Abstract
The genus Passiflora includes economically important passion fruits and over 600 other wild species. Micropropagation of Passiflora species is far from routine due to low regeneration frequencies and multiplication rates, so there is great interest in understanding the molecular control of the in vitro regeneration processes of these species. Here, we report the characterization of a Passiflora morifolia gene encoding PmTCP1, a putative TCP transcription factor that showed high sequence similarity to Arabidopsis class I TCPs. The expression patterns of the PmTCP1 gene during in vitro organogenesis and callus growth revealed differential expression of PmTCP1 modulated by different combinations of auxin and cytokinin concentrations in the culture medium. At a constant auxin concentration, cytokinin increased PmTCP1 expression, and at a constant cytokinin concentration, auxin repressed PmTCP1 expression. We also observed a correlation between PmTCP1 expression and the in vitro organogenesis of roots and shoots. We expect that these results will increase our understanding of the molecular networks and environmental signals that modulate the processes of organogenesis during plant development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Passiflora includes over 600 species. Among these are species of economic interest that produce passion fruits, a commercially important product in some countries (Vieira and Carneiro 2004; Zerbini et al. 2008). Although in vitro tissue culture-based techniques have been used to propagate and regenerate a number of Passiflora species (Dornelas et al. 2006a; Zerbini et al. 2008), micropropagation of Passiflora species is not routine due to low regeneration frequencies and multiplication rates (Vieira and Carneiro 2004; Zerbini et al. 2008; Silva et al. 2009). The effects of hormones, explant sources, and other factors on in vitro regeneration of Passiflora species show somewhat variable results. Here, we identify and characterize a putative class I TCP transcription factor in Passiflora morifolia that is potentially involved in the in vitro organogenesis of this species.
The TCP family of transcription factors was named after TEOSINTE BRANCHED 1 (TB1 from maize; Doebley et al. 1997), CYCLOIDEA (CYC from snapdragon; Luo et al. 1996), and PROLIFERATING CELL FACTOR (PCF1 and 2 from rice; Kosugi and Ohashi 1997). This plant-specific gene family controls multiple developmental traits in diverse species, generally integrating identity and growth (Dornelas et al. 2011) and including flower symmetry, embryo growth, leaf morphogenesis, and senescence (Martín-Trillo and Cubas 2009; Manassero et al. 2013). The TCP proteins share a common modified bHLH DNA-binding motif known as the TCP domain (Cubas et al. 1999). Members of the TCP family are divided into two subfamilies: class I (also known as PCF or TCP-P; Cubas et al. 1999; Navaud et al. 2007) and class II TCPs (CYC/TB1 or TCP-C; Cubas et al. 1999; Navaud et al. 2007). Class I and class II TCP proteins appear to act antagonistically, affecting local patterns of cell proliferation, modulating development, and acting upon traits that are under evolutionary scrutiny such as flower and leaf shape and shoot branching patterns (Martín-Trillo and Cubas 2009; Danisman et al. 2012; Manassero et al. 2013). Nevertheless, knowledge of the mechanism through which TCPs integrate environmental signals with plant developmental responses is just emerging (Manassero et al. 2013).
With the aim of clarifying the role of TCPs during in vitro regeneration processes, we identified and characterized PmTCP1, a putative class I TCP gene in P. morifolia that responds to different auxin and cytokinin concentrations and is differentially expressed during in vitro organogenesis.
Materials and Methods
Plant material.
For total RNA extraction, flower bud, leaf, and shoot apex tissues were obtained from mature plants of P. morifolia L. (Passifloraceae) from the Passiflora collection maintained at the Plant Biology Department, Biology Institute, State University of Campinas, Campinas-SP, Brazil. Root tissues were obtained from hydroponic plants. Callus samples were collected after 7, 14, 21, 28, and 35 d of culture in all tested media.
Tissue culture.
Callus tissues of P. morifolia L. were produced from leaf explants from plants maintained in vitro according to Dornelas and Vieira (1994) and Dornelas et al. (2006a). The cultures were induced in MS medium (Murashige and Skoog 1962) supplemented with 30 g L−1 sucrose and different combinations of 0, 0.54, 5.4, or 10.8 μM 1-naphthaleneacetic acid (NAA) and 0, 4.5, or 9.0 μM 6-benzylaminopurine (BAP). All media were adjusted to pH 5.8 and solidified with 2 g L−1 Phytagel (Sigma, St. Louis, MO). All media were autoclaved at 121°C, 22 PSI for 15 min, and then dispensed into Petri dishes. The cultures were kept at 26°C under 16 h photoperiod, photon flux density of 150 μmol m−2 s−1 (daylight fluorescent lamp).
