Abstract
Amplified fragment length polymorphism (AFLP) analysis of 24 in vitro regenerated rye plants was performed in order to evaluate the somaclonal variation rate in this species and to identify rye genomic regions where mutations are preferentially promoted by in vitro culture processes. Regenerated plants were obtained from cell lines derived from immature embryos and plants were regenerated by somatic embryogenesis. Twenty-three regenerants showed variation when compared against sibling plants obtained from the same cell line. A total number of 887 AFLP markers were scored, and 8.8% identified the same polymorphism in plants obtained independently from different cell lines, revealing putative mutational hot spots. Using controlled crossings and analysis of the corresponding progenies, we were able to verify the genetic stability in the next generation for only five of these polymorphisms. The nucleotide sequence of the AFLP amplicon of four of the polymorphic markers was obtained, but only the sequence of two markers was clearly identified in the databases. The sequence of marker A1-303 was identified as part of a tandemly repeated sequence, the 120-bp family, which is located at telomeric regions and is widely distributed among rye chromosomes. The marker A5-375 showed high similarity with regions of Angela retrotransposons.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The regeneration of whole plants using in vitro culture methods should generate individuals genetically identical to the plant from which the explants were initially obtained. However, variable proportions of regenerated plants showing genetic polymorphisms have been described in a large number of plant species, a phenomenon defined as somaclonal variation (Larkin and Scowcroft 1981). Somaclonal variation could be useful for plant breeding as a source of beneficial variation, but it is an unwelcome phenomenon if transformation or clonal propagation is the objective and true-to-type plants are required.
The causes of such variation are not well established, although proposed mechanisms include chromosomal and punctual mutations, somatic recombination, sister-chromatid exchanges, somatic genetic rearrangements, transposable-elements activity, tandemly repeated sequences instability, as well as epigenetic processes such as DNA methylation (Philips et al. 1994). The activation of mobile elements is one of the mechanisms that have frequently been considered a cause of somaclonal variation. Insertion of plant retrotransposons into coding regions after protoplast or cell culture has been demonstrated in tobacco (Grandbastien et al. 1989) and rice (Hirochika et al. 1996). Sequence amplifications or deletions are another cause of somaclonal mutations (Chowdari et al. 1998; Rostiana et al. 1999). Somaclonal variation is genotype-specific and its rate is variable between and within species. Therefore, it is very important to know the frequency of and to understand the mechanisms underlying this variation for each species, variety, and cultivar used for in vitro culture applications.
Rye (Secale cereale L.) is a species for which somaclonal variation has been reported at high rates. Phenotypic analysis as well as biochemical, cytogenetic, or molecular techniques such as random amplified polymorphic DNA (RAPD) markers have been used previously to detect somaclonal variation in this cereal (Linacero and Vázquez 1992a, b; 1993). High rates of mutation have been reported for particular loci, which are considered hypervariable genomic regions (Linacero et al. 2000). It is presumed that the above mutation-inducing phenomena that occur during plant tissue culture must increase the genetic instability of these hypervariable regions in the rye genome.
The analysis of polymorphisms between clonally regenerated plants at a level of high molecular discrimination would help to recognize hypervariable rye sequences as well as to understand the processes involved in such variation. Amplified fragment length polymorphism (AFLP) markers (Vos et al. 1995) present various advantages over the different methodologies available for genetic polymorphism screening at the genomic level, including the identification of a large number of polymorphisms distributed across the genome and the reproducibility of the amplicons. In addition, the AFLP technique is relatively easy to perform, uses small amounts of DNA, and does not require prior knowledge of sequences. AFLP markers are being widely applied for the analysis of somaclonal variants (Chen et al. 2006; Saker et al. 2006; Prado et al. 2007; Gagliardi et al. 2007).
This study presents the first AFLP analysis of in vitro regenerated rye plants in order to evaluate the somaclonal variation rate at a high-resolution level and to identify hypervariable regions. Cloning and sequencing of four hypervariable AFLP markers was performed in order to determine the sequences involved in such polymorphisms. Our data indicate that mobile elements and tandem-repeat regions are related to hypervariable regions of the rye genome.
Materials and Methods
Plant material and DNA extraction.
