Technol 16:271–278
Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O (2000) A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626
Borin S, Brusetti L, Mapelli F, D’Auria G, Brusa T, Marzorati M, Rizzi A, Yakimov M, Marty D, De Lange GJ, Van der Wielen P, Bolhuis H, McGenity TJ, Polymenakou PN, Malinverno E, Giuliano L, Corselli C, Daffonchio D (2009) Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc Natl Acad Sci USA 106:9151–9156
Brindley AA, Raux E, Leech HK, Schubert HL, Warren MJ (2003) A story of chelatase evolution: identification and characterisation of a small 13–15 kDa ‘ancestral’ cobaltochelatase (CbiXS) in the Archaea. J Biol Chem 278:22388–22395
Bubb JM, Lester JN (1991) The impact of heavy metals on lowland rivers and the implications for man and the environment. Sci Total Environ 100:207–233
Caldwell SL, Laidler JR, Brewer EA, Eberly JO, Sandborgh SC, Colwell FS (2008) Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ Sci Technol 42:6791–6799
Chellapandi P (2011) Molecular evolution of methanogens based on their metabolic facets. Front Biol. doi:10.1007/s11515-011-1154-2
Cunningham SD, Berti WR (1993) Remediation of contaminated soils with green plants: an overview. In Vitro Cell Dev Biol 29:207–212
Dupont CL, Butcher A, Valas RE, Bourne PE, Caetano-Anollése G (2010) History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc Natl Acad Sci USA 107:10567–10572
Florencio L, Field JA, Lettinga G (1994) Importance of cobalt for individual trophic groups in an anaerobic methanol-degrading consortium. Appl Environ Microbiol 60:227–234
Francke C, Siezen RJ, Teusink B (2005) Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol 13:550–558
Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H (2008) CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc IEEE 96:1254–1265
Henderson G, Naylor GE, Leahy SC, Janssen PH (2010) Presence of novel, potentially homoacetogenic bacteria in the rumen as determined by analysis of formyltetrahydrofolate synthetase sequences from ruminants. Appl Environ Microbiol 76:2058–2066
Itoh T (2003) Taxonomy of nonmethanogenic hyperthermophilic and related thermophilic archaea. J Biosci Bioeng 96:203–212
Kessler PS, McLarnan J, Leigh JA (1997) Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J Bacteriol 179:541–543
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kletzin A, Adams MW (1996) Tungsten in biological systems. FEMS Microbiol Rev 18:5–63
Lacerda CMR, Reardon KF (2009) Environmental proteomics: applications of proteome profiling in environmental microbiology and biotechnology. Brief Funct Genomic Proteomic 8:75–87
Laskowski RA, Watson JD, Thornton JM (2005) ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res 33:W89–W93
Lee C, Kim J, Shin SG, Hwang S (2008) Monitoring bacterial and archaeal community shifts in a mesophilic anaerobic batch reactor treating a high-strength organic wastewater. FEMS Microbiol Ecol 65:544–554
Lorowitz WH, Nagle DP Jr, Tanner RS (1992) Anaerobic oxidation of elemental metals coupled to methanogenesis by Methanobacterium thermoautotrophicum. Environ Sci Technol 26:1606–1610
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:327–331
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki C, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH (2005) CDD: a conserved domain database for protein classification. Nucleic Acids Res 33:192–196
Marrero J, Gonzalez LJ, Sanchez A (2004) Effect of high concentration of Co (II) on Enterobacter liquefaciens strain C-1: a bacterium highly resistant to heavy metals with an unknown genome. Proteomics 4:1265–1279
Mergeay M, Monchy S, Vallaeys T (2003) Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev 27:385–410
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Nies DH (2003) Efflux mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339
Orphan VJ, Hinrichs KU, Ussler W III, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF (2001) Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol 67:1922–1934
Orphan VJ, House CH, Hinrichs KU, Mc Keegan KD, DeLong EF (2003) Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci USA 99:7663–7668
Park C, Bega A, Unlu C, Chadderton RA, McKean WR, Kohl PM, Hunt JA, Keaney J, Willis JL, Duran M (2010) Acetoclastic methanogens in an anaerobic digester could be susceptible to trace metal supplementation. Water Sci Technol 62:2905–2911
Pedone E, Bartolucci S, Fiorentino G (2004) Sensing and adapting to environmental stress: the archaeal tactic. Front Biosci 9:2909–2926
Ranea JAG, Buchan DWA, Thornton JM, Orengo CA (2004) Evolution of protein superfamilies and bacterial genome size. J Mol Biol 336:871–887
Razia M, Raja KR, Padmanaban K, Sivaramakrishnan S, Chellapandi P (2010) Phylogenetic approach for assigning function of hypothetical proteins in Photorhabdus luminescens. subsp. laumondii T101 genome. J Comp Sci Syst Biol 3:21–29
Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T (2006) Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J Bacteriol 188:317–327
Rohlin L, Gunsalus RP (2010) Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol 10:62
Silver S, Phung LT (1996) Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol 50:753–789
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Teske A, Dhillon A, Sogin ML (2003) Genomic markers of ancient anaerobic microbial pathways: sulfate reduction, methanogenesis, and methane oxidation. Biol Bull 204:186–191
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thomsen TR, Finster K, Ramsing NB (2001) Biogeochemical and molecular signatures of anaerobic methane oxidation in marine sediment. Appl Environ Microbiol 67:1646–1656
Vothknecht UC, Tumbula DL (1999) Archaea: from genomics to physiology and the origin of life. Trends Cell Biol 9:159–161
Acknowledgments
Author is thankful to the University Grants Commission, New Delhi, India, for financial assistance (32-559/2006) to carry out the work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chellapandi, P. In silico description of cobalt and nickel assimilation systems in the genomes of methanogens. Syst Synth Biol 5, 105–114 (2011). https://doi.org/10.1007/s11693-011-9087-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11693-011-9087-2