Microscopy.
Callus samples were collected after 0, 14, 21, 28, and 35 d of culture, fixed in 4% paraformaldehyde buffered with 0.05 M phosphate buffer, pH 6.8, at 4°C for 24 h, and dehydrated in an ethanol series. For light microscopy, the samples were embedded in Historesin (hydroxyethylmethacrylate; Leica, Wetzlar, Germany). After polymerization at room temperature for 48 h, serial sections 5–8 μm thick were obtained and stained with 0.05% toluidine blue. Permanent slides were mounted in Entellan solution (Fluka, Buchs, Switzerland). Microtome sections were observed and photographed under an Axioskop-40 Zeiss microscope (Zeiss, Oberkochen, Germany). For in situ hybridization, fixed and dehydrated samples were paraffin-embedded. Thin slices (8 μm) were obtained with a rotary microtome and baked (50°C) for at least 16 h on silanized [2% 3-aminopropyltriethoxysilane (Pierce, Rockford, IL) in acetone] glass slides. For scanning electron microscopy (SEM), the material was critical point-dried in CO2 (CPD 020 Critical Point Dryer; Balzers, Schalksmühle, Germany), fixed in metallic stubs with double-sided adhesive tape (3M, Campinas-SP, Brazil), sputter coated (40 nm) with colloidal gold, and observed at 10–20 kV under a LEO 435 VP scanning electron microscope (Zeiss) at NAP/MEPA-ESALQ/USP, Piracicaba-SP, Brazil.
Sequence analysis.
We searched the PASSIOMA expressed sequence tag (EST) database (Dornelas et al. 2006b; Cutri and Dornelas 2012) using the Arabidopsis thaliana TCP20 (At3g27010) and TCP4 (At3g15030) protein sequences as baits to identify putative P. morifolia class I or class II TCP sequences. The searches were performed using keyword searching and a local BLAST (Altschul et al. 1997) tool. We found a single EST clone, PACEPM1001B7, showing high similarity (e-value = e–98) to the Arabidopsis TCP20 sequence and chose this clone for further characterization. Electro-competent Escherichia coli DH5-alpha cells were transformed with the PACEPM1001B7 construct, and ten positive clones were sequenced (3100 Genetic Analyzer, Applied Biosystems, Grand Island, NY) using T7 and SP6 primers. The obtained sequences were processed using the CAP3 (Huang and Madan 1999) algorithm available in the BioEdit software program (http://www.mbio.ncsu.edu/bioedit/bioedit.html) (Carlsbad, CA). Contig sequences were compared to public databases at NCBI using the BLAST algorithm (Altschul et al. 1997; http://www.ncbi.nlm.nih.gov/BLAST/). The presence of conserved amino acid motifs in the deduced protein sequences derived from the obtained sequences was investigated using Pfam (Finn et al. 2010), and putative subcellular localization of the deduced proteins was investigated using WoLF PSORT (Horton et al. 2007). Theoretical predictions of molecular weight and pI were performed using ExPASy (Artimo et al. 2012; www.expasy.org). A single contig was obtained using CAP3, which we named PmTCP1. Multiple sequence alignments of the deduced amino acid sequence of PmTCP1 and all TCP family members from Arabidopsis were performed using CLUSTALX (Thompson et al. 1994). Distance trees were obtained from neighbor-joining matrices (Saitou and Nei 1987), with bootstrap values calculated from 1,000 replicates and visualized with TreeView (Page 1996). Parsimony trees were obtained using hand-corrected sequence alignments with MEGA (Tamura et al. 2011; http://www.megasoftware.net).
RT-PCR.