Twenty-four regenerated plants obtained from five cell lines derived from immature embryos through somatic embryogenesis of the rye cultivar Ailés were obtained according to Linacero and Vázquez (1993) and were kindly donated by Dr. Ana M. Vázquez (Univ. Complutense de Madrid, Spain). Cell lines were established for all embryogenic calluses derived from the same embryo and were named CLa, CLd, CLe, CLh, and CLi. They included five plants each (CLd1–CLd5; CLe5–CLe9; CLh7–CLh11; CLi8–CLi12) except cell line CLa that included four regenerated plants (CLa9–CLa12). Leaves of these 24 regenerated plants were collected at 30 d old in MS medium plant stage (Fig. 1) and used for total cellular DNA extraction using the DNeasy Plant Mini Kit (Qiagen Corp., Santa Clarita, CA) following the manufacturer’s protocol.
Seeds obtained from two independent and controlled crosses using CLe6 and CLh10 plants as female parents were also kindly donated by Dr. Vázquez. The seeds were germinated on wet filter paper and transplanted to soil pots when the first leaf and roots developed. The first leaves of five siblings from each cross were collected when the second leaf emerged and were used for DNA extraction, as well as mature leaves from the Ailés plants used as male parents.
AFLP analysis.
The AFLP markers were amplified using the Plant Mapping Kit (Applied Biosystems, Foster City, CA) following the manufacturer’s protocol for regular genomes (Perkin-Elmer Applied Biosystems 1997). Genomic DNA digestion using EcoRI and MseI endonucleases (New England Biolabs, Beverly, MA) and ligation to the EcoRI and MseI site-specific double-stranded adaptors were performed in a single tube for 2 h at 37°C. Pre-selective and selective amplifications were carried out in a GenAmp PCR System 9700 thermocycler (Applied Biosystems) following protocol instructions. Adaptors, pre-selective, and selective primers were used according to the Plant Mapping Kit protocol. The PCR products of selective amplifications were separated by capillary electrophoresis on an ABI Prism 310 Genetic Analyzer and detected by fluorescence as the three EcoRI site-specific selective primers used were 5′-labelled with FAM, JOE, and NED dyes, respectively. An internal size marker, Genescan Rox-500 (35–500 bp; Applied Biosystems), labelled with ROX dye was added, allowing the co-loading of three different labelled reactions.
All the 64 selective primer pairs available from Applied Biosystems were tested in order to select the EcoRI labelled primers and MseI unlabelled primers that gave the more informative set of primer pairs. The criteria for selection of primers were to use the nine combinations that produce the maximum number of markers, using three different dye-labelled primers, in order to facilitate the detection of polymorphisms. EcoRI selected primers were FAM-EcoRI + ACT, JOE-EcoRI + ACG, and NED-EcoRI + AGC. Unlabelled MseI site-specific selective primers used were MseI + CAA, MseI + CAG, and MseI + CTA. The complete AFLP procedure, including DNA extraction, was conducted with similar results at least twice for each plant analyzed.
Data of selectively amplified DNA fragments were collected by a computer using the Data Collection 2.1 software (Applied Biosystems) connected to the ABI Prism 310. Data were analyzed using GeneScan 2.2 software (Applied Biosystems), which sized and quantified the detected fragments. GeneScan software was also used to compare the electropherograms from the analyzed plants in order to detect the polymorphic AFLP markers. The markers were scored as either present (1) or absent (0) for each plant and primer combination.
Cloning and sequencing of AFLP markers.
Four selected polymorphic AFLP markers were cloned into the pGEM-T vector (Promega Corporation, Madison, WI) and transformed into competent DH5-α Escherichia coli cells following the Polanco et al. (2005) protocol for AFLP markers detected by fluorescence capillary electrophoresis. Overnight cultures of at least two positive colonies for each specific AFLP marker were sent to the Nucleic Acids Sequencing Facility of the University of León (Spain) in order to obtain the sequence of the corresponding plasmid inserts. The sequences of the AFLP primers were removed and recognition sequences for EcoRI and MseI endonucleases were completed at the ends before sending them to the EMBL-EBI database (entries AM285291, AM285292, AM285293, and AM285294). Sequence alignments were done using Jalview Multiple Alignment Editor v.2.08.1 (Clamp et al. 2004).
Results
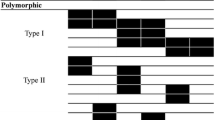
Analysis of 24 rye plants regenerated via somatic embryogenesis (Fig. 1) revealed a total of 887 AFLP markers. Nine different selective primer pairs were used and each plant produced individual markers in the range of 756 (for plant CLh11) to 840 (for plant CLa10). The average fragment length was 178 bp. The average number of fragments per primer pair and per plant was 91.4. All regenerated plants analyzed shared 607 (68.4%) non-polymorphic markers. Another 74 (8.3%) fragments were not present in any of the regenerated plants obtained from at least one cell line, indicating variability between the original embryos used to establish the cell lines. There were 116 (13.1%) singletons or AFLP fragments either present or absent in just one plant: eight (0.9%) were amplification singletons and 108 (12.2%) were non-amplified singletons. Another 12 markers (1.4%) were detected as polymorphic for more than one plant from the same cell line, and 78 (8.8%) showed the same polymorphism in regenerated plants obtained independently from different cell lines.
Comparisons within groups of regenerated plants obtained from the same cell line revealed that 23 out of the 24 regenerated plants showed at least one variation, and the number of changes per regenerated plant ranged from 2 to 90. However, the distribution of the variability was not uniform, as two plants accumulated 49.4% of the polymorphisms: plants CLe9 and CLh11 showed 84 and 90 changes, respectively, while half of the plants showed six or less changes. A summary of the variation found for each group of regenerated plants using each primer pair is indicated in Table 1.
It was not possible to perform an analysis of the progeny of the two plants that showed the highest variability (plants CLe9 and CLh11) because they were albino mutants that did not reach maturity. Therefore, of the regenerated plants that formed spikes, plants CLe6 and CLh10 were selected as female parents for controlled crosses because they showed the highest number of ALFP polymorphisms at 15 and 12, respectively. We attempted to obtain the nucleotide sequence of all the polymorphic markers from plants CLe6 and CLh10 that fulfilled the following two requirements: (1) the polymorphism was detected in various regenerated plants such that it may be linked to hypervariable genome regions and (2) the polymorphism was detected in the progeny and is therefore an inherited genome change. Five AFLP markers met the above stipulations: A1-303, A3-113, A5-212, A5-233, and A5-375. They are named according to the code of the primer pair used (see Table 1) and the size of the amplified AFLP fragment.
The A1-303 marker was detected in eight plants from cell lines CLe and CLh (four plants from each cell line), including plants CLe6 and CLh10, but not observed in plants obtained from cell lines CLa, CLd, and CLi. This indicates that A1-303 was likely polymorphic at the embryo level. However, A1-303 was undetected in two regenerated plants (CLe7 and CLh5) obtained independently from cell lines CLe and CLh. The A3-113 marker was absent only in three plants from cell line CLe (plants CLe5, CLe6, and CLe7) that may all have originated from the same cell in which the original mutation took place. The A5-212 marker was detected in six plants but from four cell lines: CLa (plant CLa12), CLe (plant CLe6), CLh (plants CLh5, CLh12, and CLh10), and CLi (plant CLi8). The A5-233 marker was absent in three plants from two cell lines: CLa (plant CLa12) and CLh (plants CLh5 and CLh10). The marker A5-375 was absent in nine plants from three cell lines: CLa (plants CLa9, CLa10, and CLa11), CLh (plants CLh8, CLh9, and CLh11), and CLi (plants CLi8, CLi9, and CLi10).
We checked that the phenotype of male parents used for controlled crosses differed from the phenotype of the CLe6 and CLh10 plants for all the above five markers. The analysis of five siblings from each of the crosses showed segregations for all of these markers.
The A1-303, A5-212, and A5-375 AFLP fragments from plant CLh10 and the A3-113 fragment from plant CLe8 were cloned and sequenced. The A5-233 marker was found to be a low intensity peak closely surrounded by high peaks from other markers and it was not possible to clone it despite several attempts. Sequence comparisons using Basic Local Alignment and Search Tool (BLAST) searches did not revealed significant similarity for marker A3-113. The first 158 bp of the A5-212 marker showed a high identity (93%) with the 3′ end of an mRNA sequence of Hordeum vulgare (accession AK249005), which is related to a family of hypothetical plant-specific proteins of unknown function.