Total RNA was extracted from P. morifolia tissues with TRIzol Reagent (Invitrogen, Carlsbad, CA) and treated with DNase (Ambion, Carlsbad, CA). First strand cDNA samples were obtained using the Superscript First-Strand Synthesis Kit (Invitrogen) with a 17-mer oligo-dT primer. Samples from the growth regulator treatment experiment (described in the tissue culture section above) were analyzed after 4 wk of in vitro culture. To access the dynamics of PmTCP1 expression during in vitro culture, samples were taken after 7, 14, 28, and 35 d of culture on MS medium supplemented with 0.54 μM NAA and 9.0 μM BAP. Samples were normalized using a pair of primers designed to detect a Passiflora ACTIN gene (PeACTIN1; Cutri and Dornelas 2012): PeAC5 (5′-GAGAAAATGACCCAGATCATG-3′) and PeAC3 (5′-TCGACATCACACTTCATGATAG-3′). RT-PCR was performed using normalized cDNA samples and gene-specific primers (PmTCPF 5′-TTTCCCTCAACCACTCCAAC-3′ and PmTCPR 5′-GGTGGGTAATGGAGGGTTTT-3′) under the following PCR conditions: initial denaturation at 94°C for 3 min, 30 cycles of 94°C for 40 s, 60°C for 40 s, and 72°C for 1 min. The obtained fragments were electrophoresed alongside a molecular weight marker in a 1.5% agarose gel visualized under UV light and photographed. The amount of PmTCP1 mRNA was calculated as the ratio of densitometric measurement (using ImageJ software, http://rsb.info.nih.gov/ij/) of the sample to that of the corresponding internal standard (PeACTIN1). Three biological replicates were used in two independent experiments.
In situ hybridization.
The in situ hybridization technique was basically as described by Dornelas et al. (2000) and Cutri and Dornelas (2012), with the following adaptations: paraffin was removed from sections by two consecutive 5 min washes in xylene followed by 5 min washes in 3:1, 1:1, and 1:3 (v/v) ethanol:xylene. The slides were then briefly rinsed in ethanol and air-dried. After drying, the slides were individually treated with proteinase K solution (10 μg mL−1 in Tris-HCl pH 7.5) at 37°C for 10 min. After the proteinase treatment, the slides were washed twice with 0.01 M Tris-HCl pH 7.5. A hybridization mix containing 50% deionized formamide, 0.1 M Tris-HCl pH 7.5, 0.05 M NaCl, 0.01 M EDTA, 100 ng mL−1 yeast tRNA, and 10% sodium dextran sulfate were used to dilute 500 ng mL−1 of DIG-labeled antisense or sense (control) PmTCP1 RNA probe [the 347-bp RT-PCR fragment cloned in the pCRII-TOPO vector (Life Technologies, Carlsbad, CA) was used as probe template]. Probe labeling was according to the labeling kit manufacturer's (Roche, Indianapolis, IN) instructions. Hybridization was carried out at 42°C for 16 h. The excess non-hybridized probe was washed out during four rinses (20 min each at 42°C): the first two rinses were 4× SSC followed by two rinses with 2× SSC. The slides were then rinsed twice with DB1 buffer (0.01 M Tris-HCl pH 7.5) at room temperature and kept in a blocking buffer [2% w/v Blocking Reagent (Roche) in 0.01 M Tris-HCl pH 7.5] for 8 min at 37°C. An alkaline phosphatase-conjugated anti-DIG antibody (Fab-fragments; Roche) was added (1:2,000 dilution), and the samples were kept at 37°C for an additional 1 h. The slides were then rinsed twice with DB1 buffer and maintained for 10 min in DB3 buffer (0.01 M MgCl2 6H2O, 0.01 M Tris-HCl pH 9.0) at room temperature. The DB3 buffer was then replaced by a phosphatase substrate (NBT/BCIP plus suppressor; Pierce), and the slides were kept overnight at room temperature in the dark. The staining reaction was stopped by two rinses with DB4 buffer (0.05 M EDTA, 0.01 M Tris-HCl pH 8.0). The hybridization signal was observed under a Zeiss Axioskope 50 microscope model equipped with a Zeiss AxioCam HRc digital camera.
Results
Isolation of a Passiflora gene encoding a class I TCP protein.
A full-length P. morifolia cDNA fragment was isolated from the EST clone found in the PASSIOMA database (Dornelas et al. 2006b; Cutri and Dornelas 2012) and designated PmTCP1 (GenBank ID: HM003688). The PmTCP1 cDNA was 1,350 bp in length and had a complete open reading frame with a deduced amino acid sequence of 374 residues. The deduced amino acid sequence of PmTCP1 had a predicted molecular weight of 39.86 kDa and a theoretical pI (isoelectric point) of 9.08. The PmTCP1 protein was predicted by WoLF PSORT (Horton et al. 2007) to be nuclear-localized and showed 42% identity with Arabidopsis TCP9, a class I TCP protein (Cubas et al. 1999; Danisman et al. 2012). In addition, PmTCP1 was 60% identical to a Populus predicted TCP domain-containing protein (NCBI locus XP_002298077) and 48% identical to a Vitis vinifera TCP9-like protein (NCBI locus XP_002282409). The PmTCP1 sequence contained all of the conserved TCP subdomains (Pfam 03634, Fig. 1a ) in its N-terminal region (residues 113 to 167). The TCP domain of PmTCP1 had a characteristic deletion of four amino acids in the basic region that is typical of class I TCPs (Cubas et al. 1999; Navaud et al. 2007; Manassero et al. 2013). Accordingly, from residues 120 to 125, PmTCP1 contained the motif EGRGRR instead of XTAKGXRDRR, the latter being typical of class II TCP basic domains (Martín-Trillo and Cubas 2009; Manassero et al. 2013; Fig. 1a ).