The sequence of marker A1-303 showed a clear pattern of internal repeats and it had identity values higher than 85% with 70 sequences of satellite DNA from 11 species of tribe Triticeae (accessions AJ517227 to AJ517293, Z75561, AY551004, AF227454, and AF354658), including Secale cereale, S. montanum, and S. vavilovii. We aligned the A1-303 sequence with the nine accessions of S. cereale which contained complete and truncated versions of a 118-bp repeat unit (Fig. 2). The alignment revealed that the sequence of marker A1-303 consists of part of a complete repeat, where the EcoRI-end of the AFLP fragment is located, followed by a truncated and a complete (with small deletions) repeat and a piece of the repeated sequence that includes the MseI-end. BLAST searches limited to the Triticeae Repeat Sequence Database (TREP) showed high identity values with telomere-associated accessions TREP107 (from Triticum aestivum) and TREP64 (from Secale cereale).
DNA sequence alignment of nine accessions of the rye 120-bp repeat family (Contento et al. 2005) with the ALFP polymorphic marker A1-303 (303A1). Complete 118-bp repeat units are indicated as _rep.1 and truncated repeat units as _rep.2 after the corresponding accession numbers. The sequence of A1-303 has been divided into four consecutive units for best alignment: _rep.Eco starting with the EcoRI site; _rep.1, _rep2, and rep.Mse ending with the MseI site. Gaps are introduced for optimal alignment. Identity scores for each base are given as bars at the bottom over the consensus sequence (larger bar represents high similarity) and are also indicated by the different grey level of shading.
The sequence of the A5-375 marker showed high BLAST identity values (more than 86%) with two regions from the Aegilops tauschii accession AF497474, with one region of the Triticum monococcum accession AY485644 and with one region of the Triticum turgidum accession AY663391. The sequences of A. tauschii corresponded to the transposon Angela_BHG4-2, the sequence of T. monococcum to the transposon Angela_AY485644-15, and the sequence of T. turgidum to the transposon Angela_AY663391-2, all of them Copia group transposons. The alignment of all these sequences with marker A5-375 is shown in the Fig. 3.
DNA sequence alignment of AFLP polymorphic marker A5-375 with regions of Aegilops tauschii accession AF497474, Triticum monococcum accession AY485644, and Triticum turgidum accession AY66339. The starting nucleotide (nt) of each region is indicated after the corresponding accession number. Gaps are introduced for optimal alignment. Identity scores for each base are given as bars at the bottom over the consensus sequence (larger bar represents high similarity) and are also indicated by the different grey level of shading.
Discussion
Rye is an allogamous species that shows a high spontaneous mutation rate in vivo. Figueiras et al. (1991) reported frequencies of 2.05 × 10−2 and 9.3 × 10−3 for chromosomal and gene mutations, respectively, for the Ailés cultivar used in this work. These rates can increase during tissue culture as indicated in other studies about somaclonal variation in rye carried out with Ailés and other cultivars. Detection techniques with less resolution than AFLPs revealed that more than half of the regenerated plants showed polymorphisms (Linacero and Vázquez 1992b; 1993). In our work with utilized AFLP markers, 95% of the regenerated plants showed at least one change when compared with plants obtained from the same cell line. But the number of mutations per plant was highly variable, from 2 to 90, and most of the polymorphic markers were scored in a few plants. Such an uneven pattern of distribution of somaclonal variation has also been found analyzing AFLP markers in both regenerated pecan (Vendrame et al. 2000) and Arabidopsis plants (Polanco and Ruiz 2002).
Although AFLPs are highly reproducible, homoplasy (co-migration of non-homologous fragments) is a major issue in the analysis and interpretation of AFLP data that results in an underestimation of genetic diversity among samples and a loss of resolution in the analysis (Meudt and Clarke 2007). We used a capillary electrophoresis system that enables precise estimates of fragment mobility (1 pb resolution) and we manually checked all the electropherogram comparisons in order to reduce the scoring errors. We analyzed AFLP patterns of clonal plants and therefore homoplasy, if present in our data, will reduce the number of somaclonal mutations identified.
The AFLP polymorphisms detected for each plant were scored as amplified or non-amplified mutations, according to the total number of regenerated plants within one cell line that showed the marker. Comparisons were made within a cell line because every embryo used as a culture explant was a genetically unique result of a cross, and the plants were obtained by somatic embryogenesis from cell lines derived from 1-mm to 2-mm long immature embryos that were too small to remove a sample from in order to obtain the AFLP pattern of the embryo. Our scoring criteria, within a cell line, should reduce the observed mutation rate rather than increase it.
We have observed that 8.8% of the analyzed AFLP markers showed the same polymorphism in regenerated plants that were obtained independently from different cell lines. Therefore, independent mutational events have occurred in the same genome regions of these plants. Mutations derived from tissue culture that appeared preferentially in some loci have been described previously in garlic (Al-Zahim et al. 1999), rice (Xie et al. 1995), and also in rye (Linacero et al. 2000). The study by Linacero et al. (2000) of regenerated rye plants obtained from immature embryos and/or immature inflorescences from different cultivars, using RAPD markers, revealed hot spots of mutation named hypervariable genome regions.
Through controlled crossings and progeny analysis, we confirmed the genetic stability of five AFLP hypervariable markers and we obtained the nucleotide sequence of four of them. The A1-303 marker was identified as part of a tandemly repeated sequence, the 120-bp family, reported to be found both in telomeres and widely distributed among rye chromosomes (Contento et al. 2005), while marker A5-375 showed a high similarity to regions of Ty1-copia retrotransposons of the subgroup Angela from three Triticeae species. Although AFLP markers are supposed to have genome-wide distribution, mapping data in rye (using ALFP markers obtained with EcoRI and MseI endonucleases) showed a clustering tendency in the proximity of centromeric and telomeric regions (Saal and Wricke 2002; Bednarek et al. 2003). The use of different enzyme combinations (Bednarek et al. 2007; Li et al. 2007) in future work will help to reveal the presence of hypervariable markers in other regions of the rye genome.
The 120-bp repeat family has been described as the major component of the rye heterochromatin (Cuadrado et al. 1995) and it is also present in many species of the Triticeae tribe. Homologous sequences have also been found in the tribe Avenae. Contento et al. (2005) analyzed the diversity and organization of 90 members of the 120-bp family obtained from 11 Triticeae species and concluded that individual sites of multiple tandem repeats were transferred as blocks, so that they can translocate within the genome and originate the variation in the position and site number that was observed. We confirmed the loss of marker A1-303, related to the 120-bp family, in two regenerated plants obtained from different cell lines. The same mutational mechanisms that spontaneously drive the genome’s variation described by Contento et al. (2005) may be at work when the cells encounter stress in the tissue culture environment, increasing the mutational rates. However, it must also be taken into account that DNA polymerases may induce exaggerated levels of mutations by slippage while replicating repetitive sequences (Kunkel and Babenek 2000).
The majority of the genomes from species in the tribe Triticeae consist of repetitive DNA sequences (Flavell 1986) and many of these sequences are retroelements. A common feature of some plant retrotransposons is that they are affected by protoplast isolation or in vitro cell tissue culture and, for example, insertion into coding sequences has been demonstrated in tobacco and rice, indicating that retrotransposition might make a significant contribution to somaclonal variation (Grandbastien 1998). In rye, Alves et al. (2005) demonstrated that foldback transposon RYS1 was activated by tissue culture and was inserted in the same genome position where it was also detected in natural populations, suggesting that these preferential integration points could be part of the hypervariable regions described for the rye genome (Linacero et al. 2000). The A5-375 marker, located within the long terminal repeat (LTR) of an Angela retrotransposon, was lost in nine out of the 24 regenerated plants analyzed in this work. Those nine plants were obtained from three different cell lines, indicating that the same mutational event has occurred independently three times and revealing a mutational hot spot. It is known that Angela and related elements (mainly Wis and BARE) are generally nested within each other in the LTRs. However, the identification of a polymorphic marker exhibiting a high level of similarity to mobile elements does not necessary mean that new insertions had occurred, because loss of restriction sites and changes of selective nucleotides are the main cause of AFLP marker polymorphism (Kuiper 1998).
Since somaclonal variation may be the result of complex processes embracing genetic and epigenetic changes, more data is required to understand the mechanisms involved in the high rate of somaclonal variation reported for rye regenerated plants in this and previous studies.
References
Alves E.; Ballesteros I.; Linacero R.; Vazquez A. M. RYS1, a foldback transposon, is activated by tissue culture and shows preferential insertion points into the rye genome. Theor. Appl. Genet. 111: 431–436; 2005 doi:10.1007/s00122-005-2013-9.