Passiflora morifolia PmTCP1 protein structure and relationships to Arabidopsis TCP family proteins. (a) Sequence alignment of PmTCP1 and all Arabidopsis class I TCP proteins showing the position of the typical TCP subdomains (Basic, Helices I and II, and Loop). Dark gray areas represent highly conserved motifs; less conserved motifs are in light gray. Numbers below the alignment refer to amino acid positions in PmTCP1. (b) Parsimony tree obtained from sequence comparison of PmTCP1 and all Arabidopsis TCP family members. Only bootstrap values above 75% are shown. Bar at the lower right indicates the number of substitutions.
To assess the relationships among PmTCP1 and the TCP proteins of Arabidopsis, we conducted sequence comparison analyses using genetic distances as well as parsimony matrices based on multiple alignments of the deduced amino acid sequences of PmTCP1 and all Arabidopsis TCP proteins. The obtained tree provided additional evidence that PmTCP1 belongs to the class I TCP proteins and is most related to AtTCP9 and AtTCP19 (Fig. 1b ).
PmTCP1 is differentially expressed in response to auxin and cytokinin.
PmTCP1 transcripts were not detected in mature leaves, but were present in shoot apices, roots and, to a lesser extent, flower buds (Fig. 2). To investigate whether the transcription of PmTCP1 could be under developmental control, we determined PmTCP1 expression in leaf explants that were cultured on MS medium with different auxin (NAA) and cytokinin (BAP) concentrations. Auxin repressed the expression of PmTCP1, especially in the presence of BAP (e.g., compare the RT-PCR results between 0.54 μM NAA + 9.0 μM BAP and 10.8 μM NAA + 9.0 μM BAP in Fig. 2). In the absence of BAP in the culture medium, increasing NAA concentration by tenfold did not produce any significant change in PmTCP1 expression (e.g., compare the RT-PCR results between 0.54 μM NAA + 0 BAP and 5.4 μM NAA + 0 BAP in Fig. 2). On the other hand, the addition of cytokinin seemed to induce the transcription of PmTCP1. When the concentration of NAA was kept constant (0.54 μM), adding 4.5 μM BAP increased the amount of PmTCP1 transcripts relative to 0 B.P. Nevertheless, when the concentration of BAP was raised to 9.0 μM, there was no additional increase in PmTCP1 transcription, indicating that the expression of PmTCP1 had reached a plateau or that other factors interfered with the expression induced via exposure to growth regulators (Fig. 2).
PmTCP1 expression pattern analysis using semiquantitative RT-PCR. (a) From top to bottom: expression of PmTCP1 in mature leaves (LV), shoot apexes (SA), roots (RT), and floral buds (FB); expression of PmTCP1 in leaf explants cultured in vitro in MS basal media supplemented with 0.54, 5.4, or 10.8 μM NAA combined with 0, 4.5, or 9.0 μM BAP for 14 d; expression of PmTCP1 in leaf-derived calli cultured in vitro in MS basal media supplemented with 5.4 μM NAA and 9.0 μM BAP for 7, 14, 28, or 35 d. (b) Relative expression levels of PmTCP1 normalized to PmACTIN1 expression; panels are in same order as in a. The results are expressed as a mean of two independent experiments with three biological replicates each. Error bars indicate standard deviation.
We could not detect PmTCP1 transcripts in explants cultured in medium containing 10.8 μM NAA and 9.0 μM BAP at any of the time points. Similarly, we could not detect PmTCP1 expression in explants cultured in 0.54 μM NAA and 9.0 μM BAP after 7 d of culture, but the amount of PmTCP1 transcripts in explants cultured in this medium peaked at 14 d of culture (Fig. 2). After 28 d of culture, the expression of PmTCP1 had greatly decreased, and PmTCP1 transcripts were undetectable after 35 d of culture (Fig. 2).