Al-Zahim M. A.; Ford-Lloyd B. V.; Newbury H. J. Detection of somaclonal variation in garlic (Allium sativum L.) using RAPD and cytological analysis. Plant. Cell. Rep. 18: 473–477; 1999 doi:10.1007/s002990050606.
Bednarek K.; Masojc P.; Lewandowska R.; Myskow B. Saturating rye genetic map with amplified fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) markers. J. Appl. Genet. 44: 21–33; 2003.
Bednarek P. T.; Orlowska R.; Koebner R. M. D.; Zimny J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biology 7: 10; 2007 doi:10.1186/1471-2229-7-10.
Chen J.; Henny R. J.; Devenand P. S.; Chao C. T. AFLP analysis of nephthytis (Syngonium podophyllum Schott) selected from somaclonal variants. Plant Cell Rep. 24: 743–749; 2006 doi:10.1007/s00299-005-0032-2.
Chowdari K. V.; Ramakrishna W.; Tamhankar S. A.; Hendre R. R.; Gupta V. S.; Sahasrabudhe N. A.; Ranjekar P. K. Identification of minor DNA variations in rice somaclonal variants. Plant Cell Rep. 18: 55–58; 1998 doi:10.1007/s002990050531.
Clamp M.; Cuff J.; Searle S. M.; Barton G. J. The Jalview Java Alignment Editor. Bioinformatics. 20: 426–427; 2004 doi:10.1093/bioinformatics/btg430.
Contento A.; Heslop-Harrison J. S.; Schwarzacher T. Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet. Genome. Res. 109: 34–42; 2005 doi:10.1159/000082379.
Cuadrado A.; Ceoloni C.; Jouve N. Variation in highly repetitive DNA composition of heterochromatin in rye studied by fluorescence in situ hybridization. Genome. 38: 1061–1069; 1995 doi:10.1139/gen-38-6-1061.
Figueiras A. M.; de la Peña A.; Benito M. C. High mutability in rye (Secale cereale L.). Mutat. Res. 264: 171–176; 1991 doi:10.1016/0165-7992(91)90073-D.
Flavell R. B. Repetitive DNA and chromosome evolution in plants. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 312: 227–242; 1986 doi:10.1098/rstb.1986.0004.
Gagliardi, R. F.; Hanai; Pacheco, G.; Oliveira, C. A.; Carneiro, L. A.; Montenegro Valls, J. F.; Mansur, E.; Carneiro Vieira, M. L. Assessment of genetic stability among in vitro plants of Arachis retusa using RAPID and AFLP markers for germplasm preservation. J. Integr. Plant. Biol. 49: 307–312; 2007. doi:10.1111/j.1744-7909.2007.00402.x.
Grandbastien M. A. Activation of plant retrotransposons under stress conditions. Trends. Plant. Sci. 3: 181–187; 1998 doi:10.1016/S1360-1385(98)01232-1.
Grandbastien M. A.; Spielman A.; Caboche M. Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature. 337: 376–380; 1989 doi:10.1038/337376a0.
Hirochika H.; Sugimoto K.; Otsuki Y.; Tsugawa H.; Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U. S. A. 93: 7783–7788; 1996 doi:10.1073/pnas.93.15.7783.
Kuiper M. T. R. Building a high-density genetic map using the AFLPTM technology. In: Martinez-ZapaterJ. M. ; SalinasJ. (eds) Arabidopsis Protocols, Methods in Molecular Biology. 82: Humana, Totowa, pp 157–171; 1998.
Kunkel T. A.; Babenek K. DNA replication fidelity. Ann. Rev. Biochem. 69: 497–529; 2000 doi:10.1146/annurev.biochem.69.1.497.
Larkin P. J.; Scowcroft W. R. Somaclonal variation a novel source of variability from cell culture for plant improvement. Theor. Appl. Genet. 60: 197–214; 1981 doi:10.1007/BF02342540.
Li X.; Yu X.; Wang N.; Feng Q.; Dong Z.; Liu L.; Shen J.; Liu B. Genetic and epigenetic instabilities induced by tissue culture in wild barley (Hordeum brevisubulatum (Trin.) Link). Plant. Cell. Tiss. Organ. Cult. 90: 153–168; 2007 doi:10.1007/s11240-007-9224-5.
Linacero R.; Freitas Alves E.; Vázquez A. M. Hot spots of DNA instability revealed through the study of somaclonal variation in rye. Theor. Appl. Genet. 100: 506–511; 2000 doi:10.1007/s001220050066.