The spatial expression pattern of PmTCP1 is related to organogenesis.
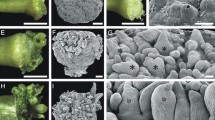
The P. morifolia explants responded differently depending on the growth regulators added to the culture medium (Fig. 3). After 35 d of culture, P. morifolia explants on medium containing either 0.54 or 5.4 μM NAA, in the absence of BAP, showed regeneration of roots, although root production was more abundant and more frequent in explants cultured in 5.4 μM NAA (Fig. 3a ). Root initials were observed in histological sections as bumps of small dividing cells with dense cytoplasm at the periphery of the callus and could be detected as early as 28 d after culture initiation (Fig. 3b ). In medium containing either 4.5 or 9.0 μM BAP in the presence of 0.54 μM NAA, shoot regeneration was observed (Fig. 3c ). Shoot buds and leaf primordia could be observed on callus surfaces after 28 d of culture (Fig. 3d, e ). When cultured in medium containing 10.8 μM NAA and 9.0 μM BAP, the explants formed only a very compact, proliferative callus composed of rapidly dividing cells (Fig. 3f–h ). No organogenesis was observed in this medium.
Morphogenic responses of leaf-derived P. morifolia explants cultured in vitro in media supplemented with different combinations of NAA and BAP. Explants after 35 d of culture (a, c, f); toluidine blue-stained histological sections (b, e, h); SEM images (d, g). P. morifolia explants cultured in medium supplemented with 5.4 μM NAA in the absence of BAP regenerated roots (a, b). The first signs of root organogenesis could be detected as early as 28 d, as peripheral groups of small cells organized in meristematic areas (b, arrow). P. morifolia explants cultured in medium supplemented with 0.54 μM NAA and 9.0 μM BAP regenerated shoots (c–e). Leaf primordia (lp; arrows in c) and shoot buds (arrow in d) could be observed after 35 d of culture. The first morphological signs of shoot organogenesis could be detected as early as 28 d, as peripheral groups of small cells organized in meristematic areas that produced leaf primordia (e). P. morifolia explants cultured for 35 d in media containing 10.8 μM NAA and 9.0 μM BAP formed only a very compact proliferative callus consisting of rapidly dividing cells (f–h). No organogenesis was observed in this callus. Bars = 1 cm (a, c, f); 300 μm (d, g); 100 μm (b, e, h).
We investigated the spatial distribution of PmTCP1 transcripts in callus tissues by means of in situ hybridization with a PmTCP1 antisense probe (Fig. 4). According to the RT-PCR results, PmTCP1 expression peaked at 14 d of culture (Fig. 2), so we focused our in situ experiments on explants cultured for 14 d. According to our preliminary tests, differences were not detected in the spatial expression patterns of PmTCP1 in explants cultured in MS medium supplemented with either 4.5 or 9.0 μM BAP in the presence of 0.54 μM NAA (data not shown). Explants cultured for 14 d in medium containing 0.54 μM NAA and 9.0 μM BAP showed a clear hybridization signal in clusters of peripheral cells of the calli giving rise to shoots (Fig. 4a, b ). Explants cultured for 28 d in the same medium showed a hybridization signal with the PmTCP1 antisense probe that was not different from background (data not shown). Similarly, explants cultured in medium supplemented with 5.4 μM NAA in the absence of BAP showed a concentration of PmTCP1 transcripts in groups of highly dividing cells in the periphery of the calli giving rise to roots (Fig. 4c, d ). Consistent with the RT-PCR results, no hybridization signal was detected in callus produced by explants cultured 14 d in medium containing 10.8 μM NAA and 9.0 μM BAP (Fig. 4e ). No hybridization signal was detected when a sense riboprobe was used (Fig. 4f )
PmTCP1 expression pattern analysis using in situ hybridization. Hybridization signal can be observed as darker areas (purple in the online version of this figure). Samples were from leaf-derived callus cultured for 1n in MS medium supplemented with 0.54 μM NAA combined with 9.0 μM BAP (a, b, f), 5.4 μM NAA in the absence of BAP (c, d), or 10.8 μM NAA and 9.0 μM BAP (e). PmTCP1 transcripts were concentrated in highly dividing cells at the periphery of the explants that are associated with regeneration of shoots (a, b) or roots (c, d). A higher magnification of the square area marked in (a) is shown in (b). In (c) and (d), clearly labeled cells (arrowhead in d) can be observed at the periphery of the callus and similar, but non-hybridized cells (arrow in d) are found at the interior of the callus. No hybridization signal was observed in callus where no organogenesis was observed (e) or in samples hybridized with a sense probe (f). The section shown in (f) was adjacent to that shown in (a). Bars = 200 μm (a, e, f); 100 μm (b, c); 50 μm (d).
Discussion
PmTCP1 likely encodes a class I TCP transcription factor.
To characterize the in vitro development of a wild passion fruit species (P. morifolia), we used a Passiflora EST database (Dornelas et al. 2006b; Cutri and Dornelas 2012) to isolate a putative TCP family gene expressed during P. morifolia development. This gene, designated PmTCP1, showed high sequence similarity to Arabidopsis TCP9. TCP9 was recently shown to be downstream of TCP20, which in turn regulates the LOX2 gene, involved in jasmonate biosynthesis (Danisman et al. 2012). Both TCP9 and TCP20 are class I TCP transcription factors (Cubas et al. 1999). The deduced protein sequence of PmTCP1 contained all of the subdomain characteristics of class I TCP proteins (Fig. 1a ). Secondary structure predictions suggest that the TCP domain, which is involved in DNA binding and dimerization, is formed by an N-terminal basic region followed by two amphipathic alpha-helices connected by a loop similar to the structure present in another family of eukaryotic transcription factor, the bHLH family (Cubas et al. 1999; Martín-Trillo and Cubas 2009; Manassero et al. 2013). Additionally, PmTCP1 contained a deletion of four amino acids in the basic domain that is typical of class I TCP proteins (Martín-Trillo and Cubas 2009; Manassero et al. 2013) and shared the same residue compositions in the loop and hydrophilic faces of helices I and II with all class I TCP proteins of Arabidopsis (Fig. 1a ; Manassero et al. 2013). Additionally, the predicted sequence of PmTCP1 did not show the 18- to 20-residue arginine-rich motif (R-domain; Cubas et al. 1999) or the glutamic acid–cysteine–glutamic acid stretch (ECE motif; Howart and Donoghue 2006; Martín-Trillo and Cubas 2009), both of unknown function, that are found only in class II TCP proteins (Martín-Trillo and Cubas 2009). Furthermore, the parsimony tree of PmTCP1 and all the Arabidopsis TCP proteins placed PmTCP1 closest to TCP9 and TCP19, which are class I TCP proteins (Fig. 1b ). These results strongly suggest that PmTCP1 encodes a class I TCP transcription factor.
PmTCP1 expression is modulated by cytokinin and auxin and correlates with organogenesis.
PmTCP1 transcripts accumulated in roots, shoot apices, and flower buds but were not detected in mature leaves (Fig. 2). Arabidopsis class I TCP genes were also expressed ubiquitously during early development of plant organs, and their expression decreased suggesting that TCPs are heterochronic regulators of plant development (Efroni et al. 2008, 2013).
A number of TCP family members function in several hormone pathways and some members are known to be modulated by growth regulators (Martín-Trillo and Cubas 2009; Manassero et al. 2013). For example, LOX2, a key enzyme in jasmonate biosynthesis, is directly and antagonistically regulated by TCP20 (class I) and TCP4 (class II) (Schommer et al. 2008; Danisman et al. 2012). TCP4 is also involved in responses to gibberellins and auxins (Sarvepalli and Nath 2011a, b). Similar to what was observed for jasmonates, a partially overlapping regulation of the auxin response pathway was also demonstrated for TCP15 (class 1) and TCP3 (class II) (Koyama et al. 2010; Uberti-Manassero et al. 2011). In addition, it was recently shown that class I TCP proteins are involved with responses to cytokinin during leaf and shoot formation (Steiner et al. 2012; Efroni et al. 2013). Accordingly, we observed that the accumulation of PmTCP1 transcripts varied depending on the concentrations of auxin (NAA) and cytokinin (BAP) in the culture medium (Fig. 2). At a constant auxin concentration, cytokinin increased PmTCP1 expression, and at a constant cytokinin concentration, auxin repressed PmTCP1 expression (Fig. 2). Nevertheless, in the absence of cytokinin in the culture medium, increasing the concentration of NAA tenfold (from 0.54 to 5.4 μM) did not significantly alter the transcription of PmTCP1. This would suggest that auxin controls PmTCP1 expression in a cytokinin-dependent manner.