Linacero R.; Vázquez A. M. Somatic embryogenesis in polyembrionic Secale cereale L. Plant. Cell. Rep. 12: 26–28; 1992a doi:10.1007/BF00232417.
Linacero R.; Vázquez A. M. Genetic analysis of chlorophyll-deficient somaclonal variants in rye. Genome. 35: 981–984; 1992b doi:10.1139/gen-35-6-981.
Linacero R.; Vázquez A. M. Somaclonal variation in rye. Mutat. Res. 302: 201–205; 1993 doi:10.1016/0165-7992(93)90105-5.
Meudt H. M.; Clarke A. C. Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant. Sci. 12: 106–117; 2007 doi:10.1016/j.tplants.2007.02.001.
Perkin-Elmer Applied Biosystems (1997) AFLPTM plant mapping protocol, P/N 4303146 Rev A. The Perkin-Elmer Corporation. Foster City, CA
Philips R. L.; Kaeppler S. M.; Olhoft P. Genetic instability of plant tissue cultures: breakdown of normal controls. Proc. Natl. Acad. Sci. U. S. A. 91: 5222–5226; 1994 doi:10.1073/pnas.91.12.5222.
Polanco C.; González A. I.; de la Puente R.; Somalo S.; Ruiz M. L. Cloning of AFLP markers detected by fluorescence capillary electrophoresis. Plant Mol. Biol. Report. 233: 271–277; 2005 doi:10.1007/BF02772757.
Polanco C.; Ruiz M. L. AFLP analysis of somaclonal variation in Arabidopsis thaliana regenerated plants. Plant. Sci. 162: 817–824; 2002 doi:10.1016/S0168-9452(02)00029-8.
Prado M. J.; Gonzalez M. V.; Romo S.; Herrera M. T. Adventitious plant regeneration of leaf explants from adult male kiwifruit and AFLP analysis of genetic variation. Plant. Cell. Tiss. Organ. Cult. 88: 1–110; 2007 doi:10.1007/s11240-006-9116-0.
Rostiana O.; Niwa M.; Marubashi W. Efficiency of inter-simple sequence repeat PCR for detecting somaclonal variation among leaf-culture-regenerated plants of horseradish. Breeding. Sci. 49: 245–250; 1999.
Saal B.; Wricke G. Clustering of amplified fragment length polymorphism markers in a linkage map of rye. Plant. Breeding. 121: 117–123; 2002 doi:10.1046/j.1439-0523.2002.00698.x.
Saker M. M.; Adawy S. S.; Mohamed A. A.; El-Itriby H. A. Monitoring of cultivar identity in tissue culture-derived date palms using RAPD and AFLP analysis. Biol. Plantarum. 50: 198–204; 2006 doi:10.1007/s10535-006-0007-3.
Vendrame W. A.; Kochert G. D.; Sparks D.; Wetzstein H. Y. Field performance and molecular evaluations of pecan trees regenerated from somatic embryogenic cultures. J. Am. Soc. Hort. Sci. 125: 542–546; 2000.
Vos P.; Hogers R.; Bleeker M.; Reijans M.; van de Lee T.; Hornes M.; Fritjers A.; Pot J.; Peleman J.; Kuiper M.; Zabeau M. AFLP: a new concept for DNA fingerprinting. Nucleic. Acids. Res. 23: 4407–4414; 1995 doi:10.1093/nar/23.21.4407.
Xie Q. J.; Oard J. H.; Rush M. C. Genetic analysis of purple-red hull rice mutation derived from tissue culture. J. Hered. 86: 154–156; 1995.
Acknowledgements
This work was supported by the Spanish DGICYT grants BMC2001-1297-C02-01 and AGL2006-14249-C02-02, and by the Junta de Castilla y León grant LEO52A6. We thank Dr. Ana M. Vázquez (Universidad Complutense de Madrid, Spain) for the kind donation of the material used in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Nigel James Taylor
Rights and permissions
About this article
Cite this article
de la Puente, R., González, A.I., Ruiz, M.L. et al. Somaclonal variation in rye (Secale cereale L.) analyzed using polymorphic and sequenced AFLP markers. In Vitro Cell.Dev.Biol.-Plant 44, 419–426 (2008). https://doi.org/10.1007/s11627-008-9152-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-008-9152-z