As the balance between auxins and cytokinins is one of the key elements determining whether shoot or root organogenesis takes place in vitro, we performed in situ hybridization using PmTCP1 riboprobes to investigate its spatial expression pattern during P. morifolia in vitro organogenesis. We detected the presence of PmTCP1 transcripts in locations associated with initial organogenesis of both roots and shoots, but PmTCP1 transcription was inhibited in tissues, cultured on media, where only callus proliferation was observed (Figs. 3 and 4). Likewise, disruption of class I TCP20 expression perturbed the development of both roots and leaves (Hervé et al. 2009; Danisman et al. 2012). Additionally, the differentiation of shoots and leaves was severely disrupted in YABBY mutants, and the observed defects were associated with altered expression of TCP genes (Sarojam et al. 2010). Two closely related Arabidopsis class I TCP proteins, TCP14 and TCP15, were shown to act together with the O-GlcNAc transferase SPINDLY (SPY) to promote cytokinin responses (Steiner et al. 2012). As mentioned above for the other growth regulator-mediated TCP responses, class II TCP4 would act antagonistically, interacting with SWI/SNF chromatin remodeling ATPase BRAHMA (BRM) and repressing cytokinin responses during organogenesis (Efroni et al. 2013). Taken together, these observations suggest that PmTCP1 might be involved with auxin- and cytokinin-mediated responses during P. morifolia in vitro organogenesis.
Conclusion
Here, we report the cloning and characterization of a P. morifolia gene encoding a putative homolog to a class I TCP family transcription factor, PmTCP1, which showed high sequence similarity to Arabidopsis class I TCP family members. We examined the expression patterns of PmTCP1 during in vitro organogenesis and callus growth and observed differential expression levels of PmTCP1 modulated by different combinations of auxin and cytokinin concentrations in the culture medium. We also observed a correlation between PmTCP1 expression and the in vitro organogenesis of roots and shoots. We expect that these results will contribute to our understanding of the molecular networks and environmental signals that modulate the processes of organogenesis during plant development.
References
Altschul S. F.; Madden T. L.; Schäffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402; 1997.
Artimo P.; Jonnalagedda M.; Arnold K.; Baratin D.; Csardi G.; de Castro E.; Duvaud S.; Flegel V.; Fortier A.; Gasteiger E.; Grosdidier A.; Hernandez C.; Ioannidis V.; Kuznetsov D.; Liechti R.; Moretti S.; Mostaguir K.; Redaschi N.; Rossier G.; Xenarios I.; Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40(W1): W597–W603; 2012.
Cubas P.; Lauter N.; Doebley J.; Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18: 215–222; 1999.
Cutri L.; Dornelas M. C. PASSIOMA: Exploring expressed sequence tags during flower development in Passiflora spp. Comp Funct Genomics 2012: 510549; 2012.
Danisman S.; Van der Wal F.; Dhondt S.; Waites R.; de Folter S.; Bimbo A.; van Dijk A. D. J.; Muino J. M.; Cutri L.; Dornelas M. C.; Angenent G. C.; Immink R. G. H. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159: 1511–1523; 2012.
Doebley J.; Stec A.; Hubbard L. The evolution of apical dominance in maize. Nature 386: 485–488; 1997.
Dornelas M. C.; Fonseca T. C.; Rodriguez A. P. M. Brazilian passionflowers and novel passionate tropical flowering gems. In: da Silva J. A. T. (ed) Floriculture, Ornamental and Plant Biotechnology, vol. 4. Global Science Books, London, pp 629–639; 2006a.
Dornelas M. C.; Patreze C. M.; Angenent G. C.; Immink R. G. H. MADS: the missing link between identity and growth? Trends Plant Sci. 16: 89–97; 2011.
Dornelas M. C.; Tsai S. M.; Rodriguez A. P. M. Expressed sequence tags of genes involved in the flowering process of Passiflora spp. In: da Silva J. A. T. (ed) Floriculture, Ornamental and Plant Biotechnology, vol. 1. Global Science Books, London, pp 483–488; 2006b.
Dornelas M. C.; Van Lammeren A. A.; Kreis M. Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J. 21: 419–429; 2000.
Dornelas M. C.; Vieira M. L. C. Tissue culture of species of Passiflora. Plant Cell Tissue Organ Cult. 36: 211–217; 1994.
Efroni I.; Blum E.; Goldshmidt A.; Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306; 2008.
Efroni I.; Han S. K.; Kim H. J.; Wu M. F.; Steiner E.; Birnbaum K. D.; Hong J. C.; Eshed Y.; Wagner D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445; 2013.
Finn R. D.; Mistry J.; Tate J.; Coggill P.; Heger A.; Pollington J. E.; Gavin O. L.; Gunasekaran P.; Ceric G.; Forslund K.; Holm L.; Sonnhammer E. L.; Eddy S. R.; Bateman A. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222; 2010.
Hervé C.; Dabos P.; Bardet C.; Jauneau A.; Auriac M. C.; Ramboer A.; Lacout F.; Tremousaygue D. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149: 1462–1477; 2009.
Horton P.; Park K. J.; Obayashi T.; Fujita N.; Harada H.; Adams-Collier C. J.; Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35: W585–W587; 2007.
Howart D. G.; Donoghue M. J. Phylogenetic analysis of the ‘ECE’ (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U. S. A. 103: 9101–9106; 2006.
Huang X.; Madan A. CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877; 1999.
Kosugi S.; Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619; 1997.
Koyama T.; Mitsuda N.; Seki M.; Shinozaki K.; Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588; 2010.
Luo D.; Carpenter R.; Vincent C.; Copsey L.; Coen E. Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799; 1996.
Manassero N. G. U.; Viola I. L.; Welchen E.; Gonzalez D. H. TCP transcription factors: architectures of plant form. BioMol Concepts 4: 111–127; 2013.
Martín-Trillo M.; Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci. 15: 31–39; 2009.
Murashige T.; Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497; 1962.
Navaud O.; Dabos P.; Carnus E.; Tremousaygue D.; Hervé C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33; 2007.
Page R. D. M. Treeview: An application to display phylogenetic trees on personal computers. Comp Appl Biosci 12: 357–358; 1996.
Saitou N.; Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425; 1987.
Sarojam R.; Sappl P. G.; Goldshmidt A.; Efroni I.; Floyd S. K.; Eshed Y.; Bowman J. L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130; 2010.
Sarvepalli K.; Nath U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67: 595–607; 2011a.
Sarvepalli K.; Nath U. Interaction of TCP4-mediated growth module with phytohormones. Plant Signal. Behav. 6: 1440–1143; 2011b.
Schommer C.; Palatnik J. F.; Aggarwal P.; Chételat A.; Cubas P.; Farmer E. E.; Nath U.; Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230; 2008.
Silva M. L.; Pinto D. L. P.; Guerra M. P.; Floh E. I. S.; Bruckner C. H.; Otoni W. C. A novel regeneration system for a wild passion fruit species (Passiflora cincinnata Mast.) based on somatic embryogenesis from mature zygotic embryos. Plant Cell Tissue Organ Cult. 99: 47–54; 2009.
Steiner E.; Efroni I.; Gopalraj M.; Saathoff K.; Tseng T. S.; Kieffer M.; Eshed Y.; Olszewski N.; Weiss D. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108; 2012.
Tamura K.; Peterson D.; Peterson N.; Stecher G.; Nei M.; Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739; 2011.
Thompson J. D.; Higgins D. G.; Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680; 1994.
Uberti-Manassero N. G.; Lucero L. E.; Viola I. L.; Vegetti A. C.; Gonzalez D. H. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63: 809–823; 2011.
Vieira M. L. C.; Carneiro M. S. Passiflora spp., passionfruit. In: Litz R. E. (ed) Biotechnology of fruit and nut crops. CABI Publishing, Oxford, pp 435–453; 2004.
Zerbini F. M.; Otoni W. C.; Vieira M. L. C. Passionfruit. In: Kole C.; Hall T. C. (eds) A compendium of transgenic crop plants – tropical and subtropical fruit and nuts, vol. 5. 1st ed. Wiley, Berlin, pp 213–234; 2008.
Acknowledgments
We acknowledge Professors E. W. Kitajima and F. Tanaka for maintaining the electron microscope facility at NAP/MEPA-ESALQ/USP, Piracicaba, Brazil. We also acknowledge the funding from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, São Paulo, Brazil), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil).
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Marco Buenrostro-Nava
Rights and permissions
About this article
Cite this article
Rosa, Y.B.C.J., Aizza, L.C.B., Bello, C.C.M. et al. PmTCP1 encodes a putative TCP transcription factor and is differentially expressed during in vitro organogenesis in Passiflora . In Vitro Cell.Dev.Biol.-Plant 50, 36–44 (2014). https://doi.org/10.1007/s11627-013-9585-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-013-9585